Suspension Culture Optimization and Transcriptome-Guided Identification of Candidate Regulators for Militarine Biosynthesis in Bletilla striata
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Capsule Collection of B. striata
2.2. Cell Suspension Culture
2.2.1. Induction Conditions and Subculture Cultivation
2.2.2. Measuring Growth Indicators
2.2.3. Selection of Landraces and Cultivation Conditions
2.3. Metabolite Extraction and Quantitative Analysis
2.4. Sample Collection and RNA Extraction
2.5. Transcriptome Sequencing and Data Quality Control
2.6. Gene Function Annotation and Expression Level Analysis
2.7. Differential Expression Analysis and Functional Enrichment
2.8. SSR Analysis
3. Results
3.1. Diversity of B. striata Landraces
3.2. Suspension Cell Proliferation and Metabolite Accumulation Analysis
3.3. Analysis of Metabolite Accumulation in Different Landraces
3.4. Optimal Culture Condition Screening and Model Construction
3.5. Transcriptome Sequencing Data Overview and Annotation Statistics
3.6. Gene Expression Levels Between Different Landraces
3.6.1. FPKM Density Distribution and Sample Correlation
3.6.2. Shared and Unique Expression Profiles
3.6.3. Pairwise Differential Expression Analysis
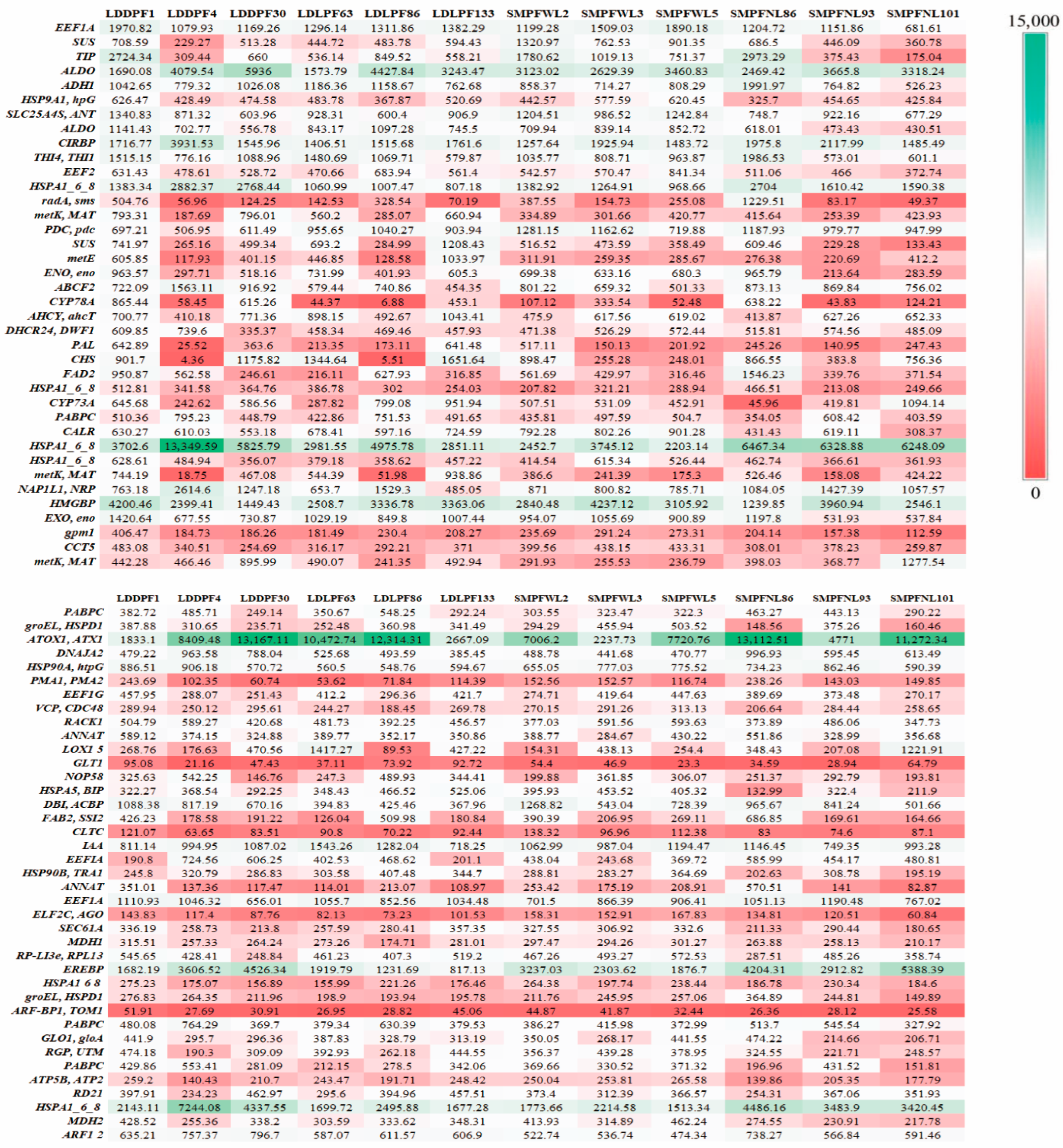
3.7. Differential Gene Analysis of Suspension Cells in Different Landraces
3.8. Functional Enrichment Analysis of DEGs
3.9. Analysis of DEGs Related to Militarine Synthesis
4. Discussion
4.1. Optimization Strategies for Efficient Suspension Culture Systems
4.2. Synergistic Regulation Promoting the Synthesis of Secondary Metabolites
4.3. Co-Regulation of Agronomic Traits on Gene Expression and Metabolic Pathways
4.4. The Role of Key Regulatory Genes in Militarine Synthesis Pathways
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| B. striata | Bletilla striata |
| RSM | Response surface methodology |
| Dpi | Days post-inoculation |
| BL | Blade length |
| BW | Blade width |
| LWRB | Length–width ratio of blade |
| PL | Plant length |
| FN | Flower number |
| FW | Fresh weight |
| DW | Dry weight |
| Rpm | Revolutions per minute |
| HPLC | High-performance liquid chromatography |
| OLC | Overlap–layout–consensus |
| PPI | Protein–protein interaction |
| SSRs | Simple sequence repeats |
| HBA | p-Hydroxybenzyl |
| SD | Standard deviation |
| CV | Coefficient of variation |
| DEG | Differentially expressed gene |
| LDDPF | LD, germplasm nursery identifier; DPF, deep purple flower |
| LDLPF | LD, germplasm nursery identifier; LPF, light purple flower |
| SMPFWL | SM, germplasm nursery identifier; PF, purple flower; WL, wide leaf |
| SMPFNL | SM, germplasm nursery identifier; PF, purple flower; NL, narrow leaf |
| TAL | Tyrosine ammonia-lyase |
| PAL | Phenylalanine ammonia-lyase |
| 4CL | 4-coumarate-CoA ligase |
References
- Li, L.; Liu, H.; Wen, W.; Huang, C.; Li, X.; Xiao, S.; Wu, M.; Shi, J.; Xu, D. Full Transcriptome Analysis of Callus Suspension Culture System of Bletilla striata. Front. Genet. 2020, 11, 995. [Google Scholar] [CrossRef]
- Xu, D.; Pan, Y.; Chen, J. Chemical Constituents, Pharmacologic Properties, and Clinical Applications of Bletilla striata. Front. Pharmacol. 2019, 10, 1168. [Google Scholar] [CrossRef]
- He, X.; Wang, X.; Fang, J.; Zhao, Z.; Huang, L.; Guo, H.; Zheng, X. Bletilla striata: Medicinal uses, phytochemistry and pharmacological activities. J. Ethnopharmacol. 2017, 195, 20–38. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.-L.; Pan, Y.-C.; Li, L.; ShangGuan, Y.-N.; Zhang, S.-B.; Liu, G.-Y.; Cheng, L.; Xiao, S.-J. Chemical constituents of Bletilla striata. J. Asian Nat. Prod. Res. 2019, 21, 1184–1189. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Li, Y.; Chen, L.; Xiao, Y.; Yang, N.; Luo, H.; Guan, J.; Xu, D. Identification and comparative genomic analysis of endophytic fungi in Bletilla striata and its potential for promoting militarine bioaccumulation. Fitoterapia 2025, 181, 106356. [Google Scholar] [CrossRef]
- Wu, T.; Kerbler, S.M.; Fernie, A.R.; Zhang, Y. Plant cell cultures as heterologous bio-factories for secondary metabolite production. Plant Commun. 2021, 2, 100235. [Google Scholar] [CrossRef]
- Pan, Y.; Li, L.; Xiao, S.; Chen, Z.; Sarsaiya, S.; Zhang, S.; ShangGuan, Y.; Liu, H.; Xu, D. Callus growth kinetics and accumulation of secondary metabolites of Bletilla striata Rchb.f. using a callus suspension culture. PLoS ONE 2020, 15, e0220084. [Google Scholar] [CrossRef]
- Murthy, H.N.; Paek, K.Y. Panax ginseng Adventitious Root Suspension Culture: Protocol for Biomass Production and Analysis of Ginsenosides by High Pressure Liquid Chromatography. Methods Mol. Biol. 2016, 1391, 125–139. [Google Scholar] [CrossRef]
- Linh, T.M.; Mai, N.C.; Hoe, P.T.; Ngoc, N.T.; Thao, P.T.H.; Ban, N.K.; Van, N.T. Development of a Cell Suspension Culture System for Promoting Alkaloid and Vinca Alkaloid Biosynthesis Using Endophytic Fungi Isolated from Local Catharanthus roseus. Plants 2021, 10, 672. [Google Scholar] [CrossRef]
- azquez-Marquez, A.M.; Bernabé-Antonio, A.; Correa-Basurto, J.; Burrola-Aguilar, C.; Zepeda-Gómez, C.; Cruz-Sosa, F.; Nieto-Trujillo, A.; Estrada-Zúñiga, M.E. Changes in Growth and Heavy Metal and Phenolic Compound Accumulation in Buddleja cordata Cell Suspension Culture under Cu, Fe, Mn, and Zn Enrichment. Plants 2024, 13, 1147. [Google Scholar] [CrossRef]
- Kim, J.-H.; Han, J.-E.; Murthy, H.N.; Kim, J.-Y.; Kim, M.-J.; Jeong, T.-K.; Park, S.-Y. Production of Secondary Metabolites from Cell Cultures of Sageretia thea (Osbeck) M.C. Johnst. Using Balloon-Type Bubble Bioreactors. Plants 2023, 12, 1390. [Google Scholar] [CrossRef]
- Lv, S.; Ding, F.; Zhang, S.; Nosov, A.M.; Kitashov, A.V.; Yang, L. Induction and Suspension Culture of Panax japonicus Callus Tissue for the Production of Secondary Metabolic Active Substances. Plants 2024, 13, 2480. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Zhang, Y.; Liang, F.; Jiang, S.; Niu, S.; Wang, X.; Zhou, Y.; Cui, B.; Yuan, X. Metabolomic and transcriptomic analyses reveal the mechanism of polysaccharide and secondary metabolite biosynthesis in Bletilla striata tubers in response to shading. Int. J. Biol. Macromol. 2024, 279 Pt 4, 135545. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Li, H.; Luo, H.; Liu, J.; Li, K.; Li, Q.; Yang, N.; Xu, D. Unveiling the Role of β-Glucosidase Genes in Bletilla striata’s Secondary Metabolism: A Genome-Wide Analysis. Int. J. Mol. Sci. 2024, 25, 13191. [Google Scholar] [CrossRef]
- Zhou, G.L.; Zhu, P. De novo transcriptome sequencing of Rhododendron molle and identification of genes involved in the biosynthesis of secondary metabolites. BMC Plant Biol. 2020, 20, 414. [Google Scholar] [CrossRef]
- Meng, K.; Lv, J.; Zhang, T.; Liu, Y.; Zhang, P.; Zhang, Y.; Hu, B.; Huang, Q.; Xie, B.; Fu, J. Chromosome-Scale Genome and Transcriptomic Analyses Reveal Differential Regulation of Terpenoid Secondary Metabolites in Hericium coralloides. J. Fungi 2024, 10, 704. [Google Scholar] [CrossRef]
- Zhu, M.; Hsu, C.-W.; Ogorek, L.L.P.; Taylor, I.W.; La Cavera, S.; Oliveira, D.M.; Verma, L.; Mehra, P.; Mijar, M.; Sadanandom, A.; et al. Single-cell transcriptomics reveal how root tissues adapt to soil stress. Nature 2025, 642, 721–729. [Google Scholar] [CrossRef]
- Lu, C.H.; Engelmann, N.J.; Lila, M.A.; Erdman, J.W., Jr. Optimization of lycopene extraction from tomato cell suspension culture by response surface methodology. J. Agric. Food Chem. 2008, 56, 7710–7714. [Google Scholar] [CrossRef]
- Zhang, L.; Kawaguchi, R.; Enomoto, T.; Nishida, S.; Burow, M.; Maruyama-Nakashita, A. Glucosinolate Catabolism Maintains Glucosinolate Profiles and Transport in Sulfur-Starved Arabidopsis. Plant Cell Physiol. 2023, 64, 1534–1550. [Google Scholar] [CrossRef]
- Xu, H.; Wang, Z.; Li, Y.; He, J.; Wu, X. Dynamic growth models for Caragana korshinskii shrub biomass in China. J. Env. Environ. Manag. 2020, 269, 110675. [Google Scholar] [CrossRef]
- Nahar, L.; Chaiwut, P.; Sangthong, S.; Theansungnoen, T.; Sarker, S.D. Progress in the analysis of phytocannabinoids by HPLC and UPLC (or UHPLC) during 2020–2023. Phytochem Anal 2024, 35, 927–989. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.R.; Koren, S.; Sutton, G. Assembly algorithms for next-generation sequencing data. Genomics 2010, 95, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Tycko, J.; Van, M.V.; DelRosso, N.; Ye, H.; Yao, D.; Valbuena, R.; Vaughan-Jackson, A.; Xu, X.; Ludwig, C.; Spees, K.; et al. Development of compact transcriptional effectors using high-throughput measurements in diverse contexts. Nat. Biotechnol. 2025, 43, 1525–1538. [Google Scholar] [CrossRef] [PubMed]
- Huckvale, E.; Moseley, H.N.B. kegg_pull: A software package for the RESTful access and pulling from the Kyoto Encyclopedia of Gene and Genomes. BMC Bioinform. 2023, 24, 78. [Google Scholar] [CrossRef]
- Depuydt, T.; De Rybel, B.; Vandepoele, K. Charting plant gene functions in the multi-omics and single-cell era. Trends Plant Sci. 2023, 28, 283–296. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Yu, X.; Xie, L.; Ge, J.; Li, H.; Zhong, S.; Liu, X. Integrating single-cell RNA-seq and spatial transcriptomics reveals MDK-NCL dependent immunosuppressive environment in endometrial carcinoma. Front. Immunol. 2023, 14, 1145300. [Google Scholar] [CrossRef]
- Weng, J.; Wu, X.F.; Shao, P.; Liu, X.P.; Wang, C.X. Medicine for chronic atrophic gastritis: A systematic review, meta- and network pharmacology analysis. Ann. Med. 2023, 55, 2299352. [Google Scholar] [CrossRef]
- Chen, J.; Amdanee, N.; Zuo, X.; Wang, Y.; Gong, M.; Yang, Y.; Li, H.; Zhang, X.; Zhang, C. Biomarkers of bipolar disorder based on metabolomics: A systematic review. J. Affect. Disord. 2024, 350, 492–503. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023, protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Sang, M.K.; Patnaik, H.H.; Park, J.E.; Song, D.K.; Jeong, J.Y.; Hong, C.E.; Kim, Y.T.; Shin, H.J.; Ziwei, L.; Hwang, H.J.; et al. Transcriptome analysis of Haemaphysalis flava female using Illumina HiSeq 4000 sequencing: De novo assembly, functional annotation and discovery of SSR markers. Parasit Vectors 2023, 16, 367. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhao, Q.; Xie, K.; Wang, M.; Li, L.; Zeng, D.; Wang, Q.; Wang, S.; Chen, A.; Xu, G. A Mycorrhiza-Induced UDP-Glucosyl Transferase Negatively Regulates the Arbuscular Mycorrhizal Symbiosis. Plant Cell Environ. 2025, 48, 1643–1655. [Google Scholar] [CrossRef]
- Li, L.; Hao, B.; Zhang, Y.; Ji, S.; Chou, G. Metabolite Profiling and Distribution of Militarine in Rats Using UPLC-Q-TOF-MS/MS. Molecules 2020, 25, 1082. [Google Scholar] [CrossRef]
- Liu, H.; Huang, C.; Li, Q.; Wang, M.; Xiao, S.; Shi, J.; He, Y.; Wen, W.; Li, L.; Xu, D. Genome-Wide Identification of Genes Related to Biosynthesis of Phenolic Acid Derivatives in Bletilla striata at Different Suspension Culture Stages. Front. Plant Sci. 2022, 13, 875404. [Google Scholar] [CrossRef]
- Wu, X.; Xia, M.; Su, P.; Zhang, Y.; Tu, L.; Zhao, H.; Gao, W.; Huang, L.; Hu, Y. MYB transcription factors in plants: A comprehensive review of their discovery, structure, classification, functional diversity and regulatory mechanism. Int. J. Biol. Macromol. 2024, 282 Pt 2, 136652. [Google Scholar] [CrossRef]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The Flavonoid Biosynthesis Network in Plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef]
- Peng, L.; Ru, M.; Liang, Z. Variation of photosynthesis, secondary metabolites and antioxidant activities in third generation of spaceflight-induced Salvia miltiorrhiza. Chin. Herb. Med. 2022, 14, 592–601. [Google Scholar] [CrossRef]
- Song, X.; Tang, S.; Liu, H.; Meng, Y.; Luo, H.; Wang, B.; Hou, X.-L.; Yan, B.; Yang, C.; Guo, Z.; et al. Inheritance of acquired adaptive cold tolerance in rice through DNA methylation. Cell 2025, 188, 4213–4224.e12. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; He, X.; Guo, Z.; Yang, N.; Bai, G.; Zhao, J.; Xu, D. The Mitochondrial Blueprint: Unlocking Secondary Metabolite Production. Metabolites 2024, 14, 711. [Google Scholar] [CrossRef]
- Song, Y.; Feng, L.; Alyafei, M.A.M.; Jaleel, A.; Ren, M. Function of Chloroplasts in Plant Stress Responses. Int. J. Mol. Sci. 2021, 22, 13464. [Google Scholar] [CrossRef]
- Smetanska, I. Production of secondary metabolites using plant cell cultures. Adv. Biochem. Eng. Biotechnol. 2008, 111, 187–228. [Google Scholar] [CrossRef]
- Zhan, X.; Chen, Z.; Chen, R.; Shen, C. Environmental and Genetic Factors Involved in Plant Protection-Associated Secondary Metabolite Biosynthesis Pathways. Front. Plant Sci. 2022, 13, 877304. [Google Scholar] [CrossRef]
- Murthy, H.N.; Yadav, G.G.; Paek, K.Y.; Park, S.Y. Production of Terpene Trilactones from Cell and Organ Cultures of Ginkgo biloba. Plants 2024, 13, 2575. [Google Scholar] [CrossRef]
- Ahanger, M.A.; Qin, C.; Maodong, Q.; Dong, X.X.; Ahmad, P.; Allah, E.F.A.; Zhang, L. Spermine application alleviates salinity induced growth and photosynthetic inhibition in Solanum lycopersicum by modulating osmolyte and secondary metabolite accumulation and differentially regulating antioxidant metabolism. Plant Physiol. Biochem. 2019, 144, 1–13. [Google Scholar] [CrossRef]
- Howat, S.; Park, B.; Oh, I.S.; Jin, Y.W.; Lee, E.K.; Loake, G.J. Paclitaxel: Biosynthesis, production and future prospects. N. Biotechnol. 2014, 31, 242–245. [Google Scholar] [CrossRef]
- Lv, B.; Feng, Q.; Shao, G.; Zuo, A.; Yan, X.; Liu, J.; Dong, J.; Ma, P. The transcription factor Dof32 coordinates salvianolic acid biosynthesis and drought tolerance in Salvia miltiorrhiza. Plant Physiol. 2025, 198, kiaf221. [Google Scholar] [CrossRef]
- Ruan, Y.L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef]
- Diniz, S.C.; Taciro, M.K.; Gomez, J.G.; da Cruz Pradella, J.G. High-cell-density cultivation of Pseudomonas putida IPT 046 and medium-chain-length polyhydroxyalkanoate production from sugarcane carbohydrates. Appl. Biochem. Biotechnol. 2004, 119, 51–70. [Google Scholar] [CrossRef]
- Titova, M.; Popova, E.; Nosov, A. Bioreactor Systems for Plant Cell Cultivation at the Institute of Plant Physiology of the Russian Academy of Sciences: 50 Years of Technology Evolution from Laboratory to Industrial Implications. Plants 2024, 13, 430. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Shen, N.; Wang, T.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant flavonoids: Classification, distribution, biosynthesis, and antioxidant activity. Food Chem. 2022, 383, 132531. [Google Scholar] [CrossRef] [PubMed]
- Dastmalchi, M.; Dhaubhadel, S. Soybean chalcone isomerase: Evolution of the fold, and the differential expression and localization of the gene family. Planta 2015, 241, 507–523. [Google Scholar] [CrossRef] [PubMed]
- Lai, S.H.; Chye, M.L. Plant Acyl-CoA-Binding Proteins-Their Lipid and Protein Interactors in Abiotic and Biotic Stresses. Cells 2021, 10, 1064. [Google Scholar] [CrossRef] [PubMed]
- Kriechbaumer, V.; Brandizzi, F. The plant endoplasmic reticulum: An organized chaos of tubules and sheets with multiple functions. J. Microsc. 2020, 280, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xu, R.; Li, Y. Molecular Networks of Seed Size Control in Plants. Annu. Rev. Plant Biol. 2019, 70, 435–463. [Google Scholar] [CrossRef]
- Tabassum, N.; Blilou, I. Cell-to-Cell Communication During Plant-Pathogen Interaction. Mol. Plant Microbe Interact. 2022, 35, 98–108. [Google Scholar] [CrossRef]
- Zhaogao, L.; Yaxuan, W.; Mengwei, X.; Haiyu, L.; Lin, L.; Delin, X. Molecular mechanism overview of metabolite biosynthesis in medicinal plants. Plant Physiol. Biochem. 2023, 204, 108125. [Google Scholar] [CrossRef]
- Vishwakarma, S.K.; Singh, N.; Kumaria, S. Genome-wide identification and analysis of the PAL genes from the orchids Apostasia shenzhenica, Dendrobium catenatum and Phalaenopsis equestris. J. Biomol. Struct. Dyn. 2023, 41, 1295–1308. [Google Scholar] [CrossRef]
- Li, K.-L.; Liang, Y.-M.; Chen, Z.; Zheng, P.-J.; Zhang, G.-Q.; Yan, B.; Elshikh, M.S.; Rizwana, H.; Chen, B.; Xu, Q. Genome-wide identification of the alkaloid synthesis gene family CYP450, gives new insights into alkaloid resource utilization in medicinal Dendrobium. Int. J. Biol. Macromol. 2024, 259 Pt 2, 129229. [Google Scholar] [CrossRef] [PubMed]
- Ojha, M.D.; Yadav, A.; P, H. Analyzing the potential of selected plant extracts and their structurally diverse secondary metabolites for α-glucosidase inhibitory activity: In vitro and in silico approach. J. Biomol. Struct. Dyn. 2023, 41, 9523–9538. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yan, Y.; Zhou, W.-H.; Feng, R.-Z.; Shuai, Y.-K.; Yang, L.; Liu, M.-J.; He, X.-Y.; Wei, Q. Transcriptome and metabolome reveal the accumulation of secondary metabolites in different varieties of Cinnamomum longepaniculatum. BMC Plant Biol. 2022, 22, 243. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.C.J.; Perkins, J.; Peakall, R. Anthocyanin and Flavonol Glycoside Metabolic Pathways Underpin Floral Color Mimicry and Contrast in a Sexually Deceptive Orchid. Front. Plant Sci. 2022, 13, 860997. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Ma, J.; Liu, Y.; Tan, X.; Li, X.; Xu, E.; Xu, L.; Luo, A. Heat Stress Alleviation by Exogenous Calcium in the Orchid Dendrobium nobile Lindl: A Biochemical and Transcriptomic Analysis. Int. J. Mol. Sci. 2023, 24, 14692. [Google Scholar] [CrossRef]
- Suttle, J.C.; Huckle, L.L.; Lu, S.; Knauber, D.C. Potato tuber cytokinin oxidase/dehydrogenase genes: Biochemical properties, activity, and expression during tuber dormancy progression. J. Plant Physiol. 2014, 171, 448–457. [Google Scholar] [CrossRef]
- Du, Y.; Zhang, Z.; Gu, Y.; Li, W.; Wang, W.; Yuan, X.; Zhang, Y.; Yuan, M.; Du, J.; Zhao, Q. Genome-wide identification of the soybean cytokinin oxidase/dehydrogenase gene family and its diverse roles in response to multiple abiotic stress. Front. Plant Sci. 2023, 14, 1163219. [Google Scholar] [CrossRef]
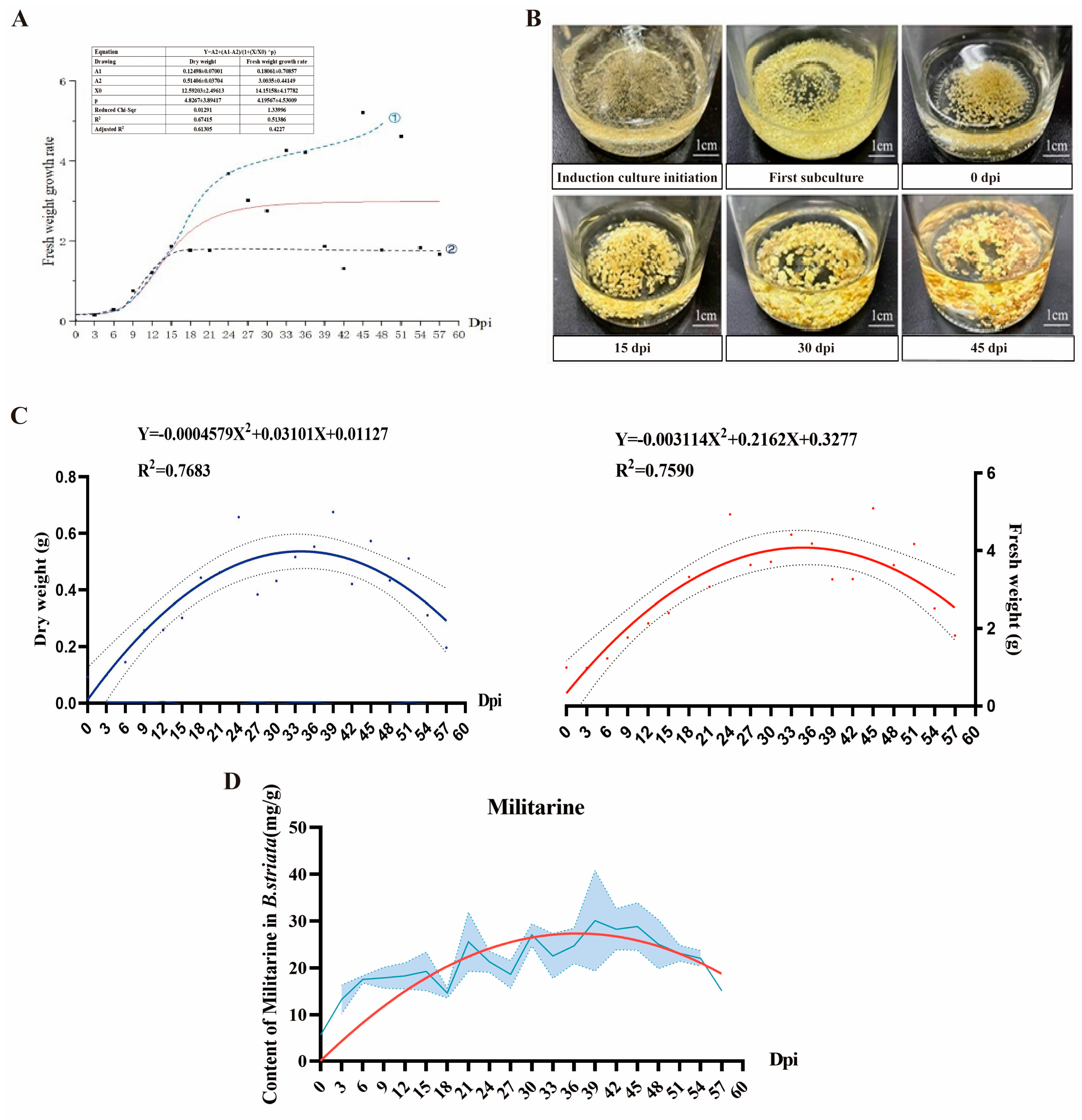
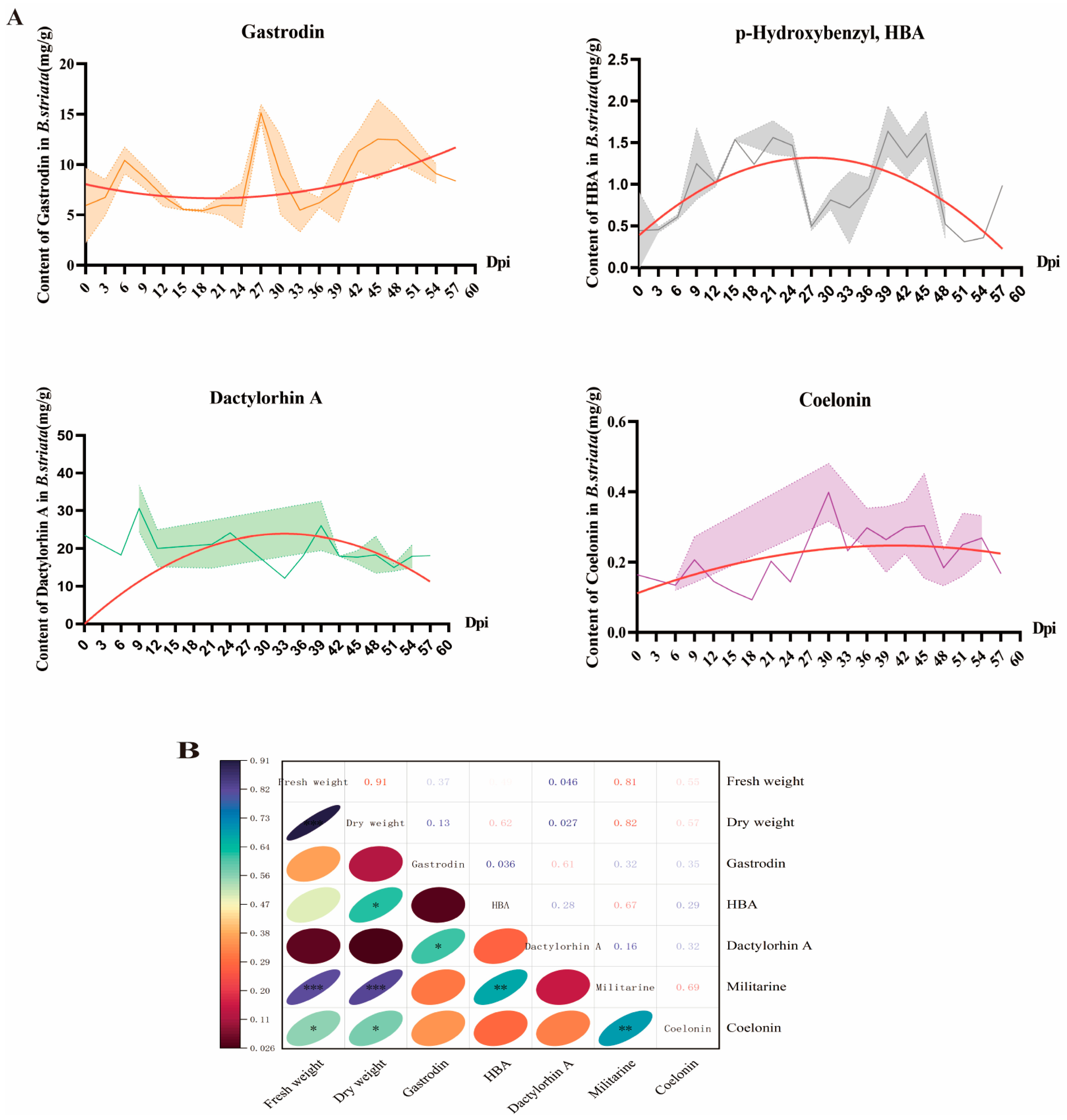
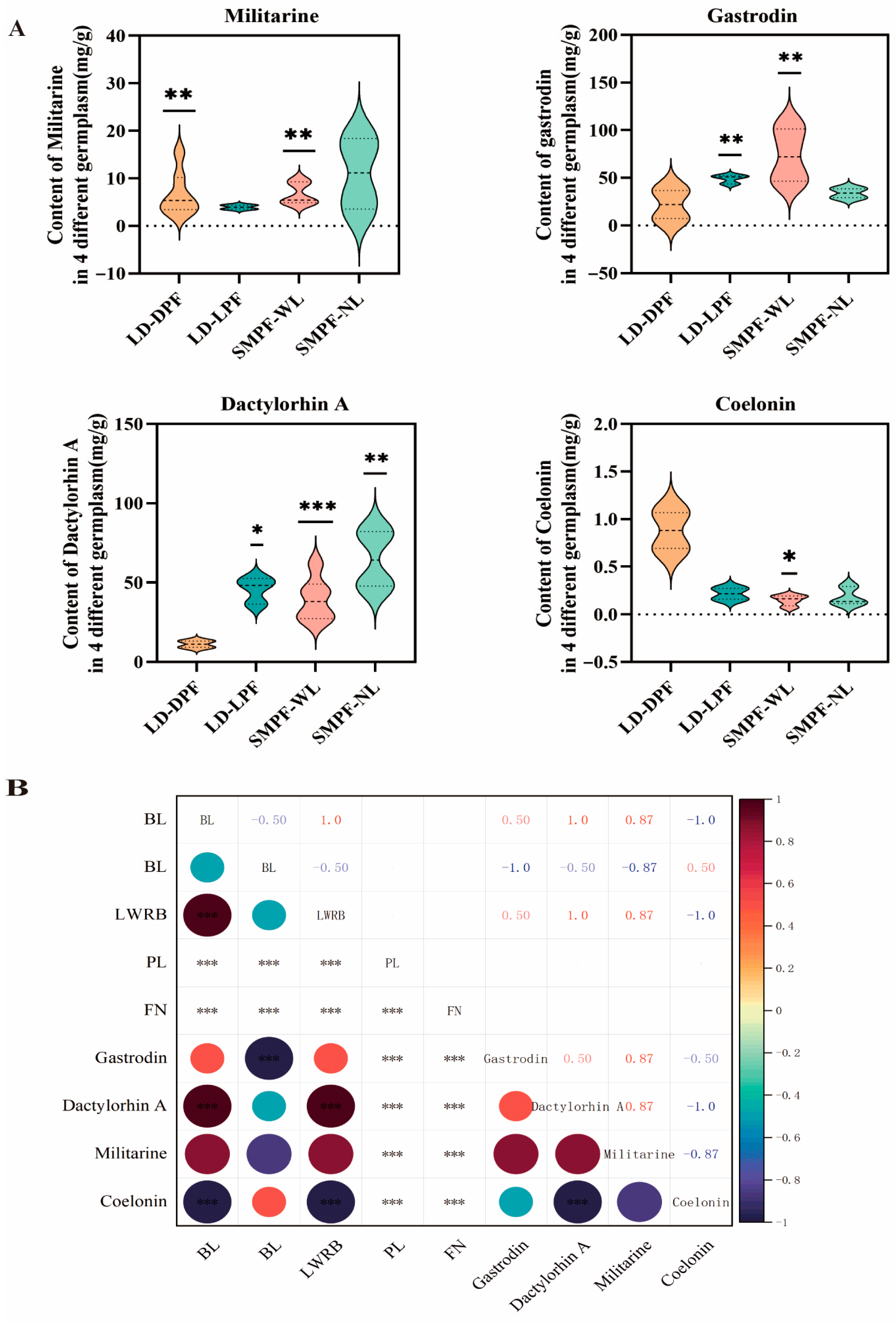
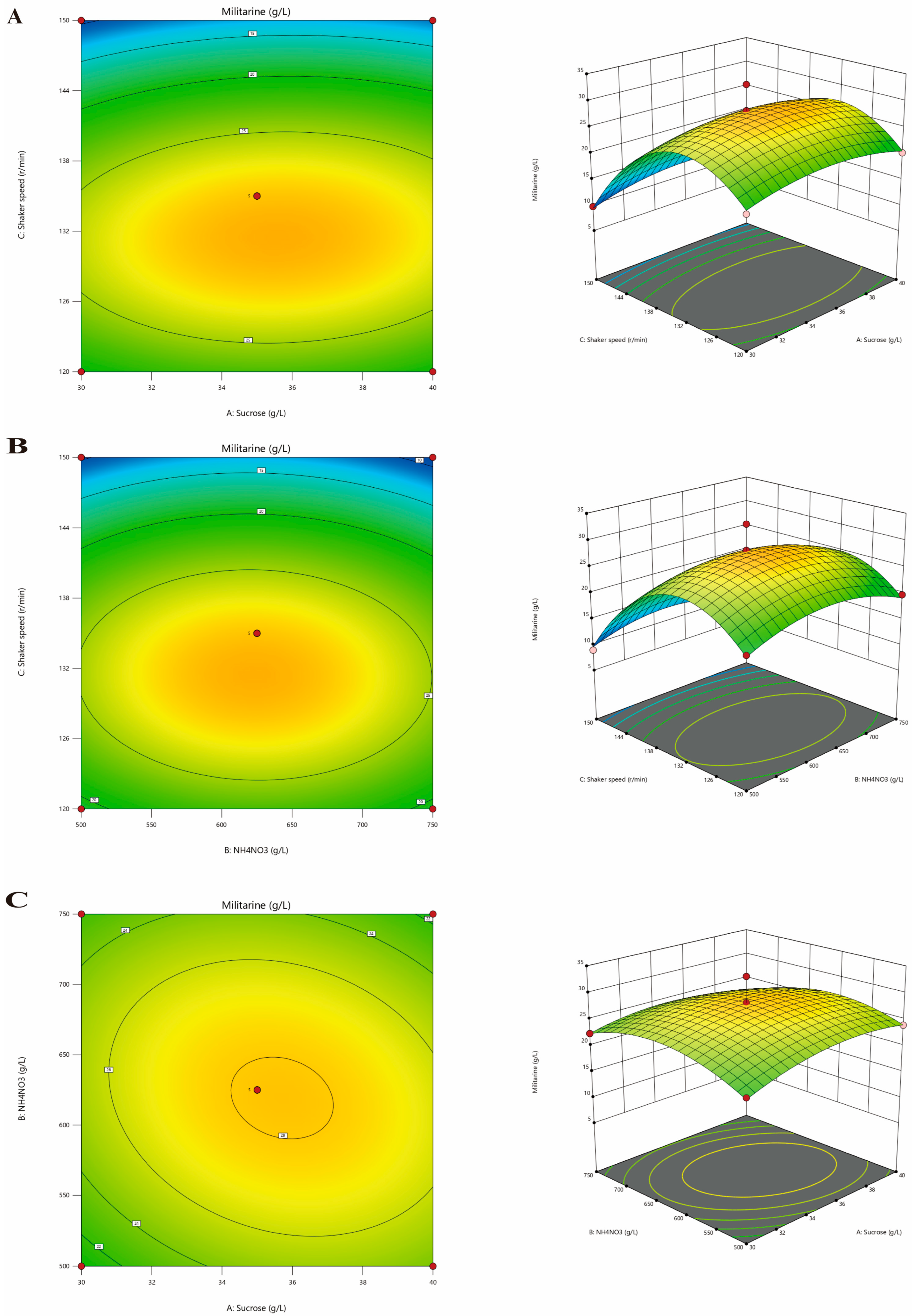
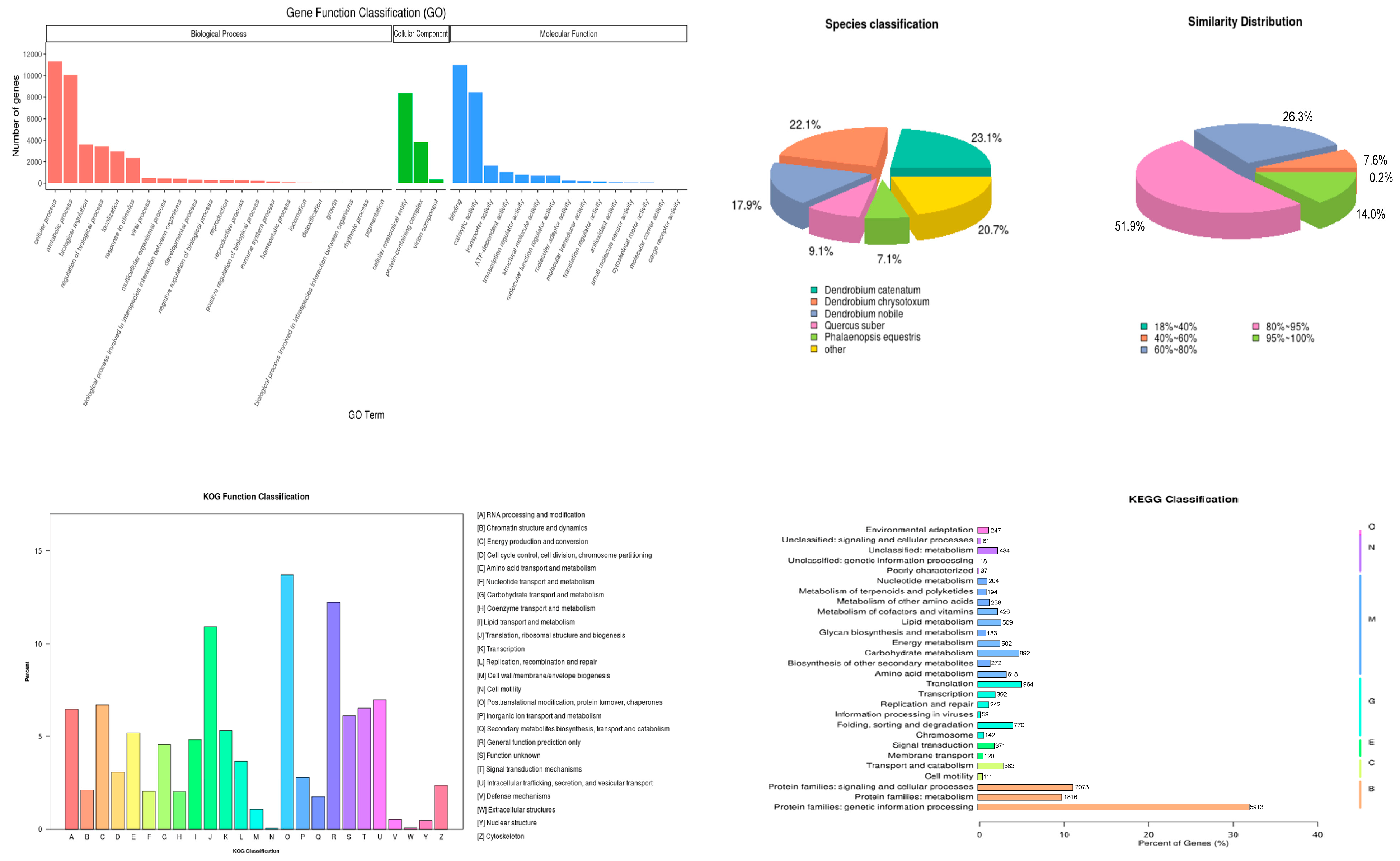

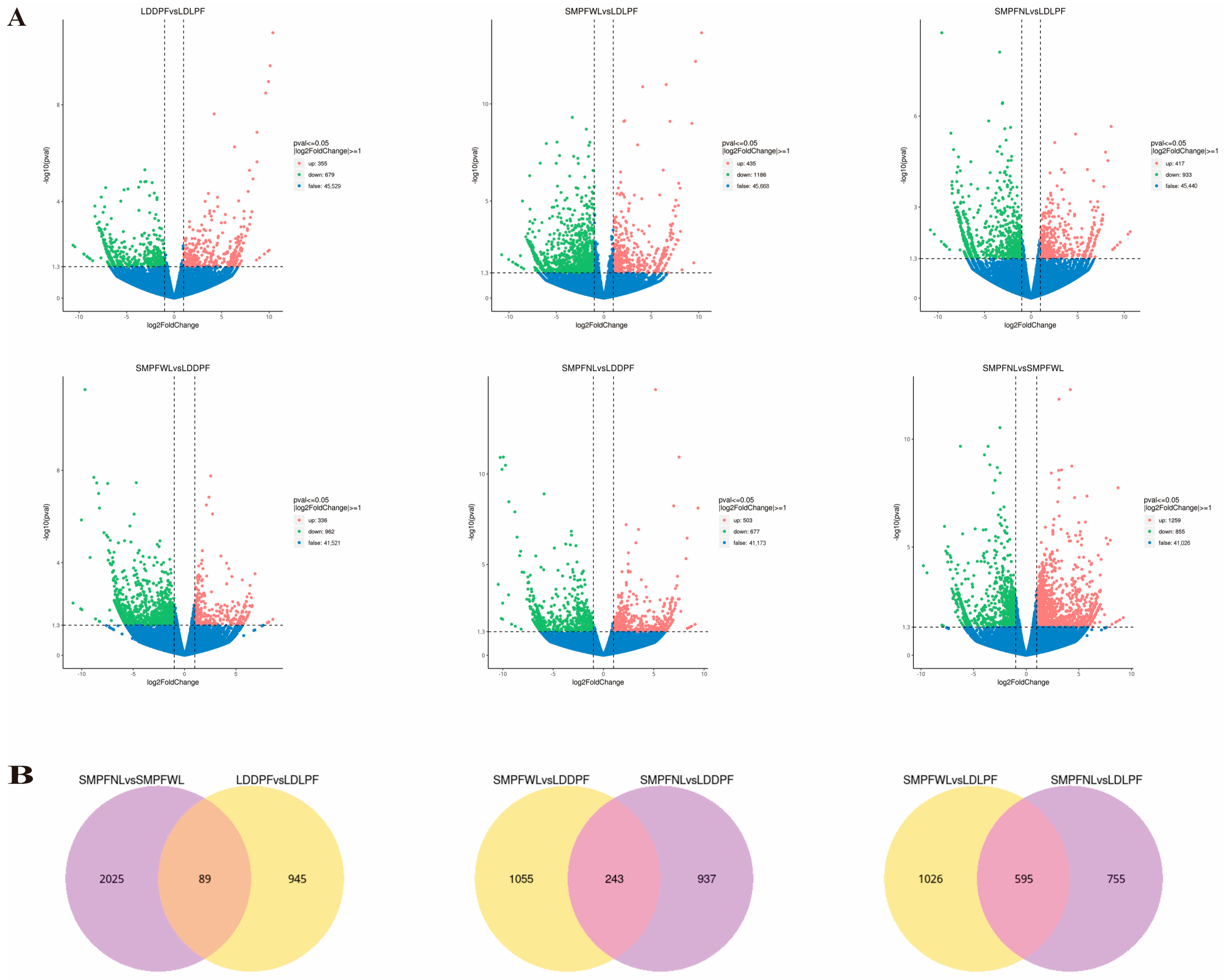
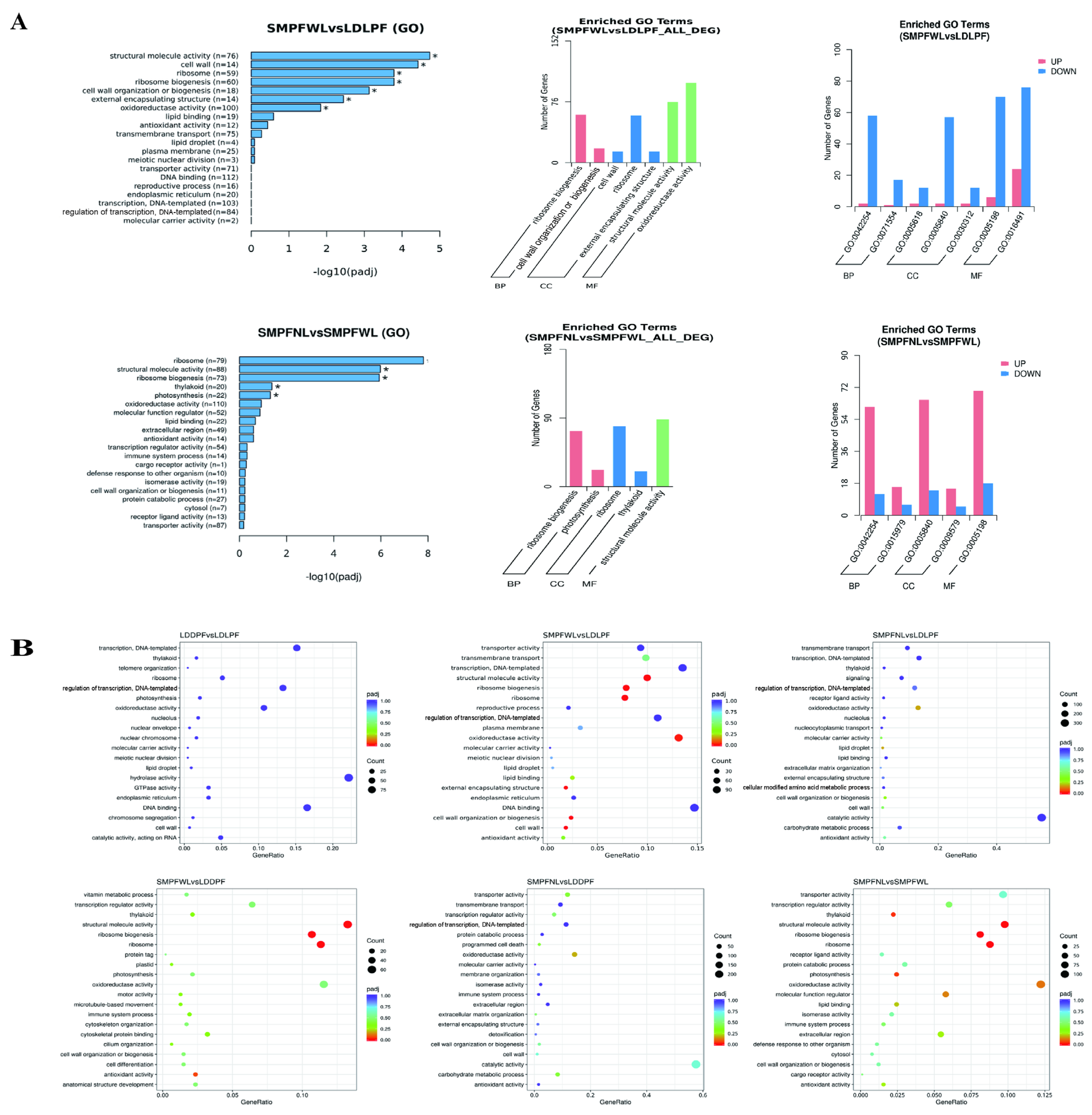
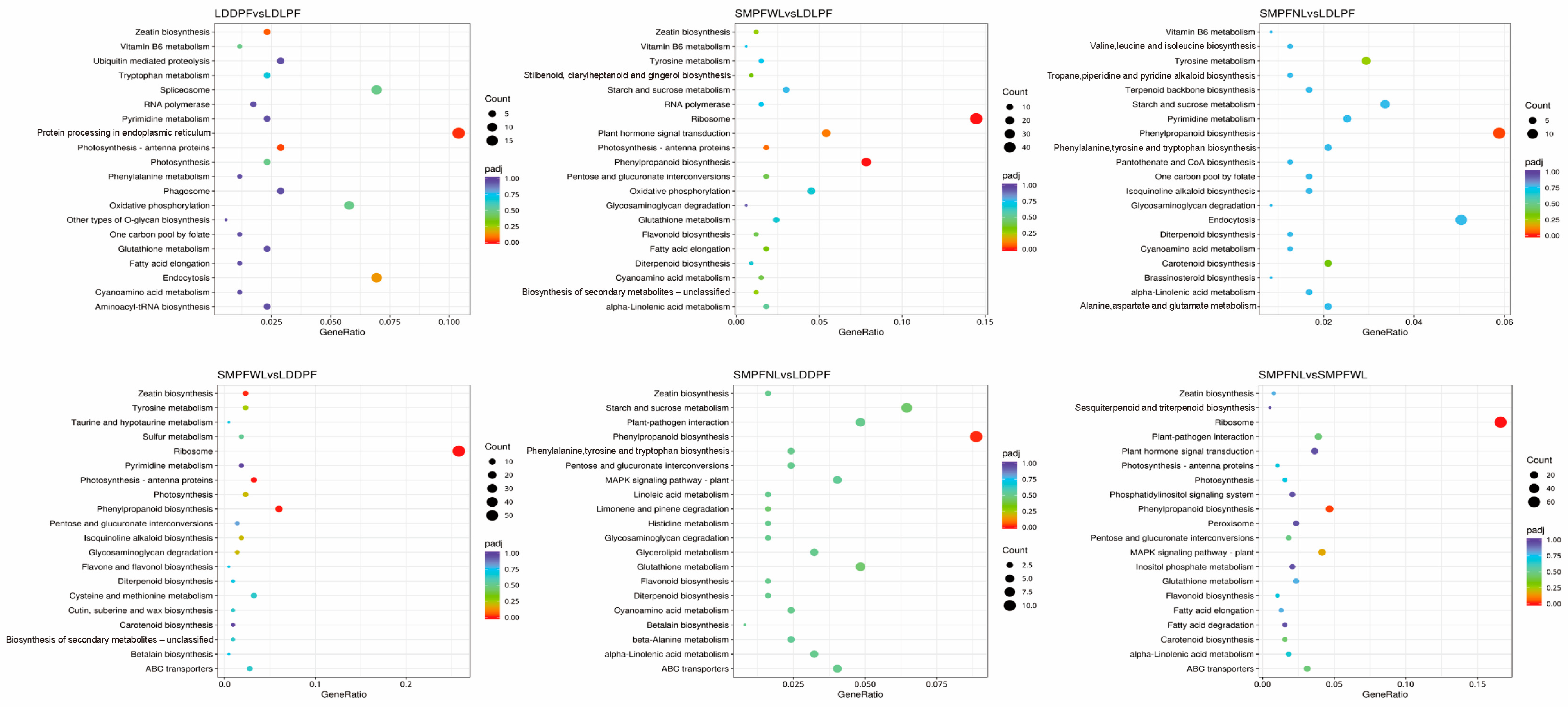
| Cultivation Period | Medium Formula | Other Cultivation Conditions |
|---|---|---|
| 0–30 days | MS + 1 mg/L 6-BA + 2 mg/L 2,4-D + 0.5 mg/L NAA + 30 g/L sucrose | 120 rpm, 25 °C, dark cultivation |
| 30–45 days | 1/2 MS + 1 mg/L 6-BA + 3 mg/L 2,4-D + 0.5 mg/L NAA + 30 g/L sucrose + 150 μmol/L NaAc | |
| 0–21 dpi | 1/2 MS + 1 mg/L 6-BA + 3 mg/L 2,4-D+ 0.5 mg/L NAA + 30 g/L sucrose + 150 μmol/L NaAc |
| Years (n) | Traits | Min | Max | Mean | SD | CV/% |
|---|---|---|---|---|---|---|
| 2023 (260) | BL | 1.50 | 36.00 | 15.98 | 3.88 | 0.24 |
| BW | 0.30 | 9.00 | 2.84 | 1.19 | 0.42 | |
| LWRB | 2.41 | 16.92 | 7.02 | 2.56 | 0.36 | |
| PL | 8.00 | 50.80 | 28.75 | 8.51 | 0.36 | |
| FN | 2.00 | 15.00 | 5.90 | 2.10 | 0.33 | |
| 2024 (318) | BL | 1.20 | 57.50 | 16.85 | 8.41 | 0.50 |
| BW | 0.30 | 16.00 | 3.90 | 2.40 | 0.62 | |
| LWRB | 0.15 | 10.95 | 4.85 | 1.54 | 0.32 | |
| PL | 4.00 | 65.00 | 30.06 | 19.06 | 0.63 | |
| FN | 0.00 | 15.00 | 6.00 | 2.59 | 0.43 |
| Sample | Library | Raw Reads | Raw Bases | Clean Reads | Clean Bases | Error Rate | Q20 | Q30 | GC Pct |
|---|---|---|---|---|---|---|---|---|---|
| LDDPF1 | FRAS240241839-1r | 23386503 | 7.02 | 22800806 | 6.84 | 0.01 | 97.96 | 94.16 | 45.54 |
| LDDPF4 | FRAS240241840-1r | 20259718 | 6.08 | 19705286 | 5.91 | 0.01 | 97.62 | 93.41 | 44.67 |
| LDDPF30 | FRAS240241841-1r | 23139646 | 6.94 | 22678057 | 6.80 | 0.01 | 97.85 | 93.96 | 45.86 |
| LDLPF63 | FRAS240241846-1r | 23841034 | 7.15 | 23313251 | 6.99 | 0.01 | 97.45 | 93.19 | 46.08 |
| LDLPF86 | FRAS240241847-1r | 22823680 | 6.85 | 22155442 | 6.65 | 0.01 | 97.50 | 93.24 | 44.80 |
| LDLPF133 | FRAS240241848-1r | 23077143 | 6.92 | 22484812 | 6.75 | 0.01 | 97.85 | 93.93 | 45.20 |
| SMPFWL2 | FRAS240241851-1r | 23324752 | 7.00 | 22807498 | 6.84 | 0.01 | 97.60 | 93.40 | 45.60 |
| SMPFWL3 | FRAS240241852-1r | 24477716 | 7.34 | 23845505 | 7.15 | 0.01 | 97.60 | 93.44 | 45.42 |
| SMPFWL5 | FRAS240241853-1r | 22866036 | 6.86 | 21999031 | 6.60 | 0.01 | 97.62 | 93.47 | 45.74 |
| SMPFNL86 | FRAS240241858-1r | 23712305 | 7.11 | 23207006 | 6.96 | 0.01 | 97.89 | 94.07 | 45.62 |
| SMPFNL93 | FRAS240241859-1r | 27960478 | 8.39 | 27250089 | 8.18 | 0.01 | 97.64 | 93.56 | 44.44 |
| SMPFNL101 | FRAS240241860-1r | 23072562 | 6.92 | 22454273 | 6.74 | 0.01 | 97.51 | 93.30 | 45.20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Xu, M.; Li, H.; Yang, N.; Wen, W.; Li, L.; Yising, L.; Vadsana, S.; Sonekeo, V.; Xu, D. Suspension Culture Optimization and Transcriptome-Guided Identification of Candidate Regulators for Militarine Biosynthesis in Bletilla striata. Horticulturae 2025, 11, 1315. https://doi.org/10.3390/horticulturae11111315
Li Y, Xu M, Li H, Yang N, Wen W, Li L, Yising L, Vadsana S, Sonekeo V, Xu D. Suspension Culture Optimization and Transcriptome-Guided Identification of Candidate Regulators for Militarine Biosynthesis in Bletilla striata. Horticulturae. 2025; 11(11):1315. https://doi.org/10.3390/horticulturae11111315
Chicago/Turabian StyleLi, Yang, Mengwei Xu, Hongwei Li, Ning Yang, Weie Wen, Lin Li, Laoxeun Yising, Sysouvong Vadsana, Vannavong Sonekeo, and Delin Xu. 2025. "Suspension Culture Optimization and Transcriptome-Guided Identification of Candidate Regulators for Militarine Biosynthesis in Bletilla striata" Horticulturae 11, no. 11: 1315. https://doi.org/10.3390/horticulturae11111315
APA StyleLi, Y., Xu, M., Li, H., Yang, N., Wen, W., Li, L., Yising, L., Vadsana, S., Sonekeo, V., & Xu, D. (2025). Suspension Culture Optimization and Transcriptome-Guided Identification of Candidate Regulators for Militarine Biosynthesis in Bletilla striata. Horticulturae, 11(11), 1315. https://doi.org/10.3390/horticulturae11111315







