Screening 60Co-γ Irradiated Camellia oleifera Lines for Anthracnose-Resistant
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials and Design
2.2. Measurement of Physiological and Biochemical Indicators
2.3. RNA Extraction and Real-Time Quantitative PCR Analysis
2.4. Statistical Analysis Methods
3. Results
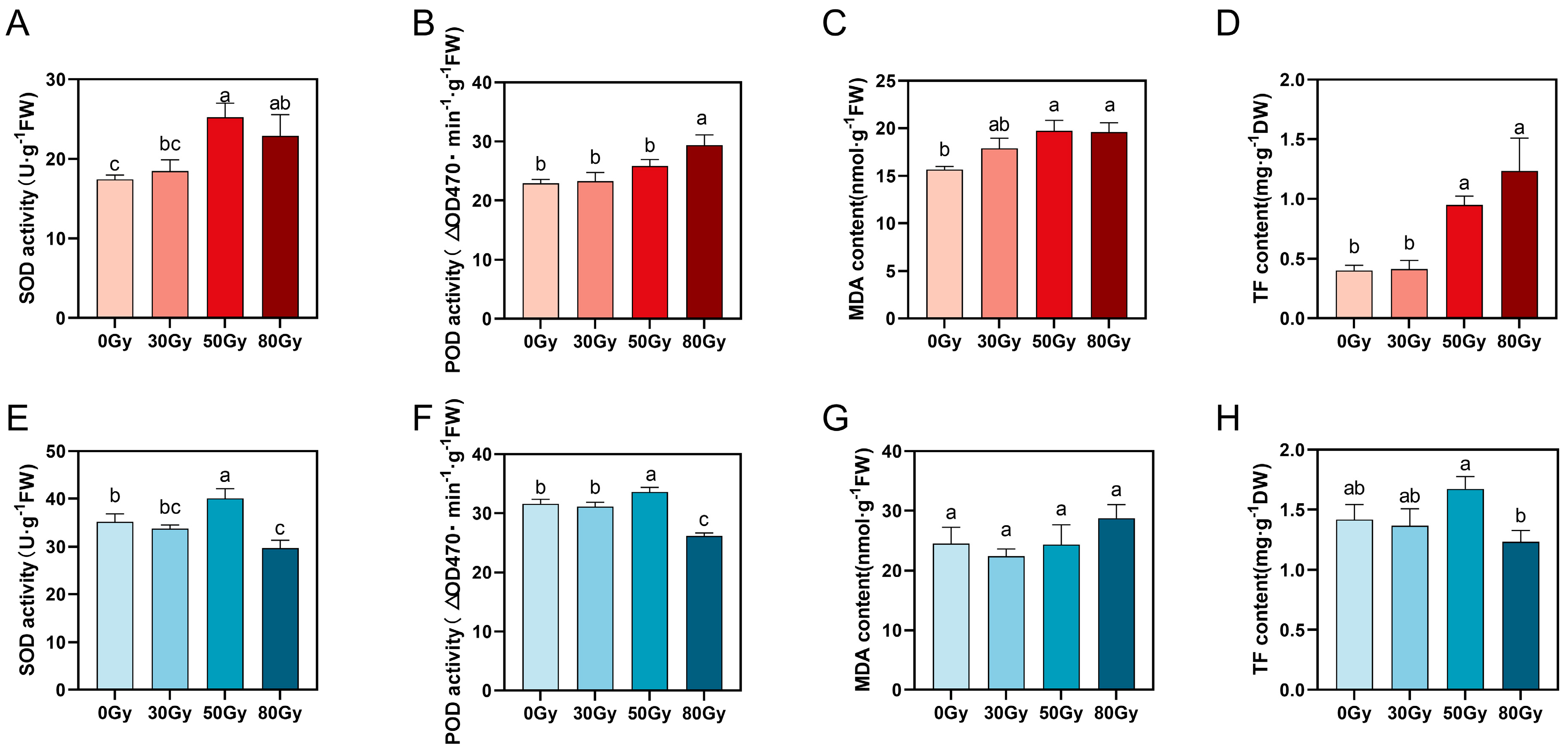
3.1. Effects of Different Doses of 60Co-γ Radiation on C. oleifera
3.1.1. Effects of Different Radiation Doses on the Germination Percentage of C. oleifera
3.1.2. Effects of Different Radiation Doses on the Physiological and Biochemical Indicators of C. oleifera
3.1.3. Effects of Different Radiation Doses on the Anthracnose Resistance of C. oleifera
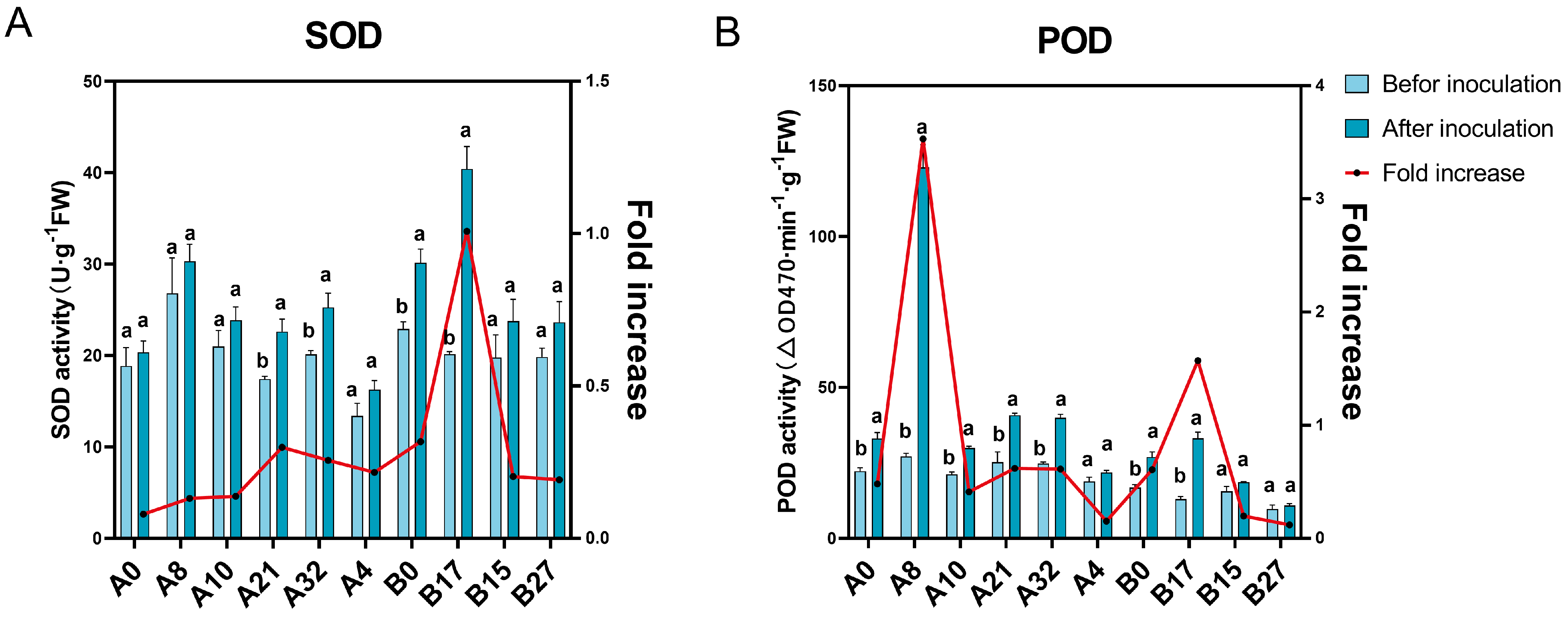
3.2. Screening of C. oleifera Mutant Seedlings with Differing Anthracnose Resistance
3.2.1. Through Phenotypic Screening
3.2.2. Physiological and Biochemical Changes in Mutants with Differing Disease Resistance before and after Inoculation
3.3. q-PCR Analysis of Gene Expression in Mutants with Differing Disease Susceptibility after C. gloeosporioides Inoculation
3.3.1. Effects on Genes Related to ROS Scavenging
3.3.2. Effects on Disease-Resistance-Related Transcription Factor Genes
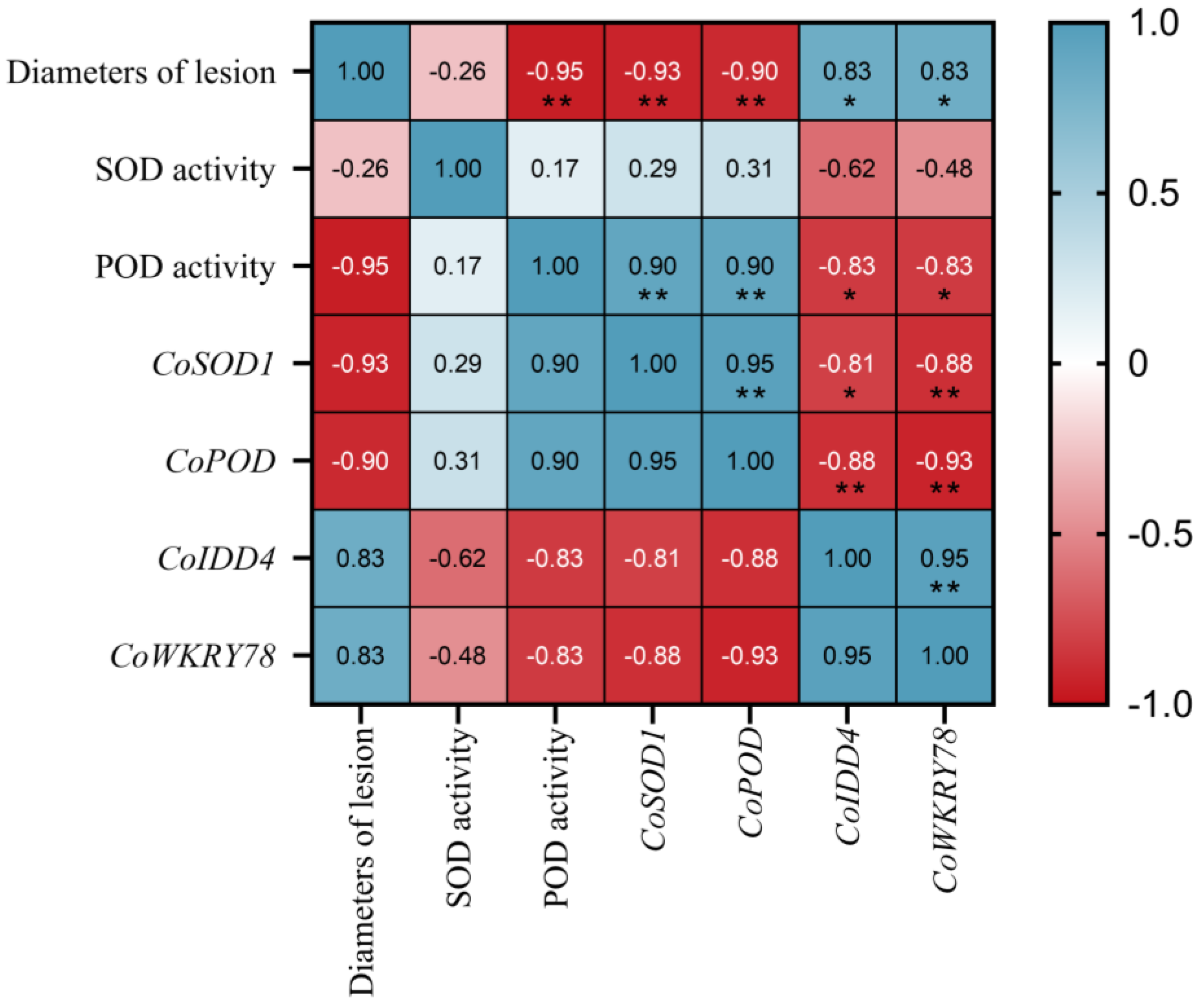
3.4. Correlation Analysis
4. Discussion
4.1. Mutagenic Effects of 60Co-γ Radiation on C. oleifera
4.2. Screening for Disease-Resistant Germplasms in C. oleifera
4.3. Gene Expression in Mutants with Differing Disease Resistance
4.4. Limitations and Future Prospects
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yang, D.; Chen, Y.; Wang, R.; He, Y.; Ma, X.; Shen, J.; He, Z.; Lai, H. Effects of Exogenous Abscisic Acid on the Physiological and Biochemical Responses of Camellia oleifera Seedlings under Drought Stress. Plants 2024, 13, 225. [Google Scholar] [CrossRef]
- Zheng, J.; Su, H.; Pu, S.; Chen, H.; EI-Kassaby, Y.A.; Yang, Z.; Feng, J. High-yield hybrid breeding of Camellia oleifolia based on ISSR molecular markers. BMC Plant Biol. 2024, 24, 517. [Google Scholar] [CrossRef]
- Choi, H.-I.; Han, S.M.; Jo, Y.D.; Hong, M.J.; Kim, S.H.; Kim, J.-B. Effects of Acute and Chronic Gamma Irradiation on the Cell Biology and Physiology of Rice Plants. Plants 2021, 10, 439. [Google Scholar] [CrossRef]
- Li, Y.; Chen, L.; Zhan, X.; Liu, L.; Feng, F.; Guo, Z.; Wang, D.; Chen, H. Biological effects of gamma-ray radiation on tulip (Tulipa gesneriana L.). PeerJ 2022, 10, e12792. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Muller, H. The Measurement of Gene Mutation Rate in Drosophila, Its High Variability, and Its Dependence upon Temperature. Genetics 1928, 13, 279–357. [Google Scholar] [CrossRef] [PubMed]
- Stadler, L. Genetic Effects of X-rays in Maize. Proc. Natl. Acad. Sci. USA 1928, 14, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Stadler, L.J. Mutations in barley induced by X-rays and radium. Science 1928, 68, 186–187. [Google Scholar] [CrossRef]
- Hazra, S.; Gorai, S.; Roy, S.; Bose, S.; Hazra, P.; Chattopadhyay, A.; Ali, M.; Jambhulkar, S.; Maji, A. Isolation of yellow vein mosaic virus (YVMV)-resistant mutants of okra (Abelmoschus esculentus L.) through applied mutagenesis. Plant Breed. 2023, 143, 232–245. [Google Scholar] [CrossRef]
- Riviello-Flores, M.d.l.L.; Cadena-Iñiguez, J.; Ruiz-Posadas, L.d.M.; Arévalo-Galarza, M.d.L.; Castillo-Juárez, I.; Soto Hernández, M.; Castillo-Martínez, C.R. Use of Gamma Radiation for the Genetic Improvement of Underutilized Plant Varieties. Plants 2022, 11, 1161. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Jiménez, M.; Pérez-Tornero, O. Mutants of Citrus macrophylla rootstock obtained by gamma radiation improve salt resistance through toxic ion exclusion. Plant Physiol. Biochem. 2020, 155, 494–501. [Google Scholar] [CrossRef]
- Truc, N.T.; Uthairatanakij, A.; Srilaong, V.; Laohakunjit, N.; Jitareerat, P. Effect of electron beam radiation on disease resistance and quality of harvested mangoes. Radiat. Phys. Chem. 2021, 180, 109289. [Google Scholar] [CrossRef]
- Sellapillai, L.; Dhanarajan, A.; Raina, A.; Ganesan, A. Gamma ray induced positive alterations in morphogenetic and yield attributing traits of finger millet (Eleusine coracana (L.) Gaertn.) in M2 generation. Plant Sci. Today 2022, 9, 939–949. [Google Scholar] [CrossRef]
- Yoon, Y.; Ameer, K.; Song, B.; Kim, J.; Park, H.; Lee, K.; Eun, J.; Park, J. Effects of X-ray irradiation on the postharvest quality characteristics of ‘Maehyang’ strawberry (Fragaria × ananassa). Food Chem. 2020, 325, 126817. [Google Scholar] [CrossRef] [PubMed]
- Ragini, R.; Murukan, N.; Sekhon, N.K.; Chugh, C.; Agarwal, P.; Yadav, P.; Mallick, N.; Jha, S.K.; Iquebal, M.A.; Tandon, G.; et al. Breaking the association between gametocidal gene(s) and leaf rust resistance gene (LrS2427) in Triticum aestivum-Aegilops speltoides derivative by gamma irradiation. Mol. Breed. 2024, 44, 54. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huang, J.; Wen, S.; Zhang, Y.; Zhong, Q.; Yang, L.; Chen, L. Abnormal megagametogenesis results in seedless-ness of a polyembryonic ‘Meiguicheng’ orange (Citrus sinensis) mutant created with gamma-rays. Sci. Hortic. 2017, 217, 73–83. [Google Scholar] [CrossRef]
- Mariana, B.D.; Arisah, H.; Yenni, Y.; Selvawajayanti, M. Seedless fruit pummelo induced by gamma ray irradiation: Fruit morphological characters and stability evaluation. Biodiversitas 2018, 19, 626–631. [Google Scholar] [CrossRef]
- Sun, Y.; Sun, H.; Li, J.; Lei, X. The photosynthetic characteristics of Camellia oleifera by Co60-γ irradiation treatment. South China For. Sci. 2017, 45, 1–3+15. [Google Scholar]
- Li, B.; Cha, Q.; Huang, Y. Effect of 60Co-y Radiation Treatments on Economic Indicators of Camellia oleifera. Guangdong For. Sci. Technol. 2015, 31, 12–16. [Google Scholar]
- Yi, L.; Li, W.; Cui, Z.; Xi, R. Effects of 60Co-γ ray irradiation on the seed germination and seedling growth of Camellia semiserrata. J. South China Agric. Univ. 2014, 35, 93–97. [Google Scholar]
- Zeng, L.; Huang, Y.; Ye, X.; Lin, W.; Li, L. The Effect of 60Co-γ Radiation on the Growth of Camellia oleifera Seedlings. Subtrop. Plant Sci. 2012, 41, 42–44. [Google Scholar]
- Sun, W.; Lei, T.; Yuan, H.; Chen, S. First Report of Anthracnose Caused by Colletotrichum kahawae and Colletotrichum horri on Tea-oil Tree in China. Plant Dis. 2022, 107, 1944. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, X.; Tan, Q.; Mo, X.; Liu, J.; Zhou, G. Recent progress on harm, pathogen classification, control and pathogenic molecular mechanism of anthracnose of oil-tea. Front. Microbiol. 2022, 13, 918339. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, J.; Xu, X.; Cheng, J.; Zheng, L.; Huang, J.; Li, D. Identification and Characterization of Colletotrichum Species Associated with Anthracnose Disease of Camellia oleifera in China. Plant Dis. 2020, 104, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wu, P.; Cao, Y.; Yang, B.; Liu, L.; Chen, J.; Zhuo, R.; Yao, X. Overexpression of dihydroflavonol 4-reductase (CoDFR) boosts flavonoid production involved in the anthracnose resistance. Front. Plant Sci. 2022, 13, 1038467. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yang, C.; Wu, P.; Yao, X.; Sheng, Y.; Zhang, C.; Lin, P.; Wang, K. Integrated Transcriptome and Metabolome Analysis Reveals Key Metabolites Involved in Camellia oleifera Defense against Anthracnose. Int. J. Mol. Sci. 2022, 23, 536. [Google Scholar] [CrossRef]
- Li, J.; Xiong, C.; Ruan, D.; Du, W.; Li, H.; Ruan, C. Identification of Camellia oleifera WRKY transcription factor genes and functional characterization of CoWRKY78. Front. Plant Sci. 2023, 14, 1110366. [Google Scholar] [CrossRef]
- Haspolat, G. Variations in Flower Color of Mutant Chrysanthemums. Horticulturae 2024, 10, 385. [Google Scholar] [CrossRef]
- Nie, L.; Cha, Q.; Huang, Y.; Chen, S. Effects of 60 Co-γ Irradiation on Seed Germination and Seedling Growth of Camellia drupifera. For. Environ. Sci. 2019, 48, 7–10. [Google Scholar]
- Wang, B.; Ye, T.; Li, C.; Li, X.; Chen, L.; Wang, G. Cell damage repair mechanism in a desert green algae Chlorella sp. against UV-B radiation. Ecotoxicol. Env. Saf. 2022, 242, 113916. [Google Scholar] [CrossRef] [PubMed]
- Morcillo, F.; Serret, J.; Beckers, A.; Collin, M.; Tisné, S.; George, S.; Poveda, R.; Louise, C.; Tranbarger, T.J. A Non-Shedding Fruit Elaeis oleifera Palm Reveals Perturbations to Hormone Signaling, ROS Homeostasis, and Hemicellulose Metabolism. Genes 2021, 12, 1724. [Google Scholar] [CrossRef]
- Zulkifli, N.A.; Izhar, K.M.A.A.; Lau, H.Y.; Azizi, M.M.F. Advances in Irradiation Technology for Plant Disease Resistance: A Review. Agric. Rev. 2024, 45, 25–34. [Google Scholar] [CrossRef]
- Ghani, M.; Sharma, S.K. Induction of powdery mildew resistance in gerbera (Gerbera jamesonii) through gamma irradiation. Physiol. Mol. Biol. Plants 2019, 25, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Jeong, R.-D.; Jeong, M.-A.; Park, M.-R. Gamma irradiation-induced disease resistance of pear (Pyrus pyrifolia “Niitaka”) against Penicillium expansum. J. Phytopathol. 2017, 165, 626–633. [Google Scholar] [CrossRef]
- Chen, X.; He, Y.; Wang, Z.; Niu, A.; Xue, Y.; Zhou, D.; Zhou, G.; Liu, J. Research progress and management strategies of fungal diseases in Camellia oleifera. Front. Microbiol. 2023, 14, 1215024. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhu, H.; He, C. Identification and Characterization of Colletotrichum Species Causing Tea-Oil Camellia (Camellia oleifera C. Abel) Anthracnose in Hainan, China. Forests 2023, 14, 1030. [Google Scholar] [CrossRef]
- Bin, L.; Qiu, W.; Chao-Hui, Z.; Zhao-Yang, H.; Hai, Y.; Jun, L.; Yan-de, L. Research on anthracnose grade of Camellia oleifera based on the combined LIBS and THz technology. Plant Methods 2022, 18, 52. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ye, J.; Zhang, Q.; Cheng, P.; Wang, Y.; Zou, J.; Lin, S.; Li, M.; Jia, M.; Chen, Y.; Jia, X.; et al. Aviation Mutagenesis Alters the Content of Volatile Compounds in Dahongpao (Camellia sinensis) Leaves and Improves Tea Quality. Foods 2024, 13, 946. [Google Scholar] [CrossRef]
- Dalvi, S.G.; Tawar, P.N.; Suprasanna, P.; Dixit, G.B.; Prasad, D.T. EMS-Based In Vitro Mutagenesis and Mutant Screening for Smut Resistance with Agronomic Traits in Sugarcane. Sugar Tech. 2021, 23, 854–864. [Google Scholar] [CrossRef]
- Tiryaki, I.; Sari, U.; Cetin, S.; Acar, O. Improved drought tolerance of EMS mutagenized Alfalfa (Medicago sativa L.) mutants by in vitro screening at germination stage. Sci. Rep. 2022, 12, 12693. [Google Scholar] [CrossRef]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.; He, S.; Xin, X. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Jarin, A.S.; Islam, M.M.; Rahat, A.; Ahmed, S.; Ghosh, P.; Murata, Y. Drought Stress Tolerance in Rice: Physiological and Biochemical Insights. Int. J. Plant Biol. 2024, 15, 692–718. [Google Scholar] [CrossRef]
- Huang, Q.; Li, F.; Meng, F. Functional Characterization of the Transcription Factor Gene CgHox7 in Colletotrichum gloeosporioides, Which Is Responsible for Poplar Anthracnose. J. Fungi 2024, 10, 505. [Google Scholar] [CrossRef]
- Jiang, S.; Li, Z.; Yuan, H.; Jin, J.; Xiao, C.; Cui, Y. Quantification Assessment of Winter Wheat Sensitivity under Different Drought Scenarios during Growth. Water 2024, 16, 2048. [Google Scholar] [CrossRef]
- Zipfel, C.; Oldroyd, G.E. Plant signalling in symbiosis and immunity. Nature 2017, 543, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Hou, C.; Ren, Z.; Wang, C.; Zhao, F.; Dahlbeck, D.; Hu, S.; Zhang, L.; Niu, Q.; Li, L.; et al. A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 2019, 572, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Lacombe, S.; Rougon-Cardoso, A.; Sherwood, E.; Peeters, N.; Dahlbeck, D.; van Esse, H.P.; Smoker, M.; Rallapalli, G.; Thomma, B.P.; Staskawicz, B.; et al. Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat. Biotechnol. 2010, 28, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Chinchilla, D.; Zipfel, C.; Robatzek, S.; Kemmerling, B.; Nürnberger, T.; Jones, J.D.; Felix, G.; Boller, T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 2007, 448, 497–500. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.A.; Jones, J.D.; Dangl, J.L. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 2006, 141, 373–378. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Shortt, B.J.; Lawrence, E.B.; Levine, E.B.; Fitzsimmons, K.C.; Shah, D.M. Disease resistance conferred by expression of a gene encoding H2O2-generating glucose oxidase in transgenic potato plants. Plant Cell 1995, 7, 1357–1368. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ghorbel, M.; Olayen, W.; Brini, F. Roles of enzymatic antioxidants in stress response and signaling in plants. In Defense-Related Proteins in Plants, 1st ed.; Upadhyay, S.K., Ed.; Academic Press: Cambridge, MA, USA, 2024; pp. 413–468. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tsuda, K.; Katagiri, F. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 2010, 13, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Völz, R.; Kim, S.K.; Mi, J.; Rawat, A.A.; Veluchamy, A.; Mariappan, K.G.; Rayapuram, N.; Daviere, J.M.; Achard, P.; Blilou, I.; et al. INDETERMINATE-DOMAIN 4 (IDD4) coordinates immune responses with plant-growth in Arabidopsis thaliana. PLoS Pathog. 2019, 15, e1007499. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rawat, A.; Völz, R.; Sheikh, A.; Mariappan, K.G.; Kim, S.K.; Rayapuram, N.; Alwutayd, K.M.; Alidrissi, L.K.; Benhamed, M.; Blilou, I.; et al. Salinity stress-induced phosphorylation of INDETERMINATE-DOMAIN 4 (IDD4) by MPK6 regulates plant growth adaptation in Arabidopsis. Front. Plant Sci. 2023, 14, 1265687. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Taj, G.; Giri, P.; Tasleem, M.; Kumar, A. MAPK Signaling Cascades and Transcriptional Reprogramming in Plant–Pathogen Interactions. In Approaches to Plant Stress and Their Management; Gaur, R., Sharma, P., Eds.; Springer: New, Delhi, India, 2014; pp. 297–316. [Google Scholar] [CrossRef]
- Vidhyasekaran, P. Mitogen-Activated Protein Kinase Cascades in Plant Innate Immunity. In PAMP Signals in Plant Innate Immunity. Signaling and Communication in Plants; Springer: Dordrecht, The Netherlands, 2014; pp. 331–374. [Google Scholar] [CrossRef]
- Thulasi, K.; Li, X.; Zhang, Y. MAP kinase signalling: Interplays between plant PAMP- and effector-triggered immunity. Cell. Mol. Life Sci. 2018, 75, 2981–2989. [Google Scholar] [CrossRef]
- Vï, R.; Kim, S.K.; Mi, J.; Mariappan, K.G.; Siodmak, A.; Al-Babili, S.; Hirt, H. A Chimeric IDD4 Repressor Constitutively Induces Immunity in Arabidopsis via the Modulation of Salicylic Acid and Jasmonic Acid Homeostasis. Plant Cell Physiol. 2019, 60, 1536–1555. [Google Scholar] [CrossRef] [PubMed]
- Erokhin, M.; Vassetzky, Y.; Georgiev, P.; Chetverina, D. Eukaryotic enhancers: Common features, regulation, and participation in diseases. Cell Mol. Life Sci. 2015, 72, 2361–2375. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nostro, A.; Papalia, T. Antimicrobial activity of carvacrol: Current progress and future prospectives. Recent Pat. Anti Infect. Drug Discov. 2012, 7, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Schoenfelder, S.; Fraser, P. Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 2019, 20, 437–455. [Google Scholar] [CrossRef]




| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| CoGAPDH | CAGGTCGAGCATCTTTGATTCC | CCACCAACTTAACAAAGAAATCATTC |
| CoSOD1 | TCAGTGGCACCGTACACTTC | TTAAGCCCAGAGACGCTTCC |
| CoPOD | TCATACATTCGGACGGGCTG | AGTTGTAAAGGCGTGGGGTC |
| CoIDD4 | AACACGAAAGCCTCCCACCA | GCAGGAGAAGTGGCATTGGC |
| CoWRKY78 | GGTCCTACTCAACTTCGGTTC | GGTAGTGGTGGTTGGGAAATA |
| Variety | Radiation Dose (Gy) | Relative Emergence Percentage | Relative Mortality Percentage |
|---|---|---|---|
| XL210 | 0 | 100% a | 0 d |
| 30 | 75.78% b | 24.22% c | |
| 50 | 9.09% c | 90.91% b | |
| 80 | 1.28% d | 98.72% a | |
| XL1 | 0 | 100% a | 0 c |
| 30 | 30.80% b | 69.20% b | |
| 50 | 29.93% b | 70.07% b | |
| 80 | 2.53% c | 97.47% a |
| Sample (Variety, Dosage) | Diameters of Lesion (mm) | Resistance |
|---|---|---|
| A0 (XL210, 0 Gy) | 5.06 ± 0.36 | / |
| A3 (XL210, 80 Gy) | 3.11 ± 0.26 * | R |
| A8 (XL210, 50 Gy) | 3.15 ± 0.22 * | R |
| A10 (XL210, 50 Gy) | 2.95 ± 0.35 * | R |
| A19 (XL210, 50 Gy) | 2.97 ± 0.21 * | R |
| A21 (XL210, 50 Gy) | 2.95 ± 0.23 * | R |
| A32 (XL210, 30 Gy) | 2.96 ± 0.16 * | R |
| A35 (XL210, 30 Gy) | 3.16 ± 0.33 * | R |
| A4 (XL210, 80 Gy) | 7.07 ± 0.54 * | S |
| B0 (XL1, 0 Gy) | 4.99 ± 0.32 | / |
| B17 (XL1, 50 Gy) | 3.21 ± 0.35 * | R |
| B15 (XL1, 50 Gy) | 8.03 ± 1.01 * | S |
| B27 (XL1, 30 Gy) | 7.84 ± 1.14 * | S |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, J.; Xun, C.; Ma, X.; Zhang, Y.; Zhang, Z.; He, Z.; He, Y.; Yang, D.; Lai, H.; Wang, R.; et al. Screening 60Co-γ Irradiated Camellia oleifera Lines for Anthracnose-Resistant. Horticulturae 2024, 10, 940. https://doi.org/10.3390/horticulturae10090940
Shen J, Xun C, Ma X, Zhang Y, Zhang Z, He Z, He Y, Yang D, Lai H, Wang R, et al. Screening 60Co-γ Irradiated Camellia oleifera Lines for Anthracnose-Resistant. Horticulturae. 2024; 10(9):940. https://doi.org/10.3390/horticulturae10090940
Chicago/Turabian StyleShen, Jiancai, Chengfeng Xun, Xiaofan Ma, Ying Zhang, Zhen Zhang, Zhilong He, Yimin He, Dayu Yang, Hanggui Lai, Rui Wang, and et al. 2024. "Screening 60Co-γ Irradiated Camellia oleifera Lines for Anthracnose-Resistant" Horticulturae 10, no. 9: 940. https://doi.org/10.3390/horticulturae10090940
APA StyleShen, J., Xun, C., Ma, X., Zhang, Y., Zhang, Z., He, Z., He, Y., Yang, D., Lai, H., Wang, R., & Chen, Y. (2024). Screening 60Co-γ Irradiated Camellia oleifera Lines for Anthracnose-Resistant. Horticulturae, 10(9), 940. https://doi.org/10.3390/horticulturae10090940






