The Role of BrKS in Leafy Head Formation Was Confirmed by Two Allelic Mutants of Chinese Cabbage (Brassica rapa L. ssp. pekinensis)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Genetic Characteristics of nhm2
2.3. Allelic Testing of nhm1 and nhm2
2.4. BrKS Clone in Mutant nhm2
2.5. Determination of Endogenous GA Content and Application of Exogenous GA3
2.6. RNA Isolation, cDNA Library Construction, and Sequencing
2.7. RNA-Seq Data Analysis
2.8. Detection of DEG Expression Level
2.9. Statistical Analysis
3. Results
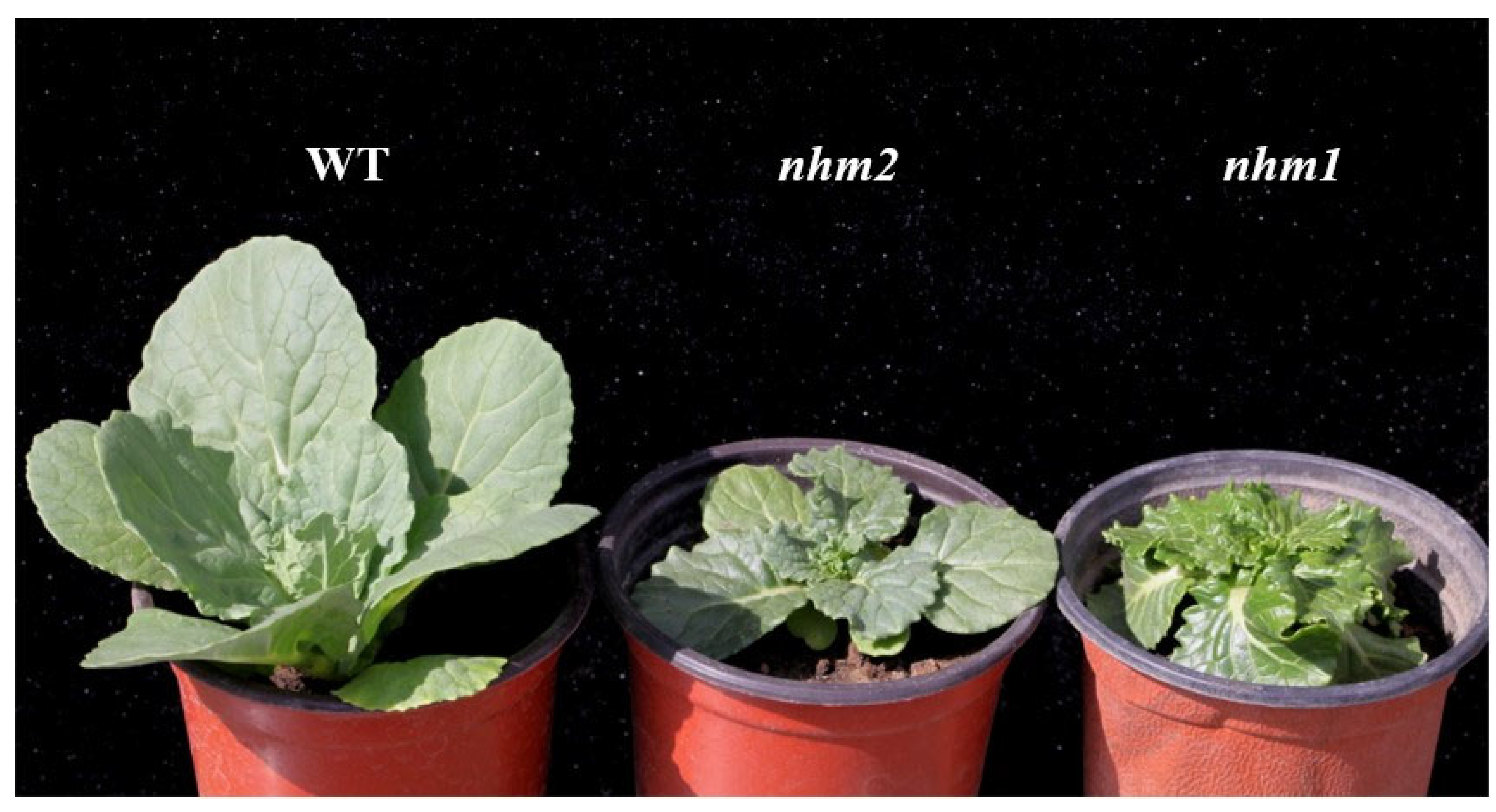
3.1. Phenotypic and Inheritance of nhm2
3.2. Allelism Testing
3.3. BrKS Clone in Mutant nhm2
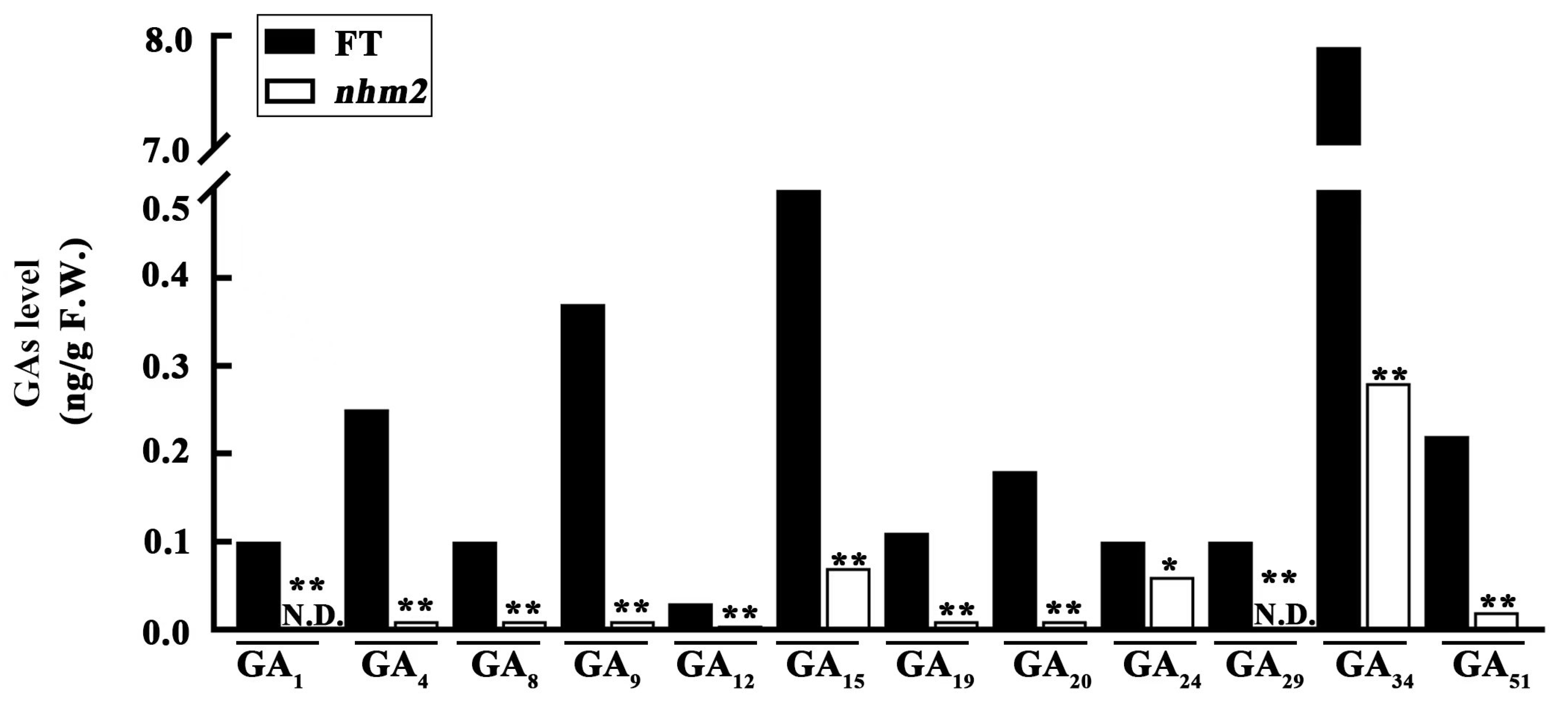
3.4. Endogenous GA Determination and Exogenous GA3 Treatment
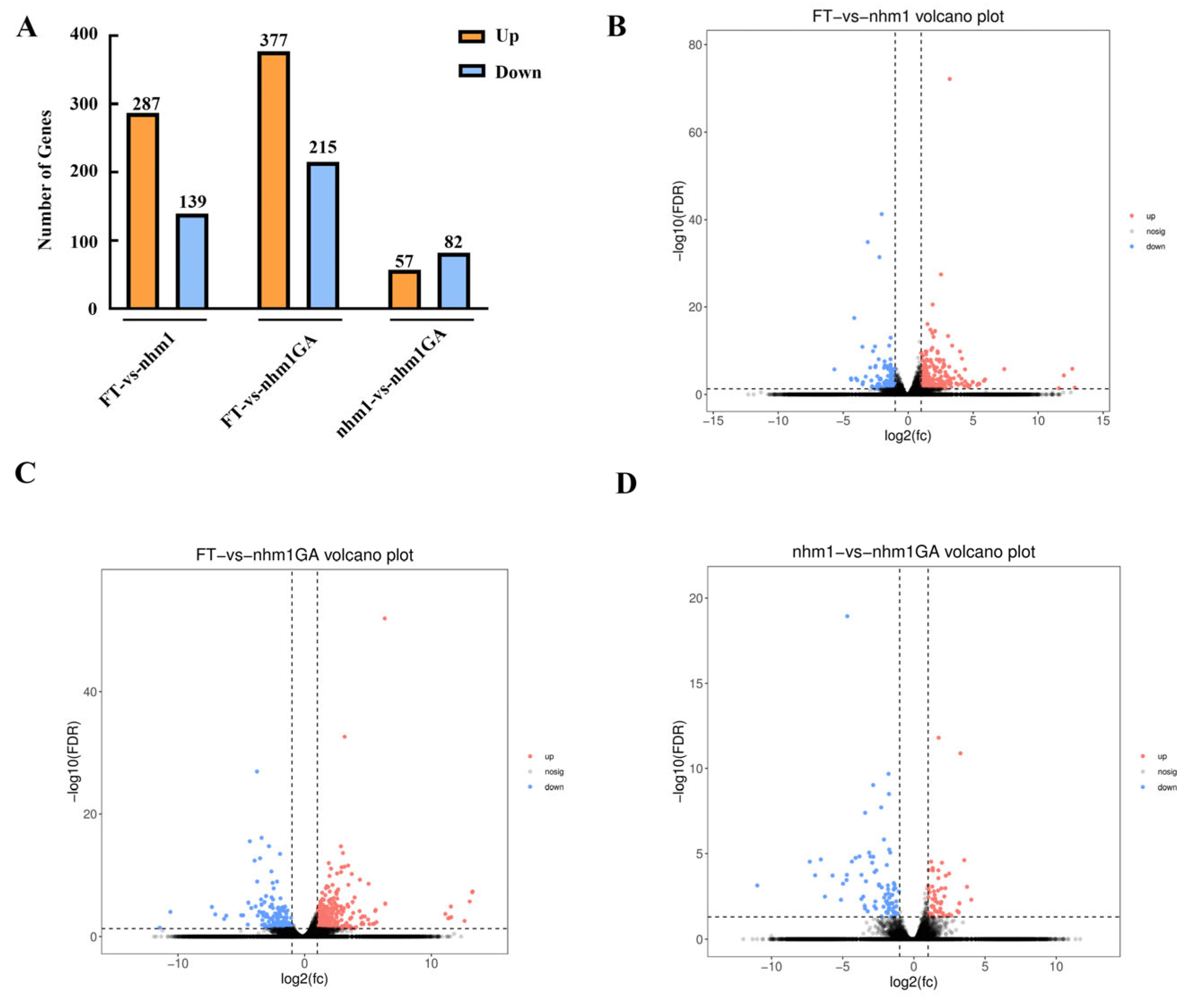
3.5. RNA-Seq Analysis
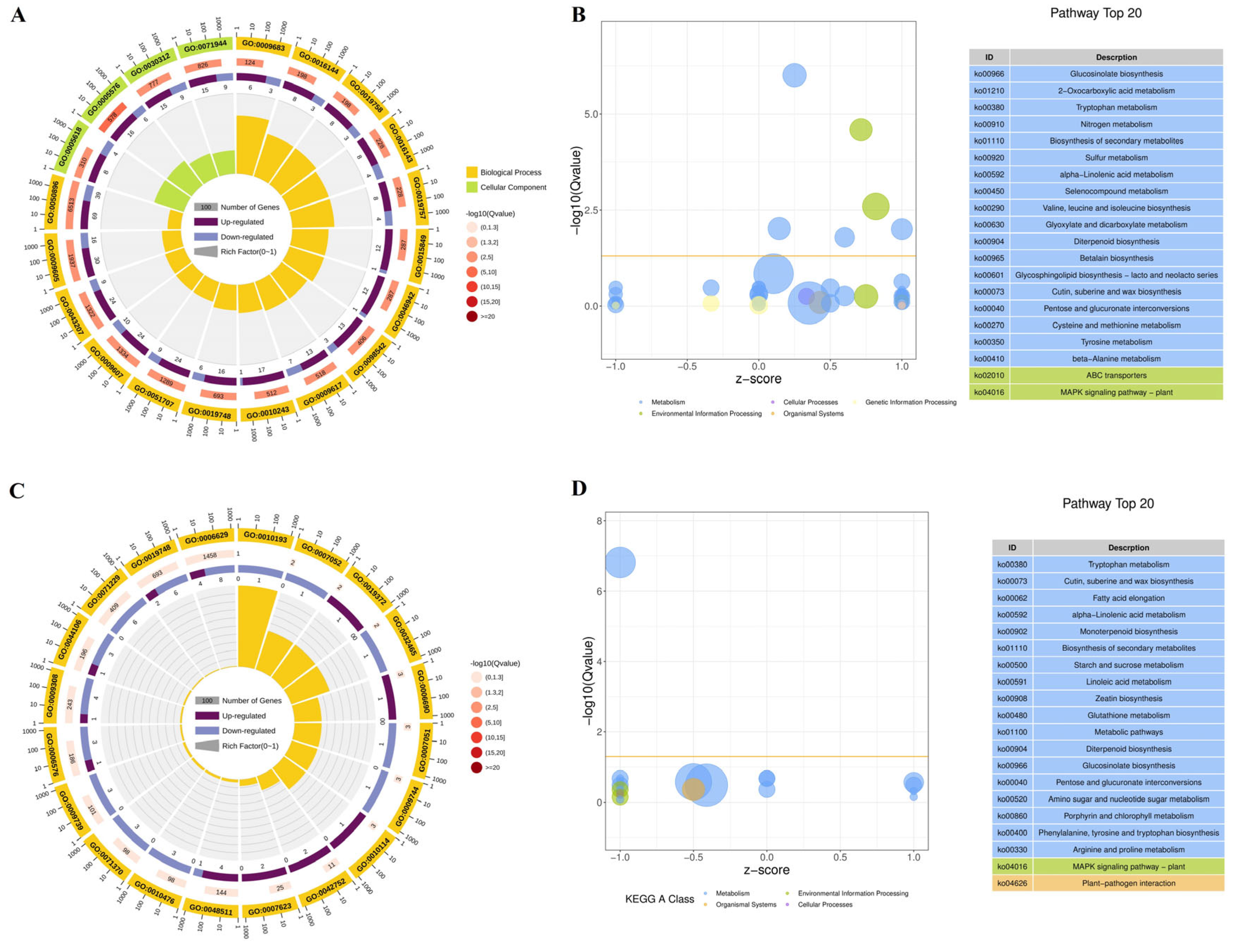
3.6. Functional Enrichment Analysis of DEGs
3.7. Analysis of the DEGs Involved in Leafy Head Formation
3.8. qRT-PCR Identification of DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yu, X.; Peng, J.; Feng, X.; Yang, S.; Zheng, Z.; Tang, X.; Shen, R.; Liu, P.; He, Y. Cloning and structural and expressional characterization of BcpLH gene preferentially expressed in folding leaf of Chinese cabbage. Sci. China C Life Sci. 2000, 43, 321–329. [Google Scholar] [CrossRef]
- Yu, X.; Wang, H.; Zhong, W.; Bai, J.; Liu, P.; He, Y. QTL mapping of leafy heads by genome resequencing in the RIL population of Brassica rapa. PLoS ONE 2013, 8, e76059. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Sun, R.; Hou, X.; Zheng, H.; Zhang, F.; Zhang, Y.; Liu, B.; Liang, J.; Zhuang, M.; Liu, Y.; et al. Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat. Genet. 2016, 48, 1218–1224. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Kato, T. Studies on the head formation of Chinese cabbage: Histological and physiological studies of head formation. J. Jpn. Soc. Hortic. Sci. 1957, 26, 154–204. [Google Scholar] [CrossRef]
- Mao, Y.; Wu, F.; Yu, X.; Bai, J.; Zhong, W.; He, Y. MicroRNA319a-targeted Brassica rapa ssp. pekinensis TCP genes modulate head shape in chinese cabbage by differential cell division arrest in leaf regions. Plant Physiol. 2014, 164, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Wang, H.; Bai, J.; Wu, F.; He, Y. Association of microRNAs with Types of Leaf Curvature in Brassica rapa. Front. Plant Sci. 2018, 9, 73. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Liu, B.; Wu, J.; Cheng, F.; Wang, X. Genetic Variation and Divergence of Genes Involved in Leaf Adaxial-Abaxial Polarity Establishment in Brassica rapa. Front. Plant Sci. 2016, 7, 94. [Google Scholar] [CrossRef]
- Cai, X.; Chang, L.; Zhang, T.; Chen, H.; Zhang, L.; Lin, R.; Liang, J.; Wu, J.; Freeling, M.; Wang, X. Impacts of allopolyploidization and structural variation on intraspecific diversification in Brassica rapa. Genome Biol. 2021, 22, 166. [Google Scholar] [CrossRef]
- Liu, Z.; Jia, L.; Wang, H.; He, Y. HYL1 regulates the balance between adaxial and abaxial identity for leaf flattening via miRNA-mediated pathways. J. Exp. Bot. 2011, 62, 4367–4381. [Google Scholar] [CrossRef]
- Ren, W.; Wu, F.; Bai, J.; Li, X.; Yang, X.; Xue, W.; Liu, H.; He, Y. BcpLH organizes a specific subset of microRNAs to form a leafy head in Chinese cabbage (Brassica rapa ssp. pekinensis). Hortic. Res. 2020, 7, 1. [Google Scholar] [CrossRef]
- Li, J.; Zhang, X.; Lu, Y.; Feng, D.; Gu, A.; Wang, S.; Wu, F.; Su, X.; Chen, X.; Li, X.; et al. Characterization of Non-heading Mutation in Heading Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Front. Plant Sci. 2019, 10, 112. [Google Scholar] [CrossRef] [PubMed]
- Horiguchi, G.; Nakayama, H.; Ishikawa, N.; Kubo, M.; Demura, T.; Fukuda, H.; Tsukaya, H. ANGUSTIFOLIA3 plays roles in adaxial/abaxial patterning and growth in leaf morphogenesis. Plant Cell Physiol. 2011, 52, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Gao, L.; Liu, W.; Song, L.; Xiao, D.; Liu, T.; Hou, X.; Zhang, C. Transcription Coactivator ANGUSTIFOLIA3 (AN3) Regulates Leafy Head Formation in Chinese Cabbage. Front. Plant Sci. 2019, 10, 520. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liang, J.; Cai, X.; Chen, H.; Wu, J.; Lin, R.; Cheng, F.; Wang, X. Divergence of three BRX homoeologs in Brassica rapa and its effect on leaf morphology. Hortic. Res. 2021, 8, 68. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.Y.; Zhuang, J.; Xiong, A.S. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef] [PubMed]
- Marsch-Martinez, N.; Greco, R.; Becker, J.D.; Dixit, S.; Bergervoet, J.H.; Karaba, A.; de Folter, S.; Pereira, A. BOLITA, an Arabidopsis AP2/ERF-like transcription factor that affects cell expansion and proliferation/differentiation pathways. Plant Mol. Biol. 2006, 62, 825–843. [Google Scholar] [CrossRef]
- Xu, Y.; Wu, H.; Zhao, M.; Wu, W.; Xu, Y.; Gu, D. Overexpression of the Transcription Factors GmSHN1 and GmSHN9 Differentially Regulates Wax and Cutin Biosynthesis, Alters Cuticle Properties, and Changes Leaf Phenotypes in Arabidopsis. Int. J. Mol. Sci. 2016, 17, 587. [Google Scholar] [CrossRef]
- Ding, A.M.; Xu, C.T.; Xie, Q.; Zhang, M.J.; Yan, N.; Dai, C.B.; Lv, J.; Cui, M.M.; Wang, W.F.; Sun, Y.H. ERF4 interacts with and antagonizes TCP15 in regulating endoreduplication and cell growth in Arabidopsis. J. Integr. Plant Biol. 2022, 64, 1673–1689. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Jin, Y.M.; Wu, T.; Hu, L.; Zhang, Y.; Jiang, W.; Du, X. OsDREB2B, an AP2/ERF transcription factor, negatively regulates plant height by conferring GA metabolism in rice. Front. Plant Sci. 2022, 13, 1007811. [Google Scholar] [CrossRef]
- He, Y.K.; Xue, W.X.; Sun, Y.D.; Yu, X.H.; Liu, P.L. Leafy head formation of the progenies of transgenic plants of Chinese cabbage with exogenous auxin genes. Cell Res. 2000, 10, 151–160. [Google Scholar] [CrossRef]
- Gao, L.W.; Lyu, S.W.; Tang, J.; Zhou, D.Y.; Bonnema, G.; Xiao, D.; Hou, X.L.; Zhang, C.W. Genome-wide analysis of auxin transport genes identifies the hormone responsive patterns associated with leafy head formation in Chinese cabbage. Sci. Rep. 2017, 7, 42229. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Yang, Y.; Wu, J.; Liang, J.; Chen, S.; Zhang, L.; Lv, H.; Yin, X.; Zhang, X.; Zhang, Y.; et al. A cluster of transcripts identifies a transition stage initiating leafy head growth in heading morphotypes of Brassica. Plant J. 2022, 11, 688–706. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Qu, G.; Huang, S.; Liu, Z.; Zhang, M.; Fu, W.; Ren, J.; Feng, H. Comparison between germinated seed and isolated microspore EMS mutagenesis in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Horticulturae 2022, 8, 232. [Google Scholar] [CrossRef]
- Gao, Y.; Huang, S.; Qu, G.; Fu, W.; Zhang, M.; Liu, Z.; Feng, H. The mutation of ent-kaurene synthase, a key enzyme involved in gibberellin biosynthesis, confers a non-heading phenotype to Chinese cabbage (Brassica rapa L. ssp. pekinensis). Hortic. Res. 2020, 7, 178. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.L.; Fu, X.M.; Liu, J.Q.; Ye, T.T.; Hou, S.Y.; Huang, Y.Q.; Yuan, B.F.; Wu, Y.; Feng, Y.Q. Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC-ESI-Q-TOF-MS analysis. J. Chromatogr. B 2012, 905, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, X.; Deng, B.; Lou, P.; Wu, J.; Sun, R.; Xu, Z.; Vromans, J.; Koornneef, M.; Bonnema, G. Genetic relationships within Brassica rapa as inferred from AFLP fingerprints. Theor. Appl. Genet. 2005, 110, 1301–1314. [Google Scholar] [CrossRef]
- Zhang, J.X.; Li, H.X.; Zhang, M.K.; Hui, M.X.; Wang, Q.; Li, L.; Zhang, L.G. Fine mapping and identification of candidate Br-or gene controlling orange head of Chinese cabbage (Brassica rapa L. ssp. pekinensis). Mol. Breed. 2013, 32, 799–805. [Google Scholar] [CrossRef]
- Zhang, X.; Su, Y.; Liu, Y.; Fang, Z.; Zhuang, M.; Zhang, Y.; Li, Z.; Lv, H. Genetic analysis and QTL mapping of traits related to head shape in cabbage (Brassica rapa ssp. pekinensis). Sci. Hortic. 2016, 207, 82–88. [Google Scholar] [CrossRef]
- Wang, F.; Li, L.; Li, H.; Liu, L.; Zhang, Y.; Gao, J.; Wang, X. Transcriptome analysis of rosette and folding leaves in Chinese cabbage using high-throughput RNA sequencing. Genomics 2012, 99, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fan, Y.; Jiao, Y.; Wu, J.; Zhang, Z.; Yu, X.; Ma, Y. Transcriptome profiling of yellow leafy head development during the heading stage in Chinese cabbage (Brassica rapa subsp. pekinensis). Physiol. Plant. 2019, 165, 800–813. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.W.; Wei, Y.P.; Xiao, D.; Gao, L.W.; Lyu, S.W.; Hou, X.L.; Bouuema, G. Transcriptomic and proteomic analyses provide new insights into the regulation mechanism of low-temperature-induced leafy head formation in Chinese cabbage. J. Proteom. 2016, 144, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Li, H.; Zhang, Y.; Li, J.; Li, L.; Liu, L.; Wang, L.; Wang, C.; Gao, J. MicroRNA expression analysis of rosette and folding leaves in Chinese cabbage using high-throughput Solexa sequencing. Gene 2013, 532, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.P.; Gubler, F. Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 2004, 55, 197–223. [Google Scholar] [CrossRef] [PubMed]
- Olszewski, N.; Sun, T.P.; Gubler, F. Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 2002, 14, S61–S80. [Google Scholar] [CrossRef] [PubMed]
- Debeaujon, I.; Koornneef, M. Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol. 2000, 122, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Yaxley, J.R.; Ross, J.J.; Sherriff, L.J.; Reid, J.B. Gibberellin biosynthesis mutations and root development in pea. Plant Physiol. 2001, 125, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Hedden, P.; Phillips, A.L. Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 2000, 5, 523–530. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Sun, T.P.; Kawaide, H.; Kamiya, Y. The GA2 locus of Arabidopsis thaliana encodes ent-kaurene synthase of gibberellin biosynthesis. Plant Physiol. 1998, 116, 1271–1278. [Google Scholar] [CrossRef]
- Margis-Pinheiro, M.; Zhou, X.R.; Zhu, Q.H.; Dennis, E.S.; Upadhyaya, N.M. Isolation and characterization of a Ds-tagged rice (Oryza sativa L.) GA-responsive dwarf mutant defective in an early step of the gibberellin biosynthesis pathway. Plant Cell Rep. 2005, 23, 819–833. [Google Scholar] [CrossRef]
- Li, Z.F.; Guo, Y.; Ou, L.; Hong, H.; Wang, J.; Liu, Z.X.; Guo, B.; Zhang, L.; Qiu, L. Identification of the dwarf gene GmDW1 in soybean (Glycine max L.) by combining mapping-by-sequencing and linkage analysis. Theor. Appl. Genet. 2018, 131, 1001–1016. [Google Scholar] [CrossRef]
- Mun, J.H.; Yu, H.J.; Shin, J.Y.; Oh, M.; Hwang, H.J.; Chung, H. Auxin response factor gene family in Brassica rapa: Genomic organization, divergence, expression, and evolution. Mol. Genet. Genom. 2012, 287, 765–784. [Google Scholar] [CrossRef]
- Lee, Y.K.; Kim, G.T.; Kim, I.J.; Park, J.; Kwak, S.S.; Choi, G.; Chung, W.I. LONGIFOLIA1 and LONGIFOLIA2, two homologous genes, regulate longitudinal cell elongation in Arabidopsis. Development 2006, 133, 4305–4314. [Google Scholar] [CrossRef]
- Rose, J.K.; Braam, J.; Fry, S.C.; Nishitani, K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomenclature. Plant Cell Physiol. 2002, 43, 1421–1435. [Google Scholar] [CrossRef]
- Mao, T.; Jin, L.; Li, H.; Liu, B.; Yuan, M. Two microtubule-associated proteins of the Arabidopsis MAP65 family function differently on microtubules. Plant Physiol. 2005, 138, 654–662. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, F.; Bai, J.; He, Y. BrpSPL9 (Brassica rapa ssp. pekinensis SPL9) controls the earliness of heading time in Chinese cabbage. Plant Biotechnol. J. 2014, 12, 312–321. [Google Scholar]
- Li, F.; Kitashiba, H.; Inaba, K.; Nishio, T. A Brassica rapa linkage map of EST-based SNP markers for identification of candidate genes controlling flowering time and leaf morphological traits. DNA Res. 2009, 16, 311–323. [Google Scholar] [CrossRef]
- Xu, L.; Xu, Y.; Dong, A.; Sun, Y.; Pi, L.; Xu, Y.; Huang, H. Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 2003, 130, 4097–4107. [Google Scholar] [CrossRef]
- Fukushima, K.; Hasebe, M. Adaxial-abaxial polarity: The developmental basis of leaf shape diversity. Genesis 2014, 52, 1–18. [Google Scholar] [CrossRef]
- Liu, Y.; Guan, X.; Liu, S.; Yang, M.; Ren, J.; Guo, M.; Huang, Z.; Zhang, Y. Genome-Wide Identification and Analysis of TCP Transcription Factors Involved in the Formation of Leafy Head in Chinese Cabbage. Int. J. Mol. Sci. 2018, 19, 847. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; He, S.P.; Xu, S.W.; Li, L.; Zheng, Y.; Li, X.B. The transcription factor ERF108 interacts with AUXIN RESPONSE FACTORs to mediate cotton fiber secondary cell wall biosynthesis. Plant Cell 2023, 35, 4133–4154. [Google Scholar] [CrossRef]





| Generations | Total Plants | Wild-Type Plants | Mutant Plants | Segregation Ratio | χ2 |
|---|---|---|---|---|---|
| P1 (‘FT’) | 50 | 50 | 0 | ||
| P2 (nhm2) | 50 | 0 | 50 | ||
| F1 (P1 × P2) | 25 | 25 | 0 | ||
| F1 (P2 × P1) | 25 | 25 | 0 | ||
| BC1 (F1 × P1) | 40 | 40 | 0 | ||
| BC1 (F1 × P2) | 60 | 31 | 29 | 1.06:1 | 0.016 |
| F2 | 300 | 228 | 72 | 3.17:1 | 0.11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qu, G.; Liu, S.; Wang, W.; Wei, S.; Liu, Y.; Gao, Y.; Feng, H. The Role of BrKS in Leafy Head Formation Was Confirmed by Two Allelic Mutants of Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Horticulturae 2024, 10, 804. https://doi.org/10.3390/horticulturae10080804
Qu G, Liu S, Wang W, Wei S, Liu Y, Gao Y, Feng H. The Role of BrKS in Leafy Head Formation Was Confirmed by Two Allelic Mutants of Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Horticulturae. 2024; 10(8):804. https://doi.org/10.3390/horticulturae10080804
Chicago/Turabian StyleQu, Gaoyang, Shihang Liu, Wei Wang, Shixiang Wei, Yuanwei Liu, Yue Gao, and Hui Feng. 2024. "The Role of BrKS in Leafy Head Formation Was Confirmed by Two Allelic Mutants of Chinese Cabbage (Brassica rapa L. ssp. pekinensis)" Horticulturae 10, no. 8: 804. https://doi.org/10.3390/horticulturae10080804
APA StyleQu, G., Liu, S., Wang, W., Wei, S., Liu, Y., Gao, Y., & Feng, H. (2024). The Role of BrKS in Leafy Head Formation Was Confirmed by Two Allelic Mutants of Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Horticulturae, 10(8), 804. https://doi.org/10.3390/horticulturae10080804






