Genome-Wide Association Study and Transcriptome Analysis Provide Candidate Genes for Agronomic Traits of Agaricus bisporus
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Phenotype Evaluation
2.2. Sequencing and Alignment
2.3. Variant Detection and Annotation
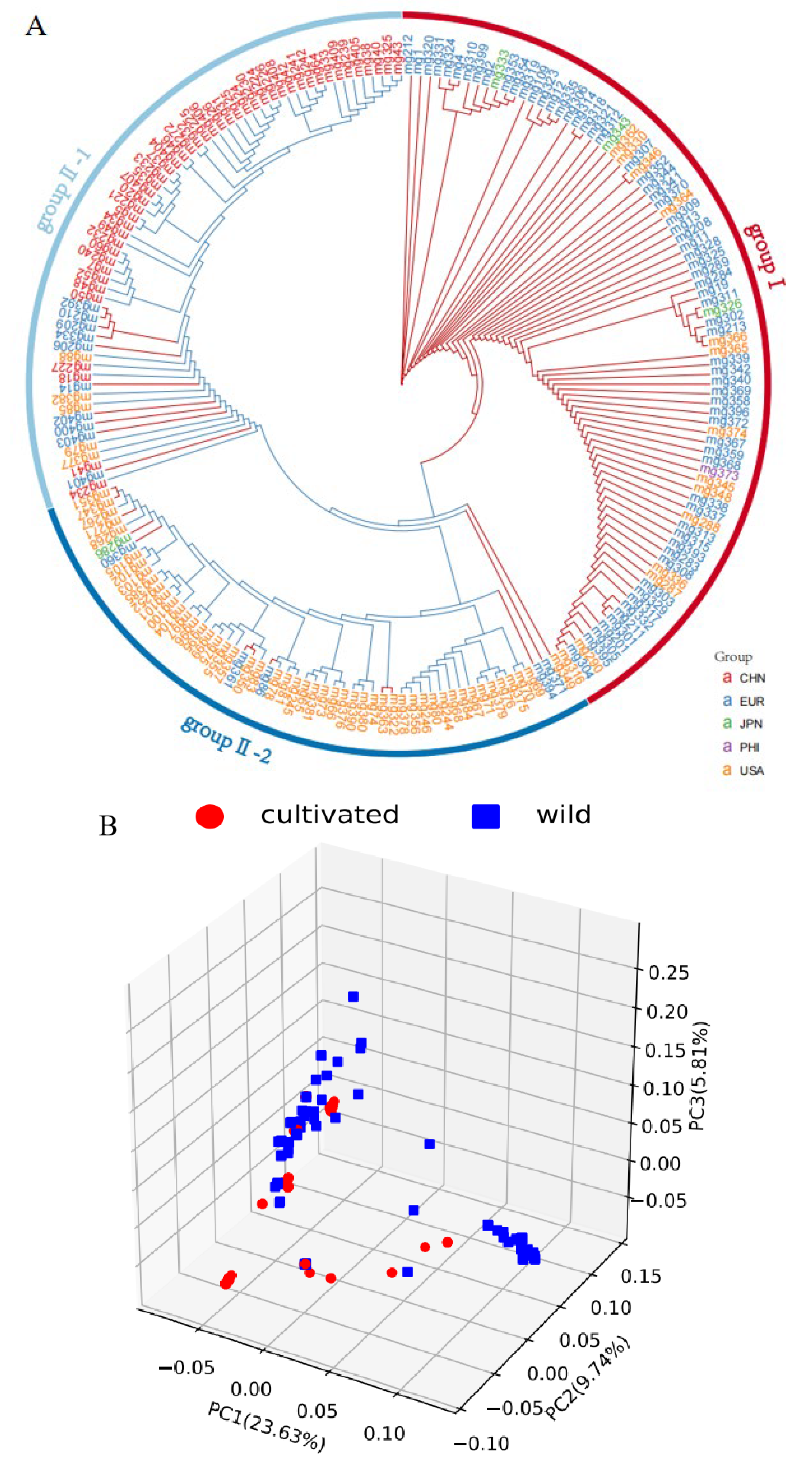
2.4. Phylogenetic Tree and Population Structure Analyses
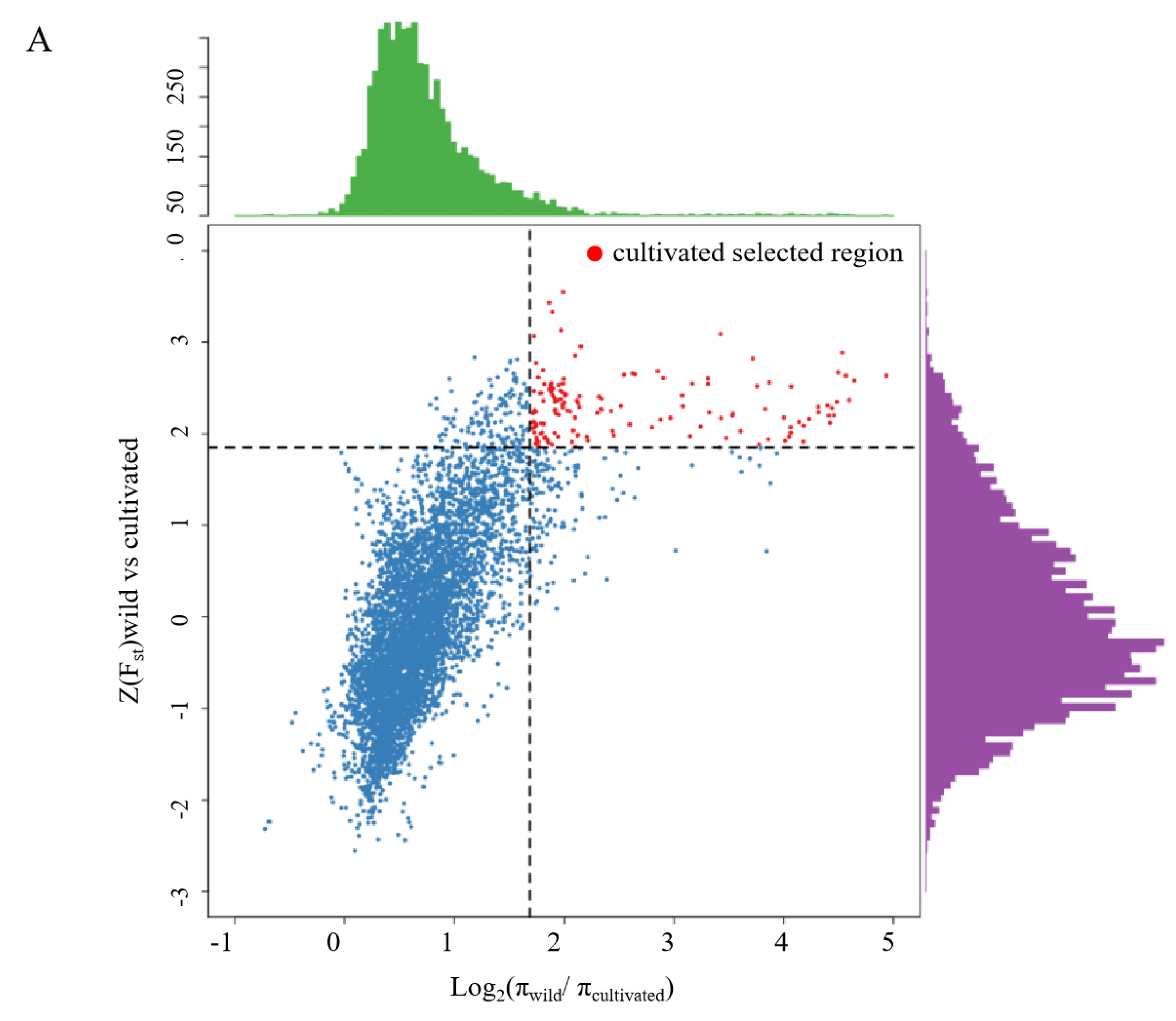
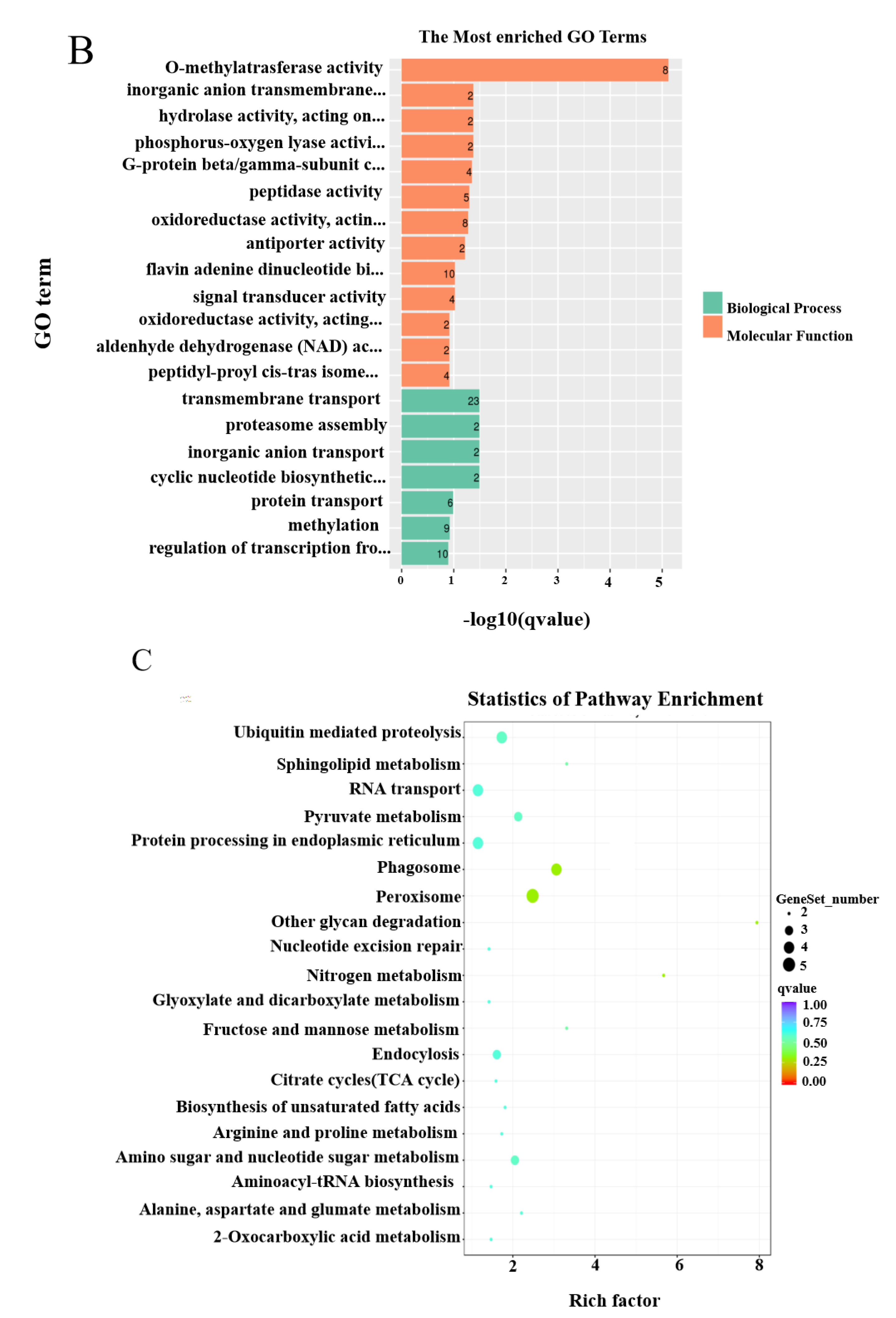
2.5. Genome Scanning for Selective Elimination Analysis and Functional Enrichment Analysis
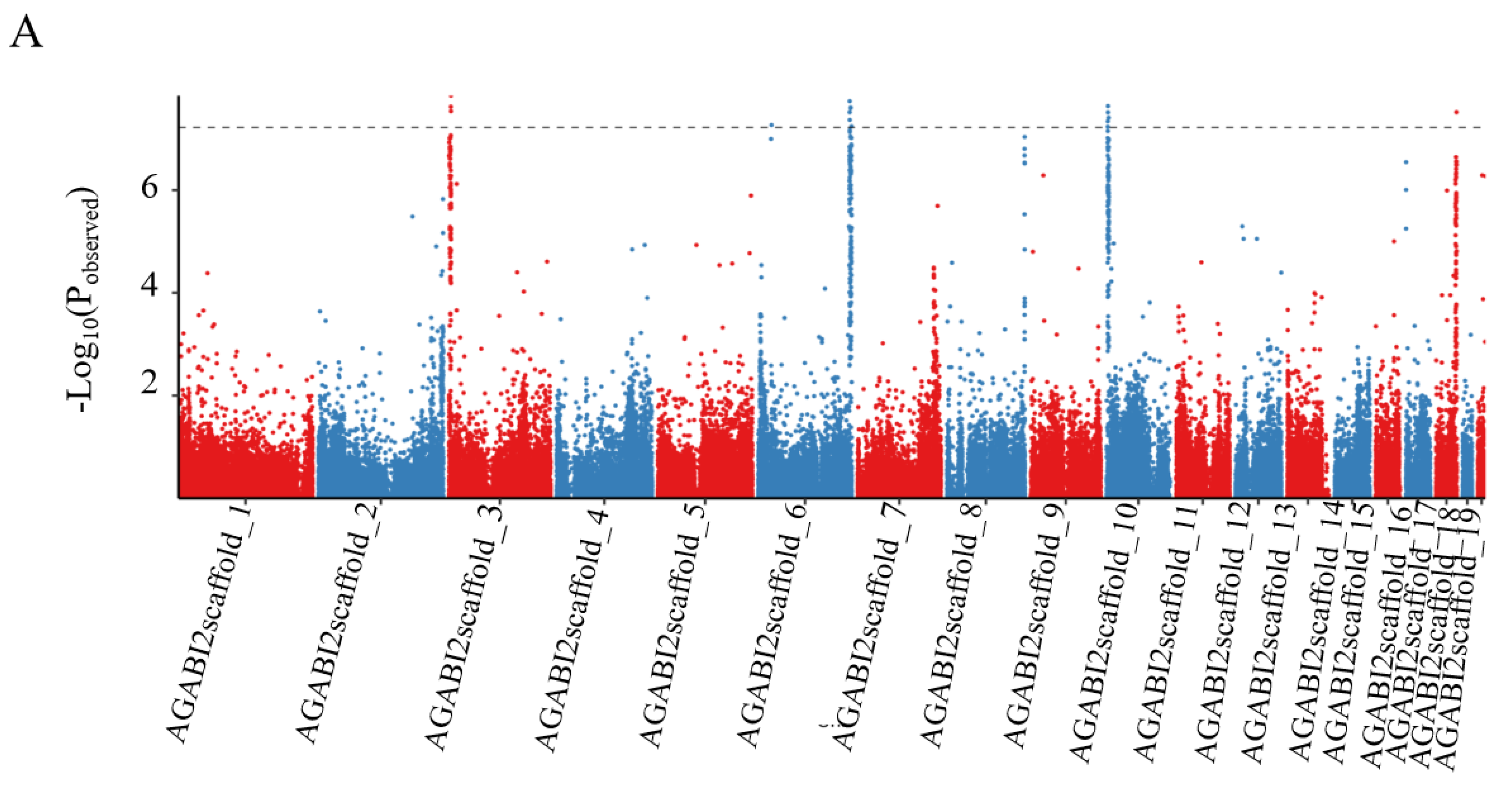
2.6. Association Analyses
2.7. Expression Analyses
3. Results
3.1. High-Quality Button Mushroom SNP
3.2. Characterization of a Population and Linkage Disequilibrium
3.3. Selection Signatures during Breeding and Expression Analysis of Candidate Genes
3.4. Identification of Candidate Genes for Esterase Isozyme Type by Integrating Genome-Wide Association Studies and RNA-Seq Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chang, S.T.; Buswell, J.A. Mushroom nutriceuticals. World J. Microbiol. Biotechnol. 1996, 12, 473–476. [Google Scholar] [CrossRef]
- Kües, U.; Liu, Y. Fruiting body production in basidiomycetes. Appl. Microbiol. Biotechnol. 2000, 54, 141–152. [Google Scholar] [CrossRef]
- Kulshreshtha, S.; Mathur, N.; Bhatnagar, P. Mushroom as a product and their role in mycoremediation. AMB Express 2014, 4, 29. [Google Scholar] [CrossRef]
- Atila, F.; Owaid, M.N.; Shariati, M.A. The nutritional and medical benefits of Agaricus bisporus: A review. J. Microbiol. Biotechnol. Food 2017, 18, 281–286. [Google Scholar] [CrossRef]
- McGee, C.F. Microbial ecology of the Agaricus bisporus mushroom cropping process. Appl. Microbiol. Biotechnol. 2018, 102, 1075–1083. [Google Scholar] [CrossRef]
- Wang, Z.; Liao, J.; Chen, M.; Cai, Z. Breeding and industrial development of Agaricus bisporus. Acta Edulis Fungi 2012, 19, 1–14. (In Chinese) [Google Scholar]
- Gao, W.; Baars, J.J.; Maliepaard, C.; Visser, R.G.; Zhang, J.; Sonnenberg, A.S. Multi-trait QTL analysis for agronomic and quality characters of Agaricus bisporus (button mushrooms). AMB Express 2016, 6, 67. [Google Scholar] [CrossRef]
- Müller-Starck, G. Isozymes. In Molecular Tools for Screening Biodiversity: Plants and Animals; Springer: Dordrecht, The Netherlands, 1998; pp. 75–81. [Google Scholar]
- Wang, H.C.; Wang, Z.S. The prediction of strain characteristecs of Agaricus bisporus by the application of isozyme electrophoresis. Fujian Mushroom J. 1991, 1, 38–48. (In Chinese) [Google Scholar]
- Wang, Z.S.; Wang, H.C. Isozyme patterns and characteristics of hybrid strains of Agaricus bisporus. Microl. Neotrop. Apl. 1990, 3, 19–29. [Google Scholar]
- Zhang, H.; Zhang, J.; Xu, Q.; Wang, D.; Di, H.; Huang, J.; Yang, X.; Wang, Z.; Zhang, L.; Dong, L.; et al. Identification of candidate tolerance genes to low-temperature during maize germination by GWAS and RNA-seq approaches. BMC Plant Biol. 2020, 20, 333. [Google Scholar] [CrossRef]
- Li, Y.; Cao, K.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Zhao, P.; Guo, J.; Ding, T.; Guan, L.; et al. Genomic analyses of an extensive collection of wild and cultivated accessions provide new insights into peach breeding history. Genome Biol. 2019, 20, 36. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Weijn, A.; Baars, J.J.; Mes, J.J.; Visser, R.G.; Sonnenberg, A.S. Quantitative trait locus mapping for bruising sensitivity and cap color of Agaricus bisporus (button mushrooms). Fungal Genet. Biol. 2015, 77, 69–81. [Google Scholar] [CrossRef]
- Pardo, A.; de Juan, A.; Alvarez-OrtÃ, M.; Pardo, J.E. Screening of Agaricus bisporus (Lange, Imbach) strains and the casing variables for quality mushroom production in Spain. Hortscience 2010, 45, 231–235. [Google Scholar] [CrossRef]
- Chen, M.; Liao, J.; Li, H.; Cai, Z.; Guo, Z.; Wach, M.P.; Wang, Z. iTRAQ-MS/MS proteomic analysis reveals differentially expressed proteins during post-harvest maturation of the white button mushroom Agaricus bisporus. Curr. Microbiol. 2017, 74, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows—Wheeler transform. Bionformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bionformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Gatk. Available online: https://gatk.broadinstitute.org/hc/en-us/articles/360035531112--How-to-Filter-variants-either-with-VQSR-or-by-hard-filtering (accessed on 22 May 2024).
- Yang, H.; Wang, K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 2015, 10, 1556–1566. [Google Scholar] [CrossRef]
- Felsenstein, J. PHYLIP (Phylogeny Inference Package), Version 3.7a; Distributed by the author; Department of Genome Sciences, University of Washington: Seattle, WA, USA, 2009. [Google Scholar]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.D.M.J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39 (Suppl. S2), W316–W322. [Google Scholar] [CrossRef]
- Reumers, J.; De Rijk, P.; Zhao, H.; Liekens, A.; Smeets, D.; Cleary, J.; Van Loo, P.; Van Den Bossche, M.; Catthoor, K.; Sabbe, B.; et al. Optimized filtering reduces the error rate in detecting genomic variants by short-read sequencing. Nat. Biotechnol. 2011, 30, 61–68. [Google Scholar] [CrossRef]
- Doebley, J.F.; Gaut, B.S.; Smith, B.D. The molecular genetics of crop domestication. Cell 2006, 127, 1309–1321. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M.; Haigh, J. The hitch-hiking effect of a favourable gene. Genet. Res. 1974, 23, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhang, C.; Zhao, X.; Fei, Z.; Wan, K.; Zhang, Z.; Pang, X.; Yin, X.; Bai, Y.; Sun, X.; et al. The jujube genome provides insights into genome evolution and the domestication of sweetness/acidity taste in fruit trees. PLoS Genet. 2016, 12, e1006433. [Google Scholar] [CrossRef]
- Yang, L.; Zhou, H.; Bo, W.; Li, Y.; Pang, X. Identification of genes related with jujube fruit size based on selective sweep analysis. J. Beijing For. Univ. 2019, 41, 30–36. (In Chinese) [Google Scholar]
- Ji, Y.S.; Wang, C.Y.; Liu, R. Exploitation of freezing-tolerant genes in pea (Pisum sativum L.) based on selective sweeping analysis. China Veg. 2020, 3, 33–42. (In Chinese) [Google Scholar]
- Cao, J.; Sun, M.; Yu, M.; Xu, Y.; Xie, J.; Zhang, H.; Chen, J.; Xu, T.; Qian, X.; Sun, S. Transcriptome analysis reveals the function of a G-Protein α subunit gene in the growth and development of Pleurotus eryngii. J. Fungi 2023, 9, 69. [Google Scholar] [CrossRef]
- Li, X.; Ke, Z.; Xu, S.; Tang, W.; Liu, Z. The G-protein alpha subunit CgGa1 mediates growth, sporulation, penetration and pathogenicity in Colletotrichum gloeosporioides. Microb. Pathog. 2021, 161, 105254. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Zhang, F.; Chong, X.; Song, A.; Guan, Z.; Fang, W.; Chen, F. Genome-wide association study identifies favorable SNP alleles and candidate genes for waterlogging tolerance in chrysanthemums. Hortic. Res. 2019, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cheng, R.; Verbitsky, M.; Kisselev, S.; Browne, A.; Mejia-Sanatana, H.; Louis, E.D.; Cote, L.J.; Andrews, H.; Waters, C.; et al. Genome-wide association study identifies candidate genes for Parkinson’s disease in an Ashkenazi Jewish population. BMC Med. Genet. 2011, 12, 104. [Google Scholar] [CrossRef]
- Lu, Y.P.; Liao, J.H.; Guo, Z.J.; Cai, Z.X.; Chen, M.Y. Genome survey and transcriptome analysis on mycelia and primordia of Agaricus blazei. Biomed. Res. Int. 2020, 2020, 1824183. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Guo, Z.; Cai, Z.; Chen, M.; Liao, J. Transcriptome analysis of Agaricus blazei fruiting bodies at different developmental stages. Mycosystema 2019, 38, 2161–2173. (In Chinese) [Google Scholar]
- Guo, J.; Li, C.; Zhang, X.; Li, Y.; Zhang, D.; Shi, Y.; Song, Y.; Li, Y.; Yang, D.; Wang, T. Transcriptome and GWAS analyses reveal candidate gene for seminal root length of maize seedlings under drought stress. Plant Sci. 2020, 292, 110380. [Google Scholar] [CrossRef] [PubMed]
- Molowitz, R.; Bahn, M.; Hock, B. The control of fruiting body formation in the ascomycete Sordaria macrospora Auersw. by arginine and biotin: A two-factor analysis. Planta 1976, 128, 143–148. [Google Scholar] [CrossRef] [PubMed]
- McHowat, J.; Creer, M.H. Catalytic features, regulation and function of myocardial phospholipase A2. Curr. Med. Chem. Cardiovasc. Hematol. Agents 2004, 2, 209–218. [Google Scholar] [CrossRef]
- Balsinde, J.; Balboa, M.A. Cellular regulation and proposed biological functions of group VIA calcium-independent phospholipase A2 in activated cells. Cell Signal 2005, 17, 1052–1062. [Google Scholar] [CrossRef] [PubMed]
- Hooks, S.B.; Cummings, B.S. Role of Ca2+-independent phospholipase A2 in cell growth and signaling. Biochem. Pharmacol. 2008, 76, 1059–1067. [Google Scholar] [CrossRef] [PubMed]
- Akiba, S.; Sato, T. Cellular function of calcium-independent phospholipase A2. Biol. Pharm. Bull. 2004, 27, 1174–1178. [Google Scholar] [CrossRef][Green Version]
- Schrevens, S.; Van Zeebroeck, G.; Riedelberger, M.; Tournu, H.; Kuchler, K.; Van Dijck, P. Methionine is required for cAMP-PKA-mediated morphogenesis and virulence of Candida albicans. Mol. Microbiol. 2018, 108, 258–275. [Google Scholar] [CrossRef]


| Gene ID | Regulated | GO Annotation | nr_Annotation |
|---|---|---|---|
| AGABI2DRAFT_196124 | down | GO:0004571, GO:0005509, GO:0016020 | Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase |
| AGABI2DRAFT_117541 | down | -- | hypothetical protein |
| AGABI2DRAFT_62224 | up | GO:0010181, GO:001649, GO:0055114 | Putative NADPH dehydrogenase |
| AGABI2DRAFT_176301 | down | -- | hypothetical protein |
| AGABI2DRAFT_190117 | down | -- | ferritin-like domain-containing protein |
| AGABI2DRAFT_203715 | up | GO:0015103, GO:0015297, GO:0015698, GO:0055085, GO:0016021 | Arsenite resistance protein ArsB, partial |
| AGABI2DRAFT_181624 | down | -- | hypothetical protein AN958_00352 |
| AGABI2DRAFT_181629 | down | GO:0008233, GO:0006508 | L-amino acid amidase |
| AGABI2DRAFT_178110 | down | -- | predicted protein |
| AGABI2DRAFT_178286 | up | -- | hypothetical protein AN958_04611 |
| AGABI2DRAFT_192454 | down | GO:0004342, GO:0016787, GO:0005975, GO:0006044 | Glucosamine-6-phosphate isomerase |
| AGABI2DRAFT_208542 | up | -- | hypothetical protein AN958_12461 |
| AGABI2DRAFT_194428 | down | GO:0005488, GO:0007165 | guanine nucleotide binding protein, alpha subunit |
| Gene ID | Regulated | GO Annotation | nr_Annotation |
|---|---|---|---|
| AGABI2DRAFT_143474 | down | -- | hypothetical protein AN958_07900 |
| AGABI2DRAFT_183745 | down | GO:0009058, GO:0016740, GO:0030170 | 8-amino-7-oxononanoate synthase |
| AGABI2DRAFT_191000 | down | GO:0000101, GO:0003333, GO:0006812, GO:0015171, GO:0016020 | High-affinity methionine permease |
| AGABI2DRAFT_123388 | down | -- | hypothetical protein AN958_01095 |
| AGABI2DRAFT_120988 | down | - | hypothetical protein BP6252_06823 |
| AGABI2DRAFT_151295 | down | -- | Calcium-independent phospholipase A2-gamma |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Y.; Guo, Z.; Ke, B.; Zheng, H.; Zeng, Z.; Cai, Z.; Zeng, H.; Liao, J.; Chen, M. Genome-Wide Association Study and Transcriptome Analysis Provide Candidate Genes for Agronomic Traits of Agaricus bisporus. Horticulturae 2024, 10, 691. https://doi.org/10.3390/horticulturae10070691
Lu Y, Guo Z, Ke B, Zheng H, Zeng Z, Cai Z, Zeng H, Liao J, Chen M. Genome-Wide Association Study and Transcriptome Analysis Provide Candidate Genes for Agronomic Traits of Agaricus bisporus. Horticulturae. 2024; 10(7):691. https://doi.org/10.3390/horticulturae10070691
Chicago/Turabian StyleLu, Yuanping, Zhongjie Guo, Binrong Ke, Huiqing Zheng, Zhiheng Zeng, Zhixin Cai, Hui Zeng, Jianhua Liao, and Meiyuan Chen. 2024. "Genome-Wide Association Study and Transcriptome Analysis Provide Candidate Genes for Agronomic Traits of Agaricus bisporus" Horticulturae 10, no. 7: 691. https://doi.org/10.3390/horticulturae10070691
APA StyleLu, Y., Guo, Z., Ke, B., Zheng, H., Zeng, Z., Cai, Z., Zeng, H., Liao, J., & Chen, M. (2024). Genome-Wide Association Study and Transcriptome Analysis Provide Candidate Genes for Agronomic Traits of Agaricus bisporus. Horticulturae, 10(7), 691. https://doi.org/10.3390/horticulturae10070691





