Effect of Phosphate-Deficiency Stress on the Biological Characteristics and Transcriptomics of Panax ginseng
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Determination of Leaflet Area, Physiological Indices, Root Morphogenesis, and Phosphorus
2.3. RNA Extraction, Illumina Sequencing, and Transcriptome Data Analysis
2.4. Identification of DEGs
2.5. Determination of Ginsenoside Content
2.6. Statistical Analysis and Drawing
3. Results
3.1. Effect of Phosphate Deficiency on the Biological Characteristics of Ginseng
3.2. Effect of Phosphate Deficiency on the Transcriptome of Ginseng
3.3. Relative Genes in the Synthesis of PHTs, Ginsenoside, and Phytohormone
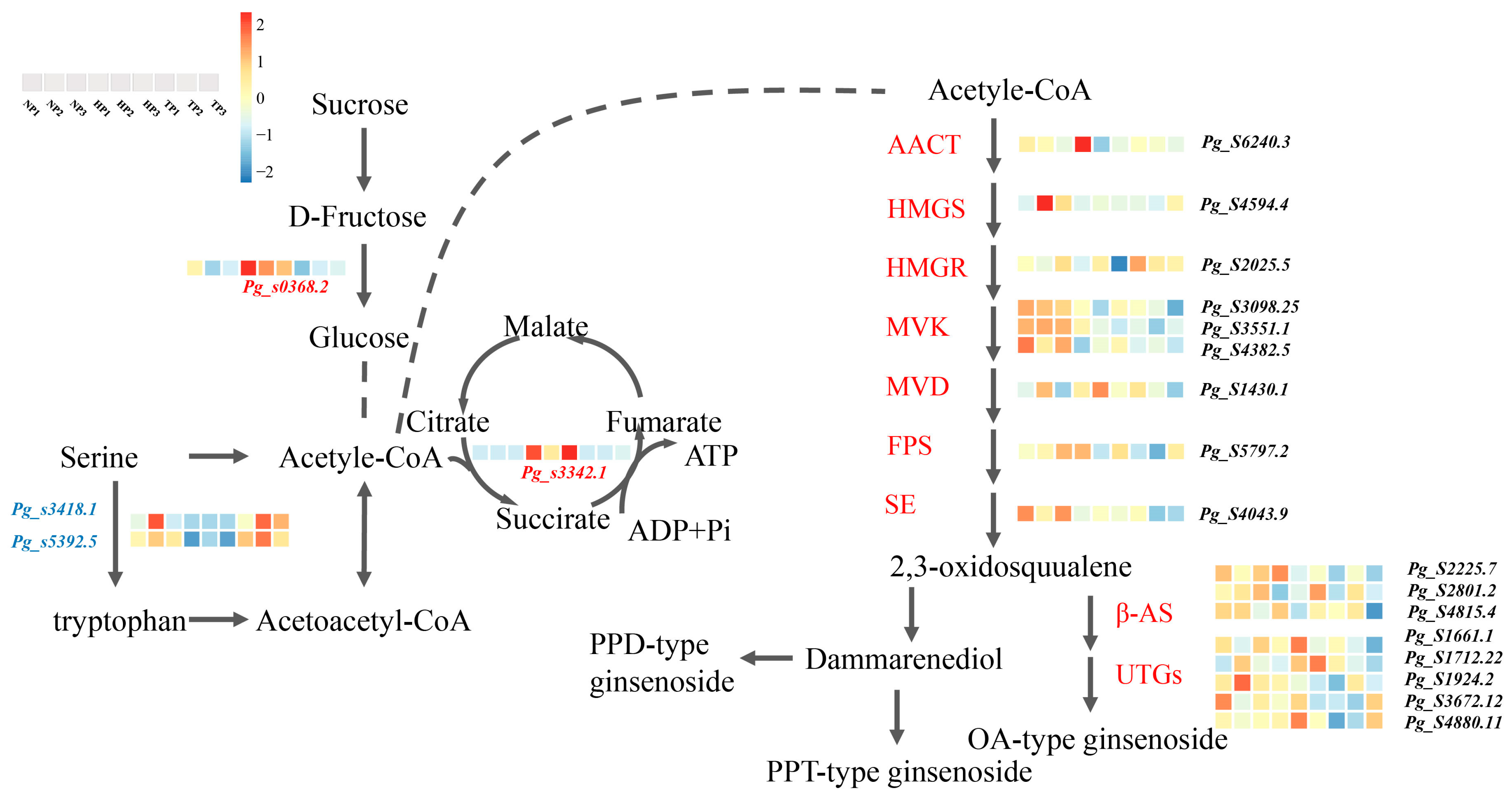
3.4. Expression Pattern of Relative Gene in the Biosynthesis Pathway of TCA and Ginsenoside Synthesis
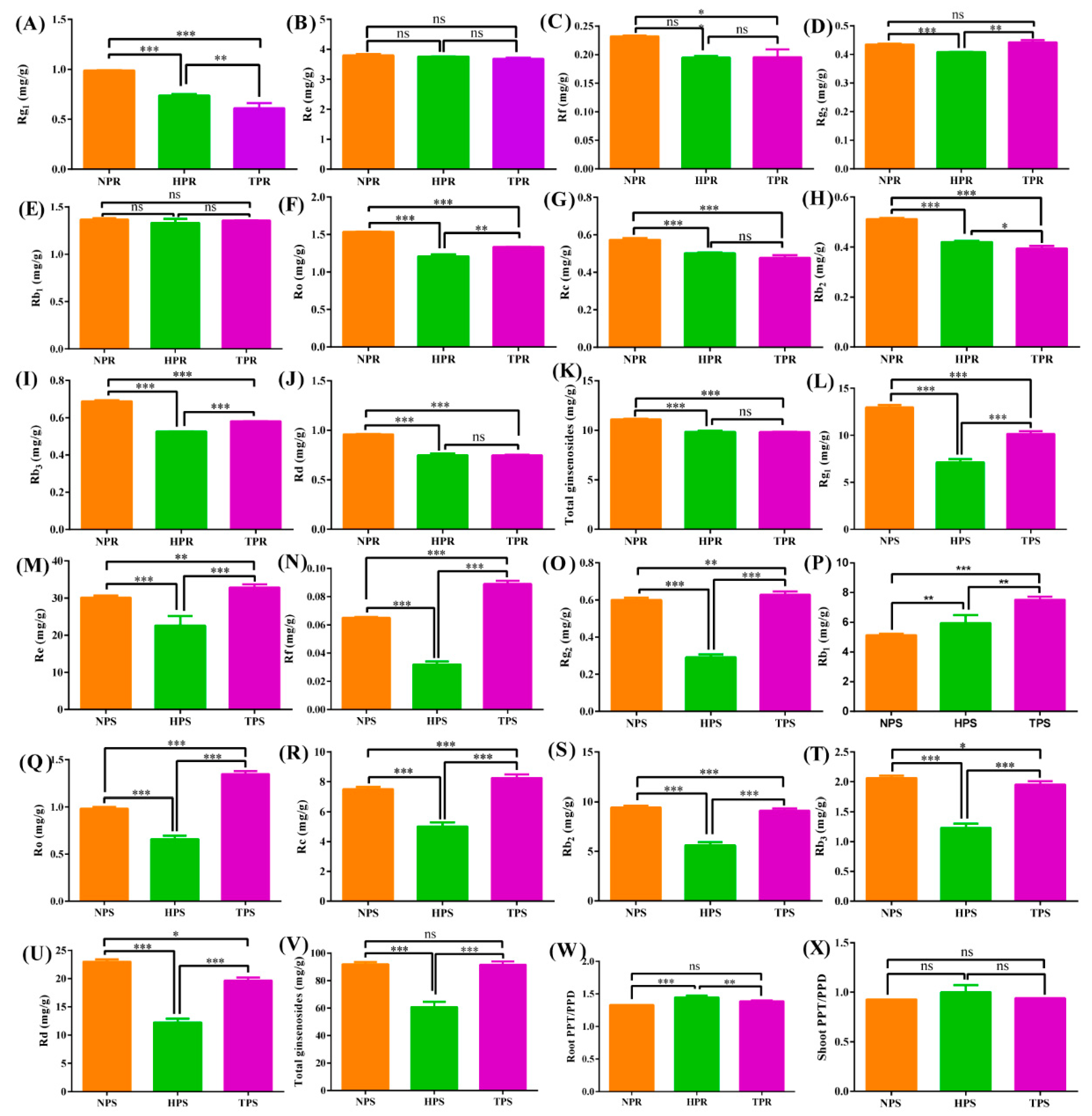
3.5. Effect of Phosphate Deficiency on Ginsenoside Accumulation
4. Discussion
4.1. Phosphate Stress Changes Ginseng Root Morphogenesis and Triggers the Expression of Different Genes
4.2. Multiple DEGs Are Involved in PHTs and Phytohormone Signal Transduction
4.3. Multiple DEGs Are Involved in TCA and Ginsenoside Synthesis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yi, Y.S. Pharmacological potential of ginseng and ginsenosides in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. J. Ginseng Res. 2024, 48, 122–128. [Google Scholar] [CrossRef]
- Cecarini, V.; Cuccioloni, M.; Gong, C.M.; Liu, Z.Q.; Bonfili, L.; Angeletti, M.; Angeloni, S.; Alessandroni, L.; Sagratini, G.; Liu, H.M.; et al. Role of Panax ginseng and ginsenosides in regulating cholesterol homeostasis. Food Biosci. 2023, 56, 103256. [Google Scholar] [CrossRef]
- Ma, R.; Yang, P.D.; Jing, C.X.; Fu, B.Y.; Teng, X.Y.; Zhao, D.Q.; Sun, L.W. Comparison of the metabolomic and proteomic profiles associated with triterpene and phytosterol accumulation between wild and cultivated ginseng. Plant Physiol. Biochem. 2023, 195, 288–299. [Google Scholar] [CrossRef]
- Kan, H.; Zhang, D.X.; Chen, W.J.; Wang, S.H.; He, Z.M.; Pang, S.F.; Qu, S.; Wang, Y.P. Identification of anti-inflammatory components in Panax ginseng of Sijunzi Decoction based on spectrum-effect relationship. Chin. Herb. Med. 2023, 15, 123–131. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Y.; Li, X.; Ruan, C.C.; Wang, L.J.; Sun, G.Z. The effects of dynamic changes of malonyl ginsenosides on evaluation and quality control of Panax ginseng C.A. Meyer. J. Pharm. Biomed. Anal. 2012, 64–65, 56–63. [Google Scholar] [CrossRef]
- Lee, S.K.; Kim, J.H.; Sohn, H.J.; Yang, J.W. Changes in aroma characteristics during the preparation of red ginseng estimated by electronic nose, sensory evaluation and gas chromatography/mass spectrometry. Sens. Actuators B Chem. 2005, 106, 7–12. [Google Scholar] [CrossRef]
- Li, L.; Yang, T.; Redden, R.; He, W.F.; Zong, X.X. Soil fertility map for food legumes production areas in China. Sci. Rep. 2016, 6, 26102. [Google Scholar] [CrossRef]
- Zhao, W.H.; Bi, X.J.; Peng, Y.Z.; Bai, M. Research advances of the phosphorus-accumulating organisms of Candidatus Accumulibacter, Dechloromonas and Tetrasphaera: Metabolic mechanisms, applications and influencing factors. Chemosphere 2022, 301 Pt 1, 135675. [Google Scholar] [CrossRef]
- Sehar, S.; Adil, M.F.; Ma, Z.X.; Karim, M.F.; Faizan, M.; Zaidi, S.S.A.; Siddiqui, M.H.; Alamri, S.; Zhou, F.R.; Shamsi, I.H. Phosphorus and Serendipita indica synergism augments arsenic stress tolerance in rice by regulating secondary metabolism related enzymatic activity and root metabolic patterns. Ecotoxicol. Environ. Saf. 2023, 256, 114866. [Google Scholar] [CrossRef]
- Zou, T.; Zhang, X.; Davidson, E.A. Global trends of cropland phosphorus use and sustainability challenges. Nature 2022, 611, 81–87. [Google Scholar] [CrossRef]
- Barra, P.J.; Duran, P.; Delgado, M.; Viscardi, S.; Claverol, S.; Larama, G.; Dumount, M.; Mora, M.D.L.L. Proteomic response to phosphorus deficiency and aluminum stress of three aluminum-tolerant phosphobacteria isolated from acidic soils. iScience 2023, 26, 207910. [Google Scholar] [CrossRef]
- Panigrahy, M.; Rao, D.N.; Sarla, N. Molecular mechanisms in response to phosphate starvation in rice. Biotechnol. Adv. 2009, 27, 389–397. [Google Scholar] [CrossRef]
- Xue, C.W.; Li, W.F.; Shen, R.F.; Lan, P. PERK13 modulates phosphate deficiency-induced root hair elongation in Arabidopsis. Plant Science 2021, 312, 111060. [Google Scholar] [CrossRef]
- Wu, Y.H.; Zhao, C.; Zhao, X.K.; Yang, L.Y.; Liu, C.; Jiang, L.Y.; Liu, G.D.; Liu, P.D.; Luo, L.J. Multi-omics-based identification of purple acid phosphatases and metabolites involved in phosphorus recycling in stylo root exudates. Int. J. Biol. Macromol. 2023, 241, 124569. [Google Scholar] [CrossRef]
- Li, D.W.; Tan, J.Z.; Li, Z.F.; Ou, L.J. Membrane lipid remodeling and autophagy to cope with phosphorus deficiency in the dinoflagellate Prorocentrum shikokuense. Chemosphere 2023, 349, 140844. [Google Scholar] [CrossRef]
- Zhang, H.C.; Wang, L.; Jin, X.L. High-throughput phenotyping of plant leaf morphological, physiological, and biochemical traits on multiple scales using optical sensing. Crop J. 2023, 11, 1303–1318. [Google Scholar] [CrossRef]
- Geng, Z.D.; Lu, Y.R.; Duan, L.F.; Chen, H.F.; Wang, Z.H.; Zhang, J.; Liu, Z.; Wang, X.M.; Zhai, R.F.; Ouyang, Y.D.; et al. High-throughput phenotyping and deep learning to analyze dynamic panicle growth and dissect the genetic architecture of yield formation. Crop Environ. 2024, 3, 1–11. [Google Scholar] [CrossRef]
- Mukul, M.M. Genetic analyses of morphological traits, and phenotypic screening of tossa jute germplasm grown under salinity stress. Heliyou 2023, 9, e12448. [Google Scholar] [CrossRef]
- Silva, P.C.; Sánchez, A.C.; Opazo, M.A.; Mardones, L.A.; Acevedo, E.A. Grain yield, anthesis-silking interval, and phenotypic plasticity in response to changing environments: Evaluation in temperate maize hybrids. Field Crops Res. 2022, 285, 108583. [Google Scholar] [CrossRef]
- Patel, M.; Rangani, J.; Kumari, A.; Parida, A.K. Mineral nutrient homeostasis, photosynthetic performance, and modulations of antioxidative defense components in two contrasting genotypes of Arachis hypogaea L. (peanut) for mitigation of nitrogen and/or phosphorus starvation. J. Biotechnol. 2020, 323, 136–158. [Google Scholar] [CrossRef]
- Zhang, S.F.; Dai, B.J.; Wang, Z.H.; Qaseem, M.F.; Li, H.L.; Wu, A.M. The key physiological and molecular responses of Neolamarckia cadamba to phosphorus deficiency stress by hydroponics. Ind. Crops Prod. 2023, 202, 117065. [Google Scholar] [CrossRef]
- Wang, S.; Zheng, S.W.; Bian, T.; Wu, T.; Li, X.X.; Fu, H.D.; Sun, Z.P.; Li, T.L. Photosynthetic characteristics combined with metabolomics analysis revealed potential mechanisms of cucumber (Cucumis sativus) yield reduction induced by different phosphorus stresses. Sci. Hortic. 2022, 302, 111156. [Google Scholar] [CrossRef]
- Hu, T.L.; Zhang, Y.H.; Wang, H.; Jin, H.Y.; Liu, B.J.; Lin, Z.B.; Ma, J.; Wang, X.J.; Liu, Q.; Liu, H.T.; et al. Biological nitrogen fixation in rice paddy soils is driven by multiple edaphic factors and available phosphorus is the greatest contributor. Pedosphere, 2023; in press. [Google Scholar] [CrossRef]
- Gu, R.L.; Chen, F.J.; Long, L.Z.; Cai, H.G.; Liu, Z.G.; Yang, J.B.; Wang, L.F.; Li, H.Y.; Li, J.H.; Liu, W.X.; et al. Enhancing phosphorus uptake efficiency through QTL-based selection for root system architecture in maize. J. Genet. Genom. 2016, 43, 663–672. [Google Scholar] [CrossRef]
- Mo, X.H.; Zhang, M.K.; Liang, C.Y.; Cai, L.Y.; Tian, J. Integration of metabolome and transcriptome analyses highlights soybean roots responding to phosphorus deficiency by modulating phosphorylated metabolite processes. Plant Physiol. Biochem. 2019, 139, 397–706. [Google Scholar] [CrossRef]
- Asif, I.; Dong, Q.; Wang, X.R.; Li, X.L.; Li, M.J.; Yang, Y.Z.; Zhou, J.; Wei, Q.P.; Zhou, B.B. Integrative physiological, transcriptome and metabolome analysis reveals the involvement of carbon and flavonoid biosynthesis in low phosphorus tolerance in cotton. Plant Physiol. Biochem. 2023, 196, 302–317. [Google Scholar]
- He, F.L.; Hu, S.Y.; Liu, R.T.; Li, X.X.; Guo, S.Q.; Wang, H.; Tian, G.; Qi, Y.T.; Wang, T.T. Decoding the biological toxicity of phenanthrene on intestinal cells of Eisenia fetida: Effects, toxicity pathways and corresponding mechanisms. Sci. Total Environ. 2023, 904, 166903. [Google Scholar] [CrossRef]
- Zhang, Z.Z.; Zhang, Z.D.; Han, X.Y.; Wu, J.H.; Zhang, L.Z.; Wang, J.R.; Wang-Pruski, G.F. Specific response mechanism to autotoxicity in melon (Cucumis melo L.) root revealed by physiological analyses combined with transcriptome profiling. Ecotoxicol. Environ. Saf. 2020, 200, 110779. [Google Scholar] [CrossRef]
- Lv, L.; Zhang, Y.Y.; Sun, H.; Zuo, X.X.; Wu, C.; Qian, J.Q. Effects of phosphorus stress on nutrients and saponins in ginseng plants. J. Jilin Agric. Univ. 2023, 45, 307–315. [Google Scholar]
- Shen, C.; Huang, B.F.; Hu, L.; Yuan, H.W.; Huang, Y.Y.; Wang, Y.B.; Sun, Y.F.; Li, Y.; Zhang, J.R.; Xin, J.L. Comparative transcriptome analysis and Arabidopsis thaliana overexpression reveal key genes associated with cadmium transport and distribution in root of two Capsicum annuum cultivars. J. Hazard. Mater. 2024, 465, 133365. [Google Scholar] [CrossRef]
- The Pharmacopoeia Commission of the People’s Republic of China. Pharmacopoeia of the People’s Republic of China, 2020th ed.; Chinese Medical Science and Technology Publishers: Beijing, China, 2015. [Google Scholar]
- Lei, H.X.; Zhang, H.F.; Zhang, Z.H.; Sun, H.; Li, M.J.; Shao, C.; Liang, H.; Wu, H.P.; Zhang, Y.Y. Physiological and transcriptomic analyses of roots from Panax ginseng C. A. Meyer under drought stress. Ind. Crops Prod. 2023, 191, 115858. [Google Scholar] [CrossRef]
- Wei, G.F.; Yang, F.; Wei, F.G.; Zhang, L.J.; Gao, Y.; Qian, J.; Chen, Z.J.; Jia, Z.W.; Wang, Y.; Su, H.; et al. Metabolomes and transcriptomes revealed the saponin distribution in root tissues of Panax quinquefolius and Panax notoginseng. J. Ginseng Res. 2020, 44, 757–769. [Google Scholar] [CrossRef]
- Herrera-Estrella, L.; López-Arredondo, D. Phosphorus: The underrated element for feeding the world. Trends Plant Sci. 2016, 21, 461–463. [Google Scholar] [CrossRef]
- Dey, P.; Santhi, R.; Maragatham, S.; Sellamuthu, K. Status of phosphorus and potassium in the Indian soils vis-à-vis world soils. Indian J. Fertil. 2017, 13, 44–59. [Google Scholar]
- Tung, A.; Levin, M. Extra-genomic instructive influences in morphogenesis: A review of external signals that regulate growth and form. Dev. Biol. 2020, 461, 1–12. [Google Scholar] [CrossRef]
- Wu, C.; Li, B.C.; Wei, Q.; Pan, Q.; Zhang, W.Y. Endophytic fungus Serendipita indica increased nutrition absorption and biomass accumulation in Cunninghamia lanceolata seedlings under low phosphate. Acta Ecol. Sin. 2019, 39, 21–29. [Google Scholar] [CrossRef]
- Akash Parida, A.P.; Srivastava, A.; Mathur, S.; Sharma, A.K.; Kumar, R. Identification, evolutionary profiling, and expression analysis of F-box superfamily genes under phosphate deficiency in tomato. Plant Physiol. Biochem. 2021, 162, 349–362. [Google Scholar] [CrossRef]
- Li, L.Y.; Yang, H.M.; Liu, P.; Ren, W.B.; Wu, X.H.; Huang, F. Combined impact of heat stress and phosphate deficiency on growth and photochemical activity of sheepgrass (Leymus chinensis). J. Plant Physiol. 2018, 231, 271–276. [Google Scholar] [CrossRef]
- Yang, Y.X.; Yao, P.P.; Song, H.; Li, Q.C. NtNCED3 regulates responses to phosphate deficiency and drought stress in Nicotiana tabacum. Gene 2023, 872, 147458. [Google Scholar] [CrossRef]
- Zheng, Q.M.; Hu, J.L.; Dong, C.F.; Hu, H.; Zhao, C.F.; Lei, K.Q.; Tian, Z.W.; Dai, T.B. Differences in membrane lipid homeostasis confer contrast tolerance to low phosphorus in two wheat (Triticum aestivum L.) cultivars. Environ. Exp. Bot. 2024, 219, 105653. [Google Scholar] [CrossRef]
- Maxwell, K.; Johnson, G.N. Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 2000, 51, 659–668. [Google Scholar] [CrossRef]
- Sayed, O.H. Chlorophyll Fluorescence as a Tool in Cereal Crop Research. Photosynthetica 2003, 41, 321–330. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, G.J. Effects of cesium accumulation on chlorophyll content fluorescence of Brassica juncea, L.J. Environ. Radioact. 2018, 195, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.F.; Tu, Z.H.; Zhang, Y.L.; Zhong, W.P.; Xia, H.; Hao, Z.Y.; Zhang, C.G.; Li, H.G. Predicting the impact of climate change on the distribution of two relict Liriodendron species by coupling the MaxEnt model and actual physiological indicators in relation to stress tolerance. J. Environ. Manag. 2022, 332, 116024. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Liu, Y.; Tian, F.P.; Wu, G.L. Soil water availability threshold indicator was determined by using plant physiological responses under drought conditions. Ecol. Indic. 2020, 118, 106740. [Google Scholar] [CrossRef]
- Ben, Y.; Cheng, M.Z.; Liu, Y.Q.; Wang, X.; Wang, L.H.; Yang, Q.; Huang, X.H.; Zhou, Q. Biomarker changes and oxidative damage in living plant cells as new biomonitoring indicators for combined heavy metal stress assessment. Ecol. Indic. 2023, 154, 110784. [Google Scholar] [CrossRef]
- Javidi, M.; Maali-Amiri, R.; Poormazaheri, H.; Niaraki, M.S.; Kariman, K. Cold stress-induced changes in metabolism of carbonyl compounds and membrane fatty acid composition in chickpea. Plant Physiol. Biochem. 2022, 192, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.X.; Xu, D.D.; Zhou, L.J.; Chen, H.; Peng, Z.W.; Chen, G.Y.; Wang, L.H.; Cao, H.S.; Peng, Y.Q.; Geng, S.Y.; et al. The critical role of CmCIPK1-CmRbohD1/D2 complexes in generating H2O2 signals for enhancing salt tolerance in pumpkins. Hortic. Plant J. 2024; in press. [Google Scholar] [CrossRef]
- Han, Y.C.; Liu, N.; Li, C.; Wang, S.W.; Jai, L.H.; Zhang, R.; Li, H.; Tan, J.F.; Xue, H.W.; Zheng, W.M. TaMADS2-3D, a MADS transcription factor gene, regulates phosphate starvation responses in plants. Crop J. 2022, 10, 243–253. [Google Scholar] [CrossRef]
- Mai NT, P.; Mai, C.D.; Ngyyen, H.V.; Le, K.Q.; Duong, L.V.; Tran, T.A.; To, H.T.M. Discovery of new genetic determinants of morphological plasticity in rice roots shoots under phosphate starvation using, G.W.A.S. J. Plant Physiol. 2021, 257, 153340. [Google Scholar] [PubMed]
- Devaiah, B.N.; Madhuvanthi, R.; Karthikeyan, A.S.; Raghothama, K.G. Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 Transcription Factor in Arabidopsis. Mol. Plant 2009, 2, 43–58. [Google Scholar] [CrossRef]
- Nazari, M.; Ghasemi-Soloklui, A.A.; Kordrostami, M.; Latef, A.A.H.A. Deciphering the response of medicinal plants to abiotic stressors: A focus on drought and salinity. Plant Stress 2023, 10, 100255. [Google Scholar] [CrossRef]
- Wu, J.J.; Liu, X.Y.; Ge, F.; Li, N. Tolerance mechanism of rice (Oryza sativa L.) seedings towards polycyclic aromatic hydrocarbons toxicity: The activation of SPX-mediated signal transduction to maintain P. homeostasis. Environ. Pollut. 2024, 341, 123009. [Google Scholar] [CrossRef] [PubMed]
- Hatem, R.; Bulak, A.A.; Yves, P. Regulation of phosphate starvation responses in plants: Signaling players and cross-talks. Mol. Plant 2010, 3, 288–299. [Google Scholar]
- Wang, L.; Deng, M.J.; Xu, J.M.; Zhu, X.L.; Mao, C.Z. Molecular mechanisms of phosphate transport and signaling in higher plants. Semin. Cell Dev. Biol. 2018, 74, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Chun, H.J.; Yun, D.J.; Kim, M.C. Cross-talk between phosphate starvation and other environmental stress signaling pathways in plants. Mol. Cells 2017, 40, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Shukla, D.; Waigel, S.; Rouchka, E.C.; Sandhu, G.; Trivedi, P.K.; Sahi, S.V. Genome-wide expression analysis reveals contrasting regulation of phosphate starvation response (PSR) in root and shoot of Arabidopsis and its association with biotic stress. Environ. Exp. Bot. 2021, 188, 104483. [Google Scholar] [CrossRef]
- Yan, H.M.; Wang, Y.L.; Chen, B.; Lv, C.W.; Li, J.Z.; Zhao, Q.Z. OsCKX2 regulates phosphate deficiency tolerance by modulating cytokinin in rice. Plant Sci. 2022, 319, 111257. [Google Scholar] [CrossRef] [PubMed]
- Prathap, V.; Kumar, S.; Meena, N.; Maheshwari, C.; Dalal, M.; Tyagi, A. Phosphorus Starvation Tolerance in Rice Through Combined Physiological, Biochemical, and Proteome Analyses. Rice Sci. 2023, 30, 613–631. [Google Scholar] [CrossRef]
- Zhu, Z.X.; Qu, K.Z.; Li, D.; Zhang, L.X.; Wang, C.Y.; Cong, L.; Bai, C.M.; Lu, X.C. SbPHO2, a conserved Pi starvation signalling gene, is involved in the regulation of the uptake of multiple nutrients in sorghum. Plant Sci. 2023, 327, 111556. [Google Scholar] [CrossRef] [PubMed]
- Ramaiah, M.; Jain, A.; Yugandhar, P.; Raghothama, K.G. ATL8, a RING E3 ligase, modulates root growth and phosphate homeostasis in Arabidopsis. Plant Physiol. Biochem. 2022, 179, 90–99. [Google Scholar] [CrossRef]
- Wang, B.; Gao, Z.Y.; Shi, Q.H.; Gong, B. SAMS1 stimulates tomato root growth and P availability via activating polyamines and ethylene synergetic signaling under low-P condition. Environ. Exp. Bot. 2022, 197, 104844. [Google Scholar] [CrossRef]
- Zhang, Y.X.; Wang, Y.Q.; Guo, Y.Y.; Chen, Y.; Sun, Y.P.; Wang, Z.L.; Guan, L.X.; Wang, L.; Chen, L.L. Multi-omics profiling reveals the effects of hydrogen peroxide treatment on carbohydrate and energy metabolism in postharvest broccoli. Postharvest Biol. Technol. 2024, 209, 112703. [Google Scholar] [CrossRef]
- Xie, L.Y.; Xu, Y.B.; Ding, X.Q.; Liang, S.; Li, D.L.; Fu, A.K.; Zhan, X.A. Itaconic acid and dimethyl itaconate exert antibacterial activity in carbon-enriched environments through the TCA cycle. Biomed. Pharmacother. 2023, 167, 115487. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.H.; Xue, B.H.; Song, G.L.; Shi, S.Q. Effects of citric acid on antioxidant system and carbon-nitrogen metabolism of Elymus dahuricus under Cd stress. Ecotoxicol. Environ. Saf. 2022, 233, 113321. [Google Scholar] [CrossRef] [PubMed]
- Shan, S.Z.; Wang, S.S.; Yan, X.; Chen, K.; Liang, L.; Li, X.H.; Zhou, C.X.; Yan, X.J.; Ruan, R.; Cheng, P.F. Mixotrophic culture of Chaetoceros sp. and the synergistic carbon and energy metabolism. Bioresour. Technol. 2023, 390, 129912. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yang, J.; Zhang, S.Y.; Gao, J.Q.; Li, X.; Zhou, J.H.; Hu, L.; Huang, L.Q. Metabolome and transcriptome analyses identify the underground rhizome growth through the regulation of rhizome apices in Panax ginseng. Ind. Crops Prod. 2023, 206, 1176354. [Google Scholar] [CrossRef]
- Kochan, E.; Sienkiewicz, M.; Szmajda-Krygier, D.; Balcerczak, E.; Szymańska, G. Thymol as a control factor of the expression of key genes of the ginsenoside biosynthesis pathway and its effect on the production of ginseng saponins in Panax quinquefolium hairy root cultures. Ind. Crops Prod. 2024, 210, 118151. [Google Scholar] [CrossRef]
- Koo, H.; Lee, Y.S.; Nguyen, V.B.; Giang, V.N.G.; Koo, H.J.; Park, H.S.; Mohanan, P.; Song, Y.H.; Ryu, B.; Kang, K.B.; et al. Comparative transcriptome and metabolome analyses of four Panax species explore the dynamics of metabolite biosynthesis. J. Ginseng Res. 2023, 47, 44–53. [Google Scholar] [CrossRef]
- Schmiderer, C.; Grausgruber-Gröger, S.; Grassi, P.; Steinborn, R.; Novak, J. Influence of gibberellin and daminozide on the expression of terpene synthases and on monoterpenes in common sage (Salvia officinalis). J. Plant Physiol. 2010, 167, 779–786. [Google Scholar] [CrossRef]
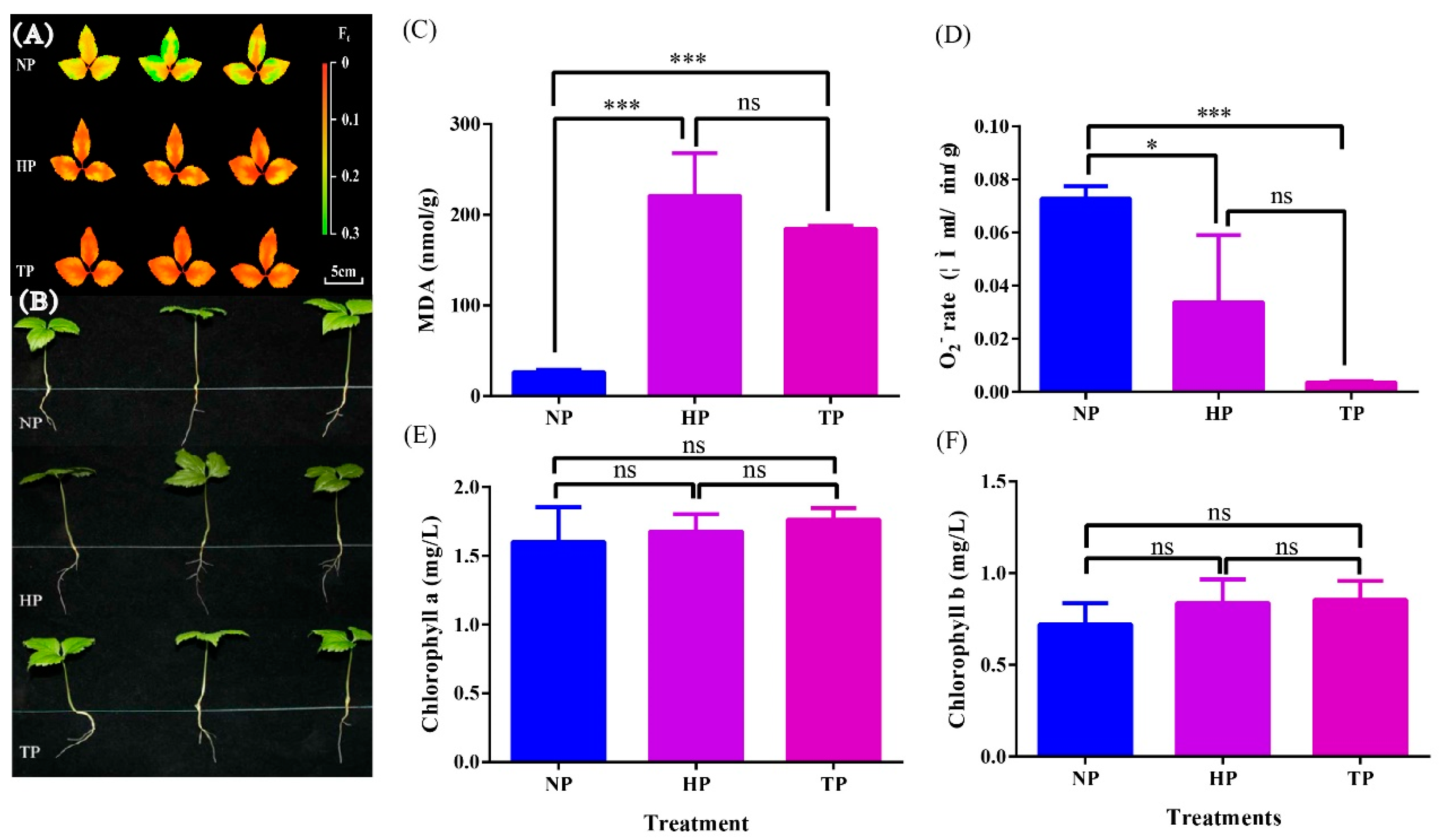

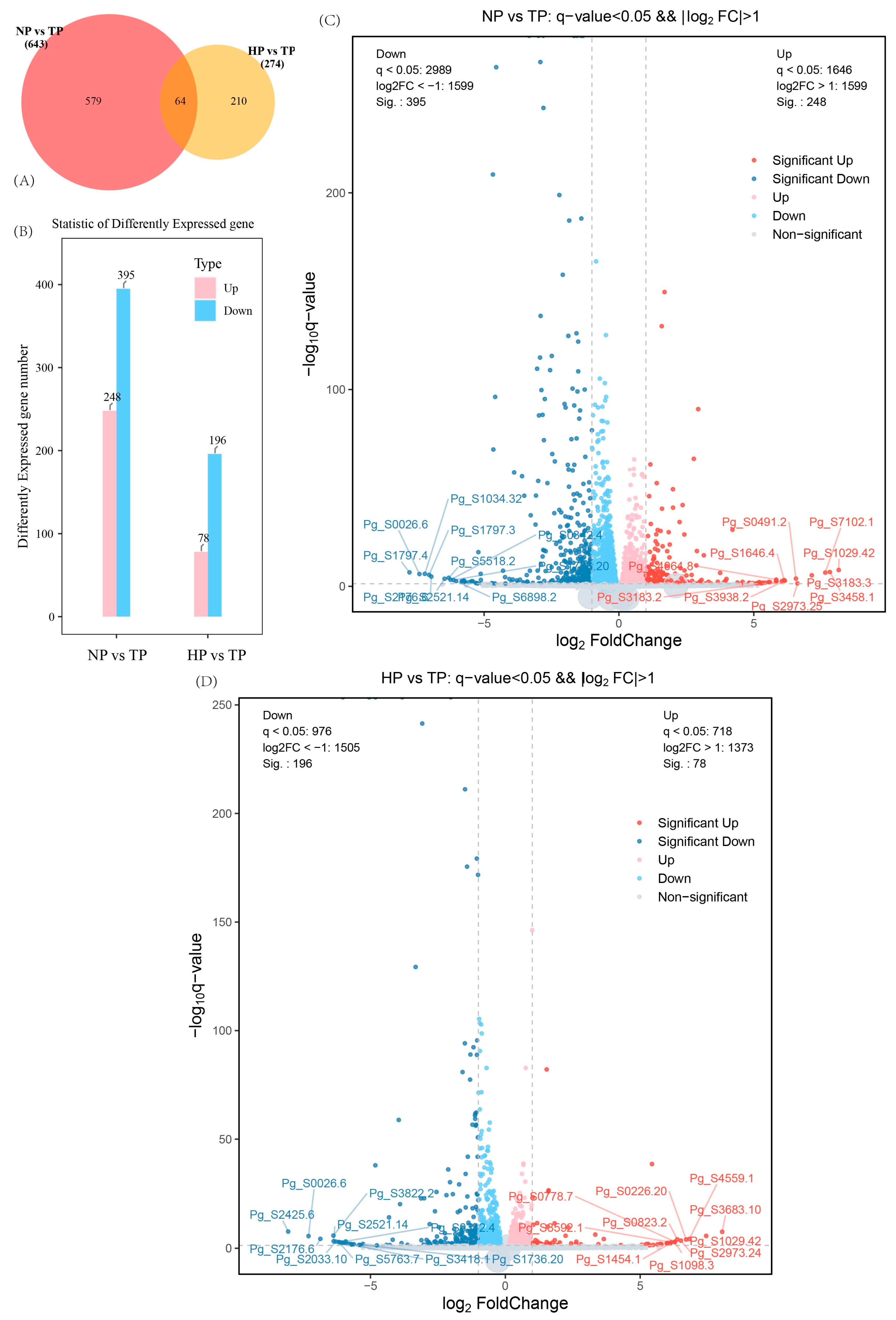
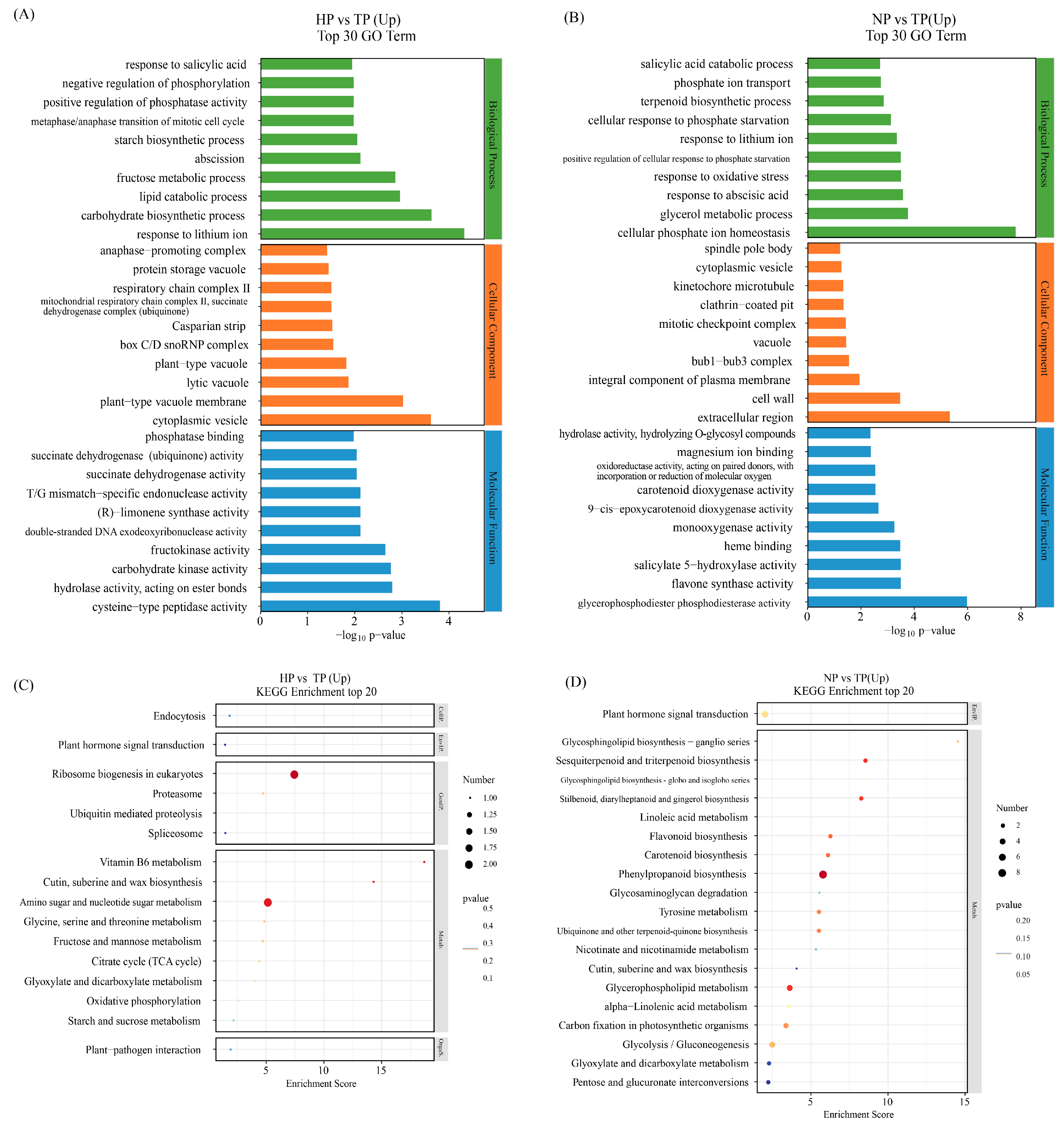
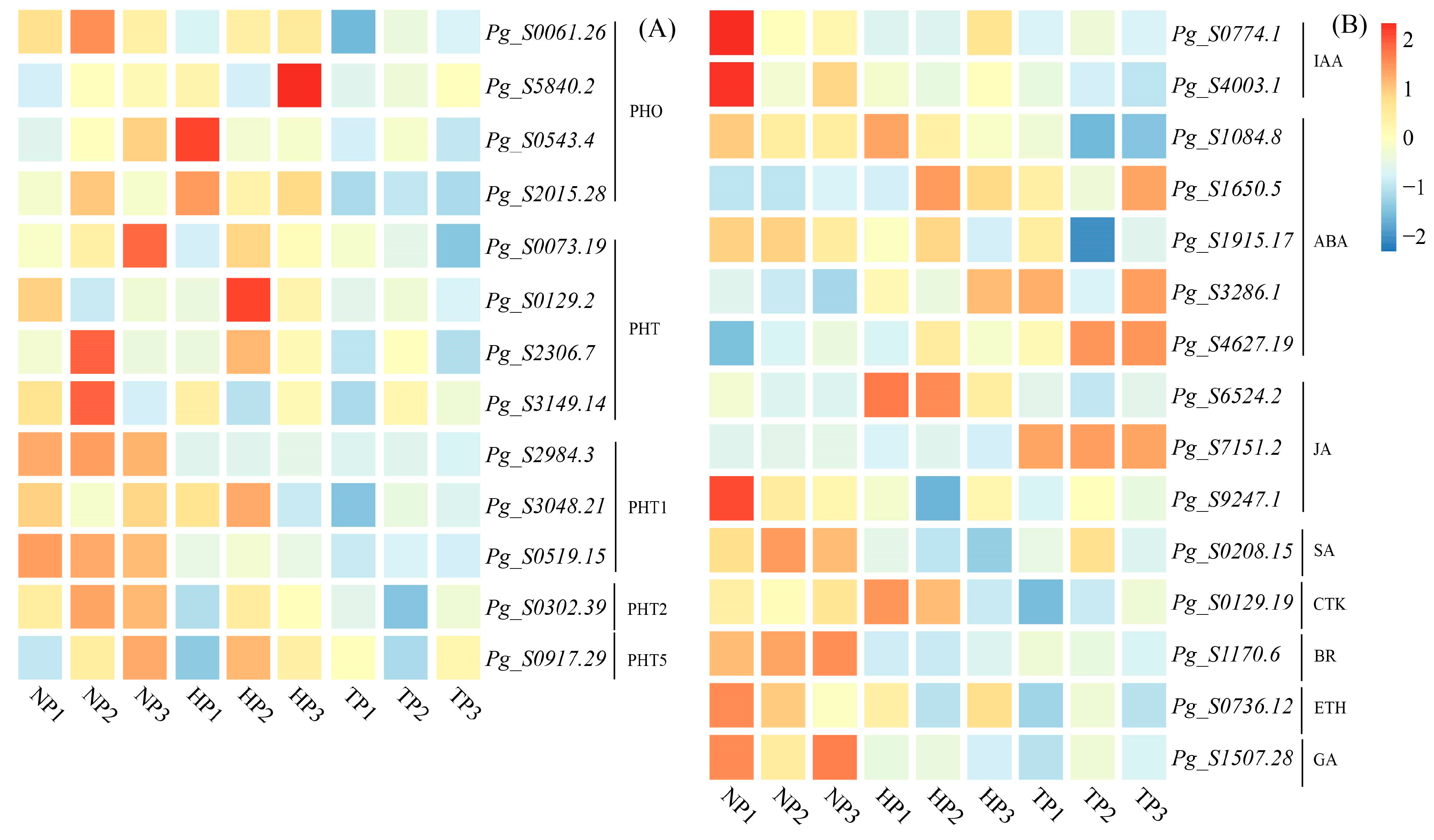
| Treatments | Root Length (cm) | Shoot Length (cm) | Average Leaflet Area (cm2) |
|---|---|---|---|
| NP | 4.36 ± 0.08 b | 5.42 ± 0.31 c | 4.53 ± 0.16 a |
| HP | 4.35 ± 0.06 b | 6.41 ± 0.22 b | 3.52 ± 0.21 a |
| TP | 5.26 ± 0.28 a | 7.90 ± 0.48 a | 4.89 ± 0.22 a |
| Treatments | Total Root Length (cm) | Root Project Area (cm2) | Root Surface Area (cm2) | Average Diameter (mm) | Later Root Amount |
|---|---|---|---|---|---|
| NP | 6.28 ± 0.68 b | 1.37 ± 0.19 b | 2.87 ± 0.58 a | 0.44 ± 0.06 b | 5.83 ± 1.61 a |
| HP | 10.29 ± 0.98 a | 2.57 ± 0.21 a | 2.97 ± 0.18 a | 0.78 ± 0.07 a | 8.17 ± 0.98 a |
| TP | 11.88 ± 1.43 a | 2.76 ± 0.42 a | 3.22 ± 0.45 a | 0.89 ± 0.13 a | 4.33 ± 1.75 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, H.; Liang, H.; Shao, C.; Qian, J.; Zhu, J.; Zhang, G.; Lv, B.; Zhang, Y. Effect of Phosphate-Deficiency Stress on the Biological Characteristics and Transcriptomics of Panax ginseng. Horticulturae 2024, 10, 506. https://doi.org/10.3390/horticulturae10050506
Sun H, Liang H, Shao C, Qian J, Zhu J, Zhang G, Lv B, Zhang Y. Effect of Phosphate-Deficiency Stress on the Biological Characteristics and Transcriptomics of Panax ginseng. Horticulturae. 2024; 10(5):506. https://doi.org/10.3390/horticulturae10050506
Chicago/Turabian StyleSun, Hai, Hao Liang, Cai Shao, Jiaqi Qian, Jiapeng Zhu, Guojia Zhang, Bochen Lv, and Yayu Zhang. 2024. "Effect of Phosphate-Deficiency Stress on the Biological Characteristics and Transcriptomics of Panax ginseng" Horticulturae 10, no. 5: 506. https://doi.org/10.3390/horticulturae10050506
APA StyleSun, H., Liang, H., Shao, C., Qian, J., Zhu, J., Zhang, G., Lv, B., & Zhang, Y. (2024). Effect of Phosphate-Deficiency Stress on the Biological Characteristics and Transcriptomics of Panax ginseng. Horticulturae, 10(5), 506. https://doi.org/10.3390/horticulturae10050506





