Identification of Candidate Genes Controlling Red Seed Coat Color in Cowpea (Vigna unguiculata [L.] Walp)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. SNP Genotyping and Data Curation
2.3. Seed Coat Phenotyping
2.4. Genetic Mapping of the Red Seed Coat Trait
2.5. Candidate Gene Identification and PCR Amplification
3. Results
3.1. Phenotypic Variation for Red Seed Coat Color
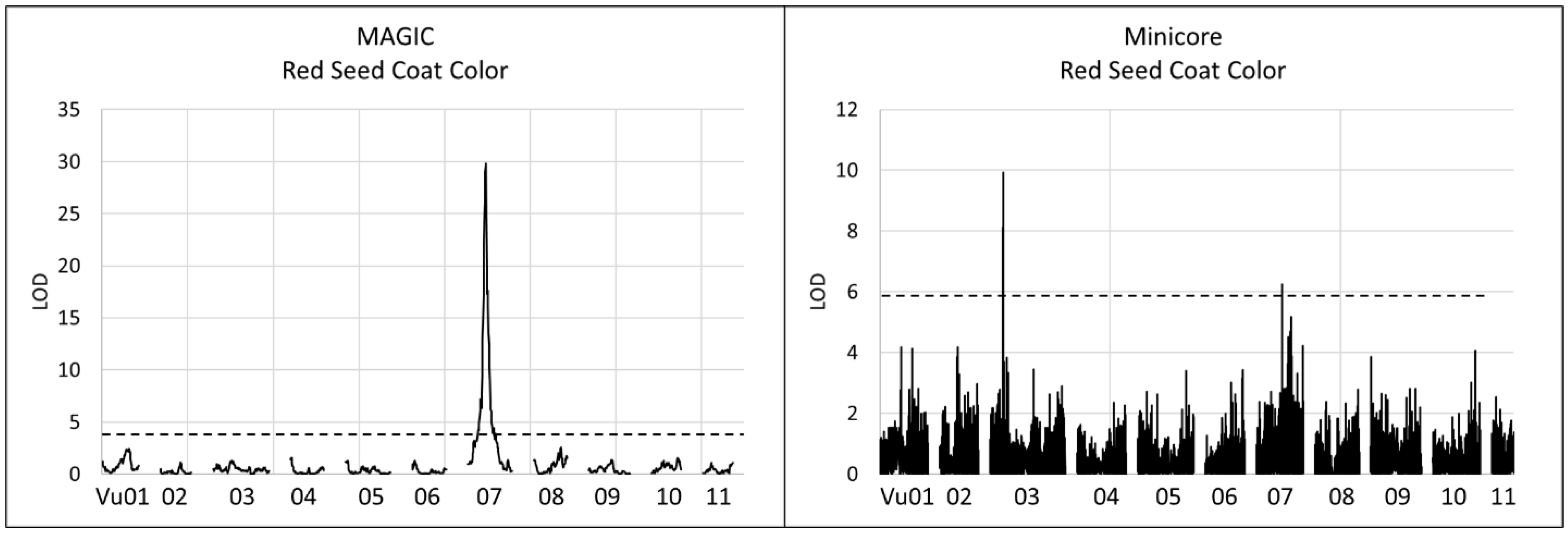
3.2. Loci Controlling Red Seed Coat Color
3.3. Identification of Candidate Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 26 February 2020).
- Ehlers, J.D.; Hall, A.E. Cowpea (Vigna unguiculata L. Walp.). Field Crops Res. 1997, 53, 187–204. [Google Scholar] [CrossRef]
- Boukar, O.; Belko, N.; Chamarthi, S.; Togola, A.; Batieno, J.; Owusu, E.; Haruna, M.; Diallo, S.; Umar, M.L.; Olufajo, O.; et al. Cowpea (Vigna unguiculata): Genetics, genomics and breeding. Plant Breed. 2019, 138, 415–424. [Google Scholar] [CrossRef]
- Singh, B.B. Cowpea: The Food Legume of the 21st Century; Crop Science Society of America: Madison, WI, USA, 2014. [Google Scholar]
- Tijjani, A.R.; Nabinta, R.T.; Muntaka, M. Adoption of innovative cowpea production practices in a rural area of Katsina State, Nigeria. J. Agric. Crop Res. 2015, 3, 53–58. [Google Scholar]
- Jaeger, S.R.; Antúnez, L.; Ares, G.; Swaney-Stueve, M.; Jin, D.; Harker, F.R. Quality perceptions regarding external appearance of apples: Insights from experts and consumers in four countries. Postharvest Biol. Technol. 2018, 146, 99–107. [Google Scholar] [CrossRef]
- Kostyla, A.S.; Clydesdale, F.M.; McDaniel, M.R. The psychophysical relationships between color and flavor. Food Sci. Nutr. 1997, 10, 303–321. [Google Scholar] [CrossRef] [PubMed]
- Langyintuo, A.S.; Lowenberg-DeBoer, J.; Faye, M.; Lambert, D.; Ibro, G.; Moussa, B.; Kergna, A.; Kushwaha, S.; Musa, S.; Ntoukam, G. Cowpea supply and demand in West and Central Africa. Field Crops Res. 2003, 82, 215–231. [Google Scholar] [CrossRef]
- Mishili, F.J.; Fulton, J.; Shehu, M.; Kushwaha, S.; Marfo, K.; Jamal, M.; Kergna, A.; Lowenberg-DeBoer, J. Consumer preferences for quality characteristics along the cowpea value chain in Nigeria, Ghana, and Mali. Agribusiness 2009, 25, 16–35. [Google Scholar] [CrossRef]
- Herniter, I.A.; Zhenyu, J.; Kusi, F. Market preferences for cowpea (Vigna unguiculata [L.] Walp) dry grain in Ghana. Afr. J. Agric. Res. 2019, 14, 928–934. [Google Scholar] [CrossRef]
- Fery, R.L. The Genetics of Cowpeas: A Review of the World Literature. In Cowpea Research, Production and Utilization; Singh, S.R., Rachie, K.O., Eds.; John Wiley & Sons, Inc.: New York, NY, USA, 1985; pp. 25–62. [Google Scholar]
- Spillman, W.J. Inheritance of the ‘eye’ in Vigna. Am. Nat. 1911, XLV, 513–523. [Google Scholar] [CrossRef]
- Harland, S.C. Inheritance of certain characters in the cowpea (Vigna sinensis). J. Genet. 1919, 8, 101–132. [Google Scholar] [CrossRef]
- Fery, R.L. Genetics of Vigna. In Horticultural Reviews; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 1980; Volume 2, pp. 311–394. [Google Scholar] [CrossRef]
- Saunders, A.R. Inheritance in the cowpea (Vigna sinensis Endb.). II: Seed coat colour pattern; flower, plant, and pod color. S. Afr. J. Agric. Sci. 1960, 3, 141–162. [Google Scholar]
- Drabo, I.; Ladeinde, T.A.O.; Smithson, J.B.; Redden, R. Inheritance of Eye Pattern and Seed Coat Colour in Cowpea (Vigna unguiculata [L.] Walp.). Plant Breed. 1988, 100, 119–123. [Google Scholar] [CrossRef]
- Kovinich, N.; Saleem, A.; Arnason, J.T.; Miki, B. Identification of two anthocyanidin reductase genes and three red-brown soybean accessions with reduced anthocyanidin reductase 1 mRNA, activity, and seed coat proanthocyanidin amounts. J. Agric. Food Chem. 2012, 60, 574–584. [Google Scholar] [CrossRef] [PubMed]
- Bassett, M.J. Genetics of Seed Coat Color and Pattern in Common Bean. In Plant Breeding Reviews, 28th ed.; Wiley: Hoboken, NJ, USA, 2007; pp. 239–315. [Google Scholar] [CrossRef]
- Campa, A.; Rodríguez Madrera, R.; Jurado, M.; García-Fernández, C.; Suárez Valles, B.; Ferreira, J.J. Genome-wide association study for the extractable phenolic profile and coat color of common bean seeds (Phaseolus vulgaris L.). BMC Plant Biol. 2023, 23, 158. [Google Scholar] [CrossRef]
- Sadohara, R.; Long, Y.; Izquierdo, P.; Urrea, C.A.; Morris, D.; Cichy, K. Seed coat color genetics and genotype × environment effects in yellow beans via machine-learning and genome-wide association. Plant Genome 2021, 15, e20173. [Google Scholar] [CrossRef]
- Herniter, I.A.; Lo, R.; Muñoz-Amatriaín, M.; Lo, S.; Guo, Y.-N.; Huynh, B.-L.; Lucas, M.; Jia, Z.; Roberts, P.A.; Lonardi, S.; et al. Seed Coat Pattern QTL and Development in Cowpea (Vigna unguiculata [L.] Walp.). Front. Plant Sci. 2019, 10, 514455. [Google Scholar] [CrossRef]
- Lonardi, S.; Muñoz-Amatriaín, M.; Liang, Q.; Shu, S.; Wanamaker, S.I.; Lo, S.; Tanskanen, J.; Schulman, A.H.; Zhu, T.; Luo, M.-C.; et al. The genome of cowpea (Vigna unguiculata [L.] Walp.). Plant J. 2019, 98, 767–782. [Google Scholar] [CrossRef]
- Muñoz-Amatriaín, M.; Mirebrahim, H.; Xu, P.; Wanamaker, S.I.; Luo, M.-C.; Alhakami, H.; Alpert, M.; Atokple, I.; Batieno, B.J.; Boukar, O.; et al. Genome resources for climate-resilient cowpea, an essential crop for food security. Plant J. 2017, 89, 1042–1054. [Google Scholar] [CrossRef]
- Muñoz-Amatriaín, M.; Lo, S.; Herniter, I.A.; Boukar, O.; Fatokun, C.; Carvalho, M.; Castro, I.; Guo, Y.-N.; Huynh, B.-L.; Roberts, P.A.; et al. The UCR Minicore: A valuable resource for cowpea research and breeding. Legum. Sci. 2021, 3, leg3.95. [Google Scholar] [CrossRef]
- Huynh, B.-L.; Ehlers, J.D.; Huang, B.E.; Muñoz-Amatriaín, M.; Lonardi, S.; Santos, J.R.P.; Ndeve, A.; Batieno, B.J.; Boukar, O.; Cisse, N.; et al. A multi-parent advanced generation inter-cross (MAGIC) population for genetic analysis and improvement of cowpea (Vigna unguiculata L. Walp.). Plant J. 2018, 93, 1129–1142. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Muñoz-Amatriaín, M.; Shu, S.; Lo, S.; Wu, X.; Carlson, J.W.; Davidson, P.; Goodstein, D.M.; Phillips, J.; Janis, N.M.; et al. A view of the pan-genome of domesticated Cowpea (Vigna unguiculata [L.] Walp.). Plant Genome 2023, e20319. [Google Scholar] [CrossRef]
- Herniter, I.A.; Muñoz-Amatriaín, M.; Lo, S.; Guo, Y.-N.; Close, T.J. Identification of Candidate Genes Controlling Black Seed Coat and Pod Tip Color in Cowpea (Vigna unguiculata [L.] Walp). G3 Genes|Genomes|Genet. 2018, 8, 3347–3355. [Google Scholar] [CrossRef]
- Huang, B.E.; George, A.W. R/mpMap: A computational platform for the genetic analysis of multiparent recombinant inbred lines. Bioinformatics 2011, 27, 727–729. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ersoz, E.; Lai, C.-Q.; Todhunter, R.J.; Tiwari, H.K.; Gore, M.A.; Bradbury, P.J.; Yu, J.; Arnett, D.K.; Ordovas, J.M.; et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Buckler, E.; Bradbury, P.; Kroon, D.; Ramdoss, R.; Fink, A.J.; Zhang, Z.; Johnson, L.; Miller, Z.; Casstevens, T. TASSEL; Version 5.2.93; Windows; Edward Buckler: Ithaca, NY, USA, 2023. [Google Scholar]
- Yao, S.; Jiang, C.; Huang, Z.; Torres-Jerez, I.; Chang, J.; Zhang, H.; Udvardi, M.; Liu, R.; Verdier, J. The Vigna unguiculata Gene Expression Atlas (VuGEA) from de novo assembly and quantification of RNA-seq data provides insights into seed maturation mechanisms. Plant J. 2016, 88, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef] [PubMed]
- Yaeno, T.; Iba, K. BAH1/NLA, a RING-type ubiquitin E3 ligase, regulates the accumulation of salicylic acid and immune responses to Pseudomonas syringae DC3000. Plant Physiol. 2008, 148, 1032–1041. [Google Scholar] [CrossRef] [PubMed]
- Bassett, M.J.; Lee, R.; Otto, C.; Mcclean, P.E. Classical and Molecular Genetic Studies of the Strong Greenish Yellow Seedcoat Color in ‘Wagenaar’ and ‘Enola’ Common Bean. J. Am. Soc. Hortic. Sci. 2002, 127, 50–55. [Google Scholar] [CrossRef]
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef]
- Ahmadiani, N.; Robbins, R.J.; Collins, T.M.; Giusti, M.M. Molar absorptivity (ε) and spectral characteristics of cyanidin-based anthocyanins from red cabbage. Food Chem. 2016, 197, 900–906. [Google Scholar] [CrossRef]
- Zhang, H.; Tian, H.; Chen, M.; Xiong, J.; Cai, H.; Liu, Y. Transcriptome analysis reveals potential genes involved in flower pigmentation in a red-flowered mutant of white clover (Trifolium repens L.). Genomics 2018, 110, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Jara-Palacios, M.J.; Santisteban, A.; Gordillo, B.; Hernanz, D.; Heredia, F.J.; Escudero-Gilete, M.L. Comparative study of red berry pomaces (blueberry, red raspberry, red currant and blackberry) as source of antioxidants and pigments. Eur. Food Res. Technol. 2019, 245, 1–9. [Google Scholar] [CrossRef]
- Asen, S.; Norris, K.H.; Stewart, R.N. Absorption spectra and color of aluminium-cyanidin 3-glucoside complexes as influenced by pH. Phytochemistry 1969, 8, 653–659. [Google Scholar] [CrossRef]
- Mol, J.; Grofewold, E.; Koes, R. How genes paint flowers and seeds. Trends Plant Sci. 1998, 3, 212–217. [Google Scholar] [CrossRef]
- Fiscus, C.J.; Herniter, I.A.; Tchamba, M.; Paliwal, R.; Muñoz-Amatriaín, M.; Roberts, P.A.; Abberton, M.; Alaba, O.; Close, T.J.; Oyatomi, O.; et al. The pattern of genetic variability in a core collection of 2021 cowpea accessions. bioRxiv 2023, 2023.12.21.572659. [Google Scholar] [CrossRef]
- Commission Internationale de l’Eclairage. Colorimetry, CIE 015:2018; CIE: Vienna, Austria, 2018. [Google Scholar] [CrossRef]
- Sarkar, S.; Shekoofa, A.; McClure, A.; Gillman, J.D. Phenotyping and Quantitative Trait Locus Analysis for the Limited Transpiration Trait in an Upper-Mid South Soybean Recombinant Inbred Line Population (‘Jackson’ × ‘KS4895′): High Throughput Aquaporin Inhibitor Screening. Front. Plant Sci. 2022, 12, 779834. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.; Zhao, Q.; Liao, H. Genetic improvement of legume roots for adaption to acid soils. Crop J. 2023, 11, 1022–1033. [Google Scholar] [CrossRef]


| Population (# of Lines) | Not Red | Red | Other | Pred. Seg. Ratio | χ2 | Probability |
|---|---|---|---|---|---|---|
| UCR Minicore (368) | 190 | 80 | 98 | -- | -- | -- |
| MAGIC (305) | 157 | 92 | 56 | 5:3 | 0.032 | 0.86 |
| Sasaque-by-Sanzi (108) | 86 | 22 | 0 | 3:1 | 1.23 | 0.27 |
| Population | Chr | Peak SNP | Flanking SNPs | QTL pos. (bp) | LOD/−log10 (p) | R2 (%) | Effect |
|---|---|---|---|---|---|---|---|
| Sasaque × Sanzi (F2) | Vu03 | -- | 2_38494–2_05272 | 7,657,076–41,457,212 | -- | -- | |
| UCR Minicore | Vu03 | 2_20787 | 2_03389–2_20787 | 10,933,603–11,068,119 | 9.82 | 16.6 | −0.56 (G) |
| UCR Minicore | Vu07 | 2_02375 | -- | 22,014,760 | 6.26 | 9.8 | −0.61 (A) |
| MAGIC | Vu07 | -- | 2_51274–2_04829 | 14,976,909–22,928,810 | 29.81 | 38.82 |
| Vigun03g118700 | ||||||
|---|---|---|---|---|---|---|
| BP | RefGen Base | Mutant Base | Genotypes Present In | AA Code | AA | Mutation Type |
| 378 | C | T | UCR779 | ATT→ATC | Ile→Ile | Silent |
| 592 | G | A | UCR779 | ACT→GCT | Thr→Ala | Polar Uncharged → hydrophobic |
| 593 | C | A | ZN016, TZ30 | GCT→GAT | Ala→Asp | hydrophobic → polar negative |
| 661 | T | G | Sasaque | ATG→AGC | Met→Ser | hydrophobic → polar uncharged |
| 672 | G | T | Sasaque | GTT→TTT | Val→Phe | hydrophobic → hydrophobic |
| Vigun07g118600 | ||||||
| BP | RefGen Base | Mutant Base | Genotypes Present In | AA Code | AA | Mutation Type |
| 761 | T | C | UCR779 | TTC→TCC | Phe→Ser | hydrophobic → polar uncharged |
| 849+ | ZN016, TZ30 | Completely different AA sequence | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Herniter, I.A.; Muñoz-Amatriaín, M.; Lo, S.; Guo, Y.-N.; Lonardi, S.; Close, T.J. Identification of Candidate Genes Controlling Red Seed Coat Color in Cowpea (Vigna unguiculata [L.] Walp). Horticulturae 2024, 10, 161. https://doi.org/10.3390/horticulturae10020161
Herniter IA, Muñoz-Amatriaín M, Lo S, Guo Y-N, Lonardi S, Close TJ. Identification of Candidate Genes Controlling Red Seed Coat Color in Cowpea (Vigna unguiculata [L.] Walp). Horticulturae. 2024; 10(2):161. https://doi.org/10.3390/horticulturae10020161
Chicago/Turabian StyleHerniter, Ira A., María Muñoz-Amatriaín, Sassoum Lo, Yi-Ning Guo, Stefano Lonardi, and Timothy J. Close. 2024. "Identification of Candidate Genes Controlling Red Seed Coat Color in Cowpea (Vigna unguiculata [L.] Walp)" Horticulturae 10, no. 2: 161. https://doi.org/10.3390/horticulturae10020161
APA StyleHerniter, I. A., Muñoz-Amatriaín, M., Lo, S., Guo, Y.-N., Lonardi, S., & Close, T. J. (2024). Identification of Candidate Genes Controlling Red Seed Coat Color in Cowpea (Vigna unguiculata [L.] Walp). Horticulturae, 10(2), 161. https://doi.org/10.3390/horticulturae10020161







