Rapid Construction and Application of a Vector for Tobacco Ringspot Virus-Induced McPDS Silencing in Bitter Gourd
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Virus Expression Vector
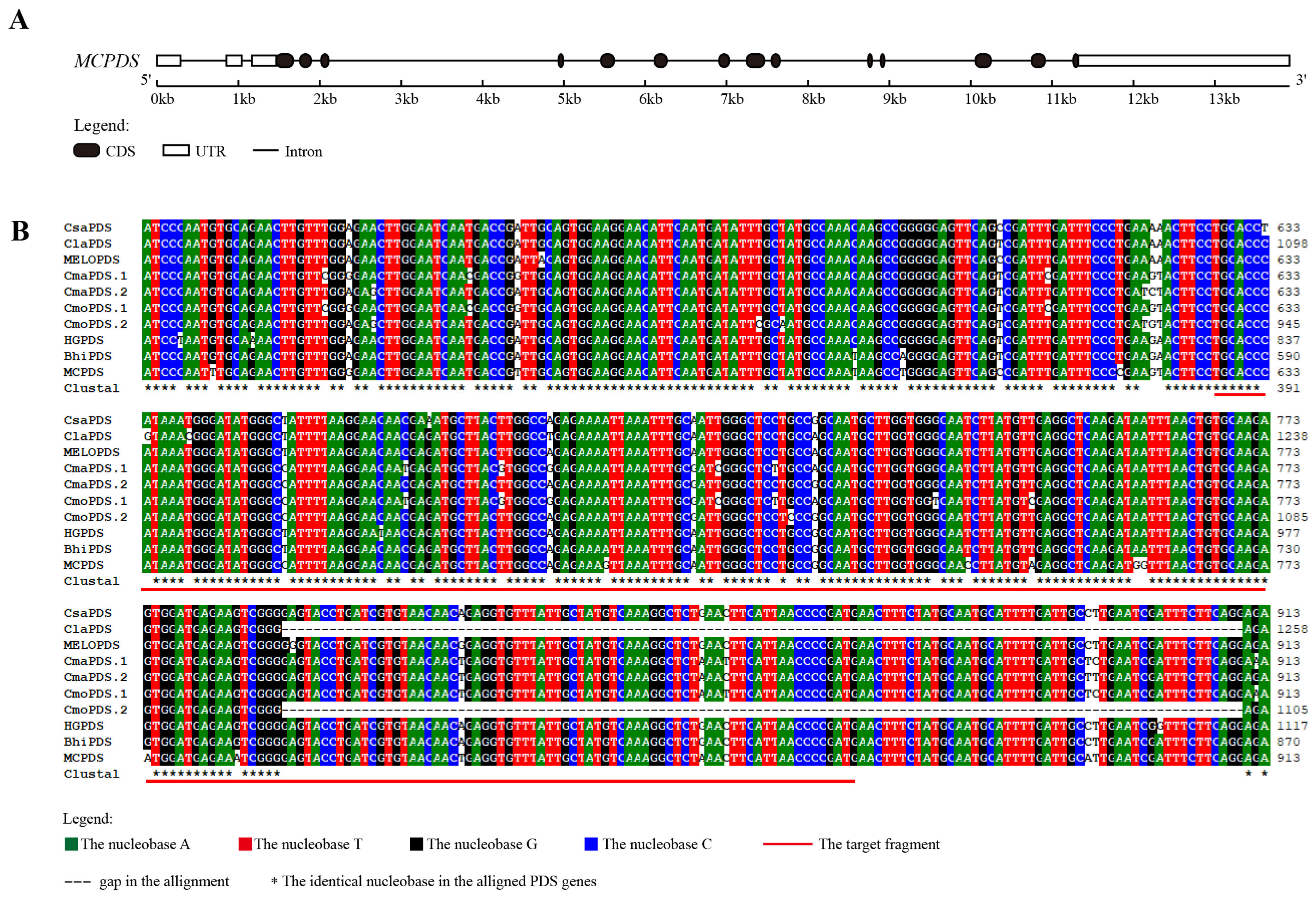
2.2. Alignment Analysis of PDS Genes in Cucurbits
2.3. Sequence and Primer Design
2.4. Constructing pTRSV2 Target (McPDS) Vector Using an Improved Method
2.5. TRSV Cultured in Host and Inoculation
2.6. TRSV Virus Assay, Gene Silencing Efficiency Assay, Chlorophyll Assay
3. Results
3.1. Target Selection via Sequence Analysis of Bitter Gourd McPDS
3.2. pTRSV2-McPDS Vector Constructed by Enzymic Ligation While Digesting
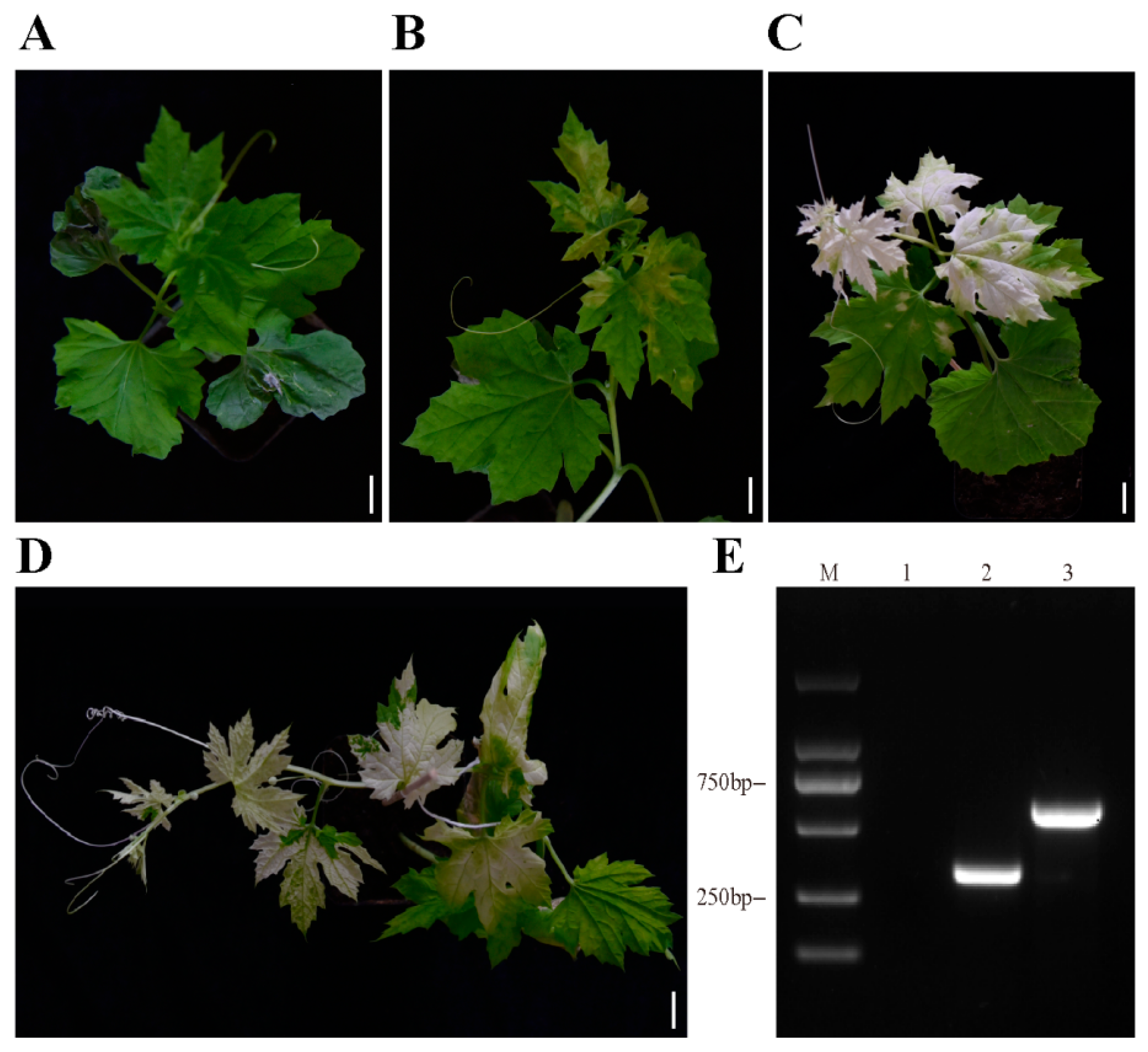
3.3. TRSV-McPDS Induced Photo-Bleaching in Bitter Gourd
3.4. TRSV Detected in Systemic Leaves of the Bitter Gourd Plants
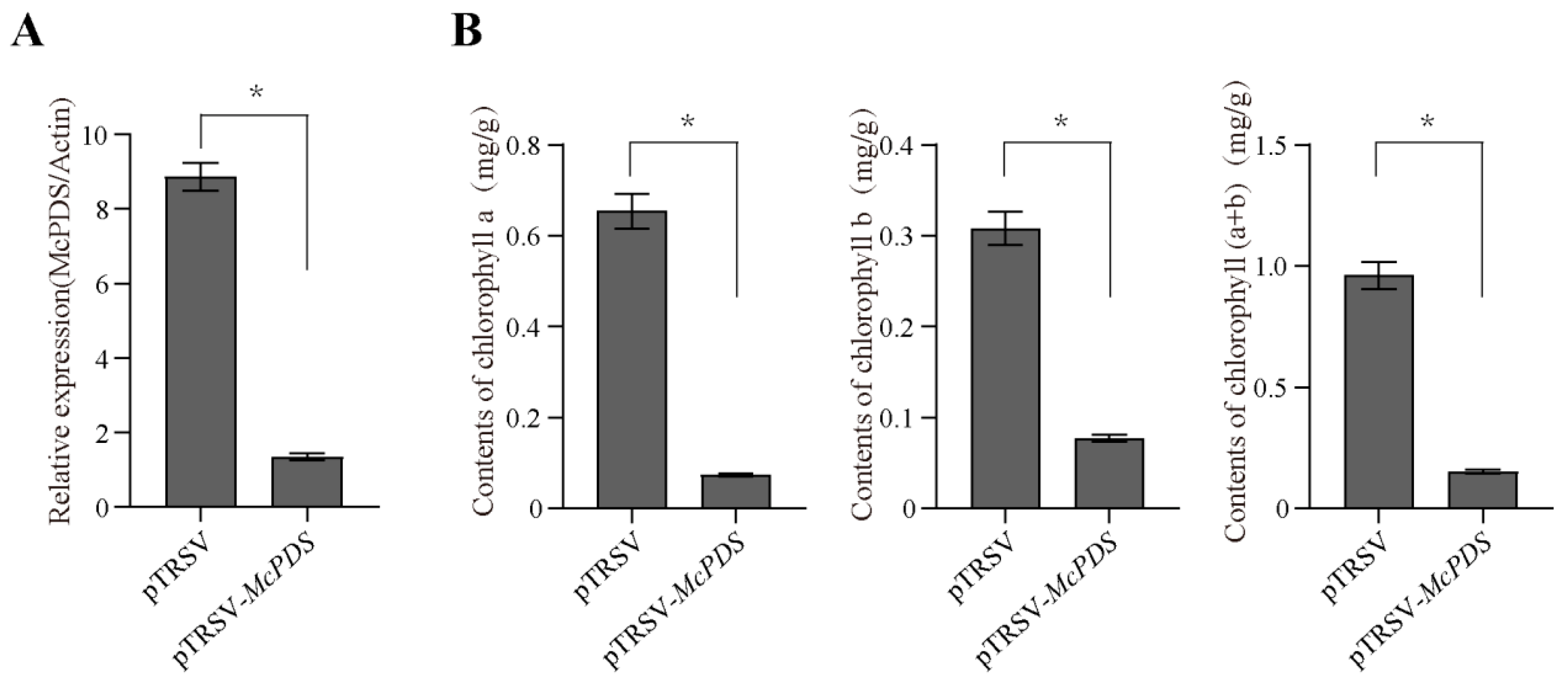
3.5. TRSV-McPDS-Induced Downregulation of McPDS Gene Expression
3.6. TRSV-McPDS-Induced Decrease in Chlorophyll Content
4. Discussion
4.1. Functional TRSV-McPDS (Gene Target) Vector Constructed Rapidly Using an Improved Method
4.2. The Inoculation Method Optimized for TRSV-Induced McPDS Silencing in Bitter Gourd
4.3. TRSV-Based Vector Suitable for VIGS in Bitter Gourd
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cui, J.J.; Yang, Y.; Luo, S.B.; Wang, L.; Huang, R.K.; Wen, Q.F.; Han, X.X.; Miao, N.S.; Cheng, J.W.; Liu, Z.J.; et al. Whole-genome sequencing provides insights into the genetic diversity and domestication of bitter gourd (Momordica spp.). Hortic. Res. 2020, 7, 85. [Google Scholar] [CrossRef] [PubMed]
- Fu, A.Z.; Zheng, Y.Y.; Guo, J.; Grierson, D.; Zhao, X.Y.; Wen, C.L.; Liu, Y.; Li, J.; Zhang, X.W.; Yu, Y.; et al. Telomere-to-telomere genome assembly of bitter melon (Momordica charantia L. var. abbreviate Ser.) reveals fruit development, composition and ripening genetic characteristics. Hortic. Res. 2023, 10, uhac228. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.A.; Cui, J.J.; Zhang, C.Y.; Yang, X.; Pan, H.R.; Du, H.; Ahmad, A.; Wu, T.Q.; Yao, C.P. Genome-wide identification and analysis of the MLO gene family for candidate powdery mildew susceptibility factors in Momordica charantia. Sci. Hortic. 2021, 283, 110119. [Google Scholar] [CrossRef]
- Urasaki, N.; Takagi, H.; Natsume, S.; Uemura, A.; Taniai, N.; Miyagi, N.; Fukushima, M.; Suzuki, S.; Trora, K.; Tamaki, M.; et al. Draft genome sequence of bitter gourd (Momordica charantia), a vegetable and medicinal plant in tropical and subtropical regions. DNA Res. 2017, 24, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Wang, N.N.; Li, Y.; Wang, H.H.; Zhang, W.N.; Wang, H.S.; Cai, S. Recent progress in genetic transformation and gene editing technology in cucurbit crops. Agronomy 2023, 13, 755. [Google Scholar] [CrossRef]
- Fang, L.; Wei, X.Y.; Liu, L.Z.; Zhou, L.X.; Tian, Y.P.; Geng, C.; Li, X.D. Erratum to: A tobacco ringspot virus-based vector system for gene and microrna function studies in cucurbits. Plant Physiol. 2021, 186, 853–864. [Google Scholar] [CrossRef]
- Xin, T.X.; Tian, H.J.; Ma, Y.L.; Wang, S.H.; Yang, L.; Li, X.T.; Zhang, M.Z.; Chen, C.; Wang, H.S.; Li, H.Z.; et al. Targeted creation of new mutants with compact plant architecture using crispr/cas9 genome editing by an optimized genetic transformation procedure in cucurbit plants. Hortic. Res. 2022, 9, b86. [Google Scholar] [CrossRef] [PubMed]
- Lange, M.; Yellina, A.L.; Orashakova, S.; Becker, A. Virus-induced gene silencing (VIGS) in plants: An overview of target species and the virus-derived vector systems. Methods Mol. Biol. 2013, 975, 1–14. [Google Scholar]
- Dhariwal, G.K.; Dhariwal, R.; Frick, M.; Laroche, A. Virus Induced Gene Silencing: A Tool to Study Gene Function in Wheat. In Genomics of Cereal Crops; Springer Protocols Handbooks; Wani, S.H., Kumar, A., Eds.; Humana: New York, NY, USA, 2022; pp. 107–155. [Google Scholar]
- Ke, X.B.; Shen, J.J.; Niu, Y.Q.; Zhao, H.J.; Guo, Y.L.; Sun, P.Y.; Yang, T.W.; Jiang, Y.X.; Zhao, B.S.; Wang, Z.; et al. Cucumber NUCLEAR FACTOR-YC2/-YC9 target translocon component CsTIC21 in chloroplast photomorphogenesis. Plant Physiol. 2023, 192, 2822–2837. [Google Scholar] [CrossRef]
- Ramegowda, V.; Mysore, K.S.; Senthil-Kumar, M. Virus-induced gene silencing is a versatile tool for unraveling the functional relevance of multiple abiotic-stress-responsive genes in crop plants. Front. Plant Sci. 2014, 5, 323. [Google Scholar] [CrossRef]
- Jagram, N.; Dasgupta, I. Principles and practice of virus induced gene silencing for functional genomics in plants. Virus Genes 2023, 59, 173–187. [Google Scholar] [CrossRef]
- Zulfiqar, S.; Farooq, M.A.; Zhao, T.; Wang, P.; Tabusam, J.; Wang, Y.; Xuan, S.; Zhao, J.; Chen, X.; Shen, S.; et al. Virus-Induced Gene Silencing (VIGS): A Powerful Tool for Crop Improvement and Its Advancement towards Epigenetics. Int. J. Mol. Sci. 2023, 24, 5608. [Google Scholar] [CrossRef]
- Bu, R.; Wang, R.; Wei, Q.; Hu, H.; Sun, H.; Song, P. Silencing of glycerol-3-phosphate acyltransferase 6 (GPAT6) gene using a newly established virus induced gene silencing (VIGS) system in cucumber alleviates autotoxicity mimicked by cinnamic acid (CA). Plant Soil 2019, 438, 329–346. [Google Scholar] [CrossRef]
- Li, C.; Yamagishi, N.; Kasajima, I.; Yoshikawa, N. Virus-induced gene silencing and virus-induced flowering in strawberry (Fragaria × ananassa) using apple latent spherical virus vectors. Hortic. Res. 2019, 6, 1428–1437. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Liang, Z.; Aranda, M.A.; Hong, N.; Liu, L.M.; Kang, B.; Gu, Q.S. A cucumber green mottle mosaic virus vector for virus-induced gene silencing in cucurbit plants. Plant Methods 2020, 16, 9. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.Y.J.Y. Identification of the pleiotropic function of TOUSLED kinase in tomato (Solanum lycopersicum L.) using a Cucumber mosaic virus-based vector. Hortic. Environ. Biotechnol. 2018, 59, 105–114. [Google Scholar] [CrossRef]
- Rhee, S.J.; Jang, Y.J.; Park, J.Y.; Ryu, J.; Lee, G.P. Virus-induced gene silencing for in planta validation of gene function in cucurbits. Plant Physiol. 2022, 190, 2366–2379. [Google Scholar] [CrossRef]
- Tzean, Y.; Lee, M.C.; Jan, H.H.; Chiu, Y.S.; Yeh, H.H. Cucumber mosaic virus-induced gene silencing in banana. Sci. Rep. 2019, 9, 11553. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Gao, L.W.; Liu, W.S.; Song, L.X.; Xiao, D.; Liu, T.K.; Hou, X.L.; Zhang, C.W. Transcription Coactivator ANGUSTIFOLIA3 (AN3) Regulates Leafy Head Formation in Chinese Cabbage. Front. Plant Sci. 2019, 10, 520. [Google Scholar] [CrossRef]
- Yamagishi, N.; Yoshikawa, N. Efficient virus-induced gene silencing system in pumpkin (Cucurbita maxima) using apple latent spherical virus vector. J. Virol. Methods 2022, 301, 114456. [Google Scholar] [CrossRef]
- Kumagai, M.H.; Donson, J.; Della, C.G.; Harvey, D.; Hanley, K.; Grill, L.K. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc. Natl. Acad. Sci. USA 1995, 92, 1679–1683. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, G.; Ji, J.; Wang, J.H. The effect of pds gene silencing on chloroplast pigment composition, thylakoid membrane structure and photosynthesis efficiency in tobacco plants. Plant Sci. 2009, 177, 222–226. [Google Scholar] [CrossRef]
- Igarashi, A.; Yamagata, K.; Sugai, T.; Takahashi, Y.; Sugawara, E.; Tamura, A. Apple latent spherical virus vectors for reliable and effective virus-induced gene silencing among a broad range of plants including tobacco, tomato, Arabidopsis thaliana, cucurbits, and legumes. Virology 2009, 386, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Lim, S.; Igori, D.; Yoo, R.H.; Kwon, S.Y.; Moon, J.S. Development of tobacco ringspot virus-based vectors for foreign gene expression and virus-induced gene silencing in a variety of plants. Virology 2016, 492, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Liu, M. Construction of a VIGS Vector Based on Cucumber Green Mottle Virus; Chinese Academy of Agricultural Sciences: Beijing China, 2019. [Google Scholar]
- Cui, H.; Wang, A. An efficient viral vector for functional genomic studies of Prunus fruit trees and its induced resistance to Plum pox virus via silencing of a host factor gene. Plant Biotechnol. J. 2017, 15, 344–356. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.J.; Wang, C.H.; Cheng, H.X.; Qiao, J.L.; Yue, P.L.; Xian, F.Q.; Yan, H. Overexpression and vigs system for functional gene validation in oriental melon (Cucumis melo var. makuwa makino). Plant Cell 2019, 137, 275–284. [Google Scholar] [CrossRef]
- Yu, J.; Wu, S.; Sun, H.H.; Wang, X.; Tang, X.M.; Guo, S.G.; Zhang, Z.H.; Huang, S.W.; Xu, Y.; Weng, Y.Q.; et al. CuGenDBv2: An updated database for cucurbit genomics. Nucleic Acids Res. 2023, 51, D1457–D1464. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.P.; Wu, Y.Y.; Nie, H.Z.; Tang, D.Z. Rpn1a, a 26s proteasome subunit, is required for innate immunity in arabidopsis. Plant J. 2012, 71, 1015–1028. [Google Scholar] [CrossRef]
- Li, G.D.; Yan, L.; Yao, X.Z.; Lu, L.T. Establishment of a Virus-Induced Gene-Silencing (VIGS) System in Tea Plant and Its Use in the Functional Analysis of CsTCS1. Int. J. Mol. Sci. 2023, 24, 392. [Google Scholar] [CrossRef]

| Primer ID | Sequence | Description |
|---|---|---|
| McPDS-F | TTTACGTATGCACCCATAAATGGGATA | PCR primer for McPDS target cloning |
| McPDS-R | CCTACGTACATCGGGGTTAATGAAGTT | |
| McActin-F | AATGGGTATGGTCTGCAAGT | qRT-PCR primer for reference gene |
| McActin-R | GGAGAATGTTTCAAGAGGGTAG | |
| q McPDS-F | TCAGCCGATTTGATTTCC | qRT-PCR primer for McPDS |
| q McPDS-R | ACATAAGATTGCCCACCA | |
| pTRSV2-F | ATTTTCTTAGGTTCTTATGCTGG | PCR primer for Specific fragment of TRSV2 |
| pTRSV2-R | TAAGGGCAGGAACCTAAAAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, L.; Zhang, H.; Guo, J.; Deng, Z.; Liu, H.; Du, H.; Zhong, Y.; Zhang, C.; Yao, C. Rapid Construction and Application of a Vector for Tobacco Ringspot Virus-Induced McPDS Silencing in Bitter Gourd. Horticulturae 2024, 10, 110. https://doi.org/10.3390/horticulturae10020110
Zeng L, Zhang H, Guo J, Deng Z, Liu H, Du H, Zhong Y, Zhang C, Yao C. Rapid Construction and Application of a Vector for Tobacco Ringspot Virus-Induced McPDS Silencing in Bitter Gourd. Horticulturae. 2024; 10(2):110. https://doi.org/10.3390/horticulturae10020110
Chicago/Turabian StyleZeng, Lingen, Hui Zhang, Jinju Guo, Zhijun Deng, Hongbiao Liu, Hu Du, Yujuan Zhong, Changyuan Zhang, and Chunpeng Yao. 2024. "Rapid Construction and Application of a Vector for Tobacco Ringspot Virus-Induced McPDS Silencing in Bitter Gourd" Horticulturae 10, no. 2: 110. https://doi.org/10.3390/horticulturae10020110
APA StyleZeng, L., Zhang, H., Guo, J., Deng, Z., Liu, H., Du, H., Zhong, Y., Zhang, C., & Yao, C. (2024). Rapid Construction and Application of a Vector for Tobacco Ringspot Virus-Induced McPDS Silencing in Bitter Gourd. Horticulturae, 10(2), 110. https://doi.org/10.3390/horticulturae10020110







