Abstract
Hydrangea macrophylla (H. macrophylla), a species in the genus Hydrangea in the family Hydrangeaceae, is widely valued for its ornamental qualities in both domestic and international markets. Notably, H. macrophylla is known for its ability to accumulate aluminum (Al). Moreover, aluminum ions (Al3+) participate in sepal bluing. However, the underlying mechanisms of Al accumulation in the sepals remain unclear. In this study, we utilized transcriptome data from two cultivars to identify genes associated with Al accumulation. In total, 154 differentially expressed isoforms between the CK and Tr groups in the sepals of both cultivars were screened. Through gene enrichment analysis and similarity identification in the CDS (coding sequence) region, 43 differentially expressed genes were identified, including 30 upregulated and 13 downregulated genes, in the sepals of the Al treatment group. Further analysis revealed that seven of these upregulated genes are related to Al accumulation in sepals. Among the seven, the gene HmALS3.1 was identified as a potential key player in Al transport within the sepals of H. macrophylla. This study lays the groundwork for further exploration into the mechanisms by which HmALS3.1 regulates Al accumulation in H. macrophylla.
1. Introduction
Al is not an essential element for plant growth and development, and it is toxic to most plants, causing stunted growth, weak stalks, leaf chlorosis, and in severe cases, leaf desiccation and plant death [1]. However, moderate amounts of Al have been shown to be beneficial to the growth and development of certain plants [2,3]. For instance, moderate Al levels can promote root system development [4], enhance the absorption of trace elements, and improve photosynthesis in plants [1,5,6,7]. Some plants can accumulate high levels of Al3+ in their aboveground tissues without exhibiting toxic symptoms; these species are commonly referred to as Al-accumulating plants. Al-accumulating plants are specifically defined as those containing more than 1000 mg/kg dry weight of Al in their stems and leaves [8], including species like H. macrophylla, Fagopyrum esculentum, and Camellia sinensis [9,10].
H. macrophylla accumulates significant levels of Al3+ in its aboveground tissues [11]. Leaves and sepals are the primary organs for Al accumulation in these tissues. Al content in leaves can reach up to 5 mg per g of dry weight over several months [11]. Al content in red sepals ranges from 0 to 10 μg/g fresh weight, while in blue sepals, it exceeds 40 μg/g fresh weight [12,13]. In H. macrophylla, appropriate Al concentrations promote the growth and development of roots, stems, and leaves, as well as the absorption of essential nutrients, including nitrogen (N), potassium (K), phosphorus (P), calcium (Ca), and magnesium (Mg) [14]. Moreover, Al alters flower color from pink to blue, thereby enhancing their ornamental value [1,15]. Al3+ is critical for the blue pigmentation of sepals [13,16,17]. Given that H. macrophylla is an Al-accumulating plant, it likely possesses a specialized mechanism for Al accumulation. As a result, the mechanisms of Al absorption and transport in H. macrophylla have become prominent research topics.
Organic acids can form complexes with Al3+ to mitigate its toxicity in H. macrophylla. In the rhizosphere, Al3+ forms complexes with oxalic acid secreted by the root system, which partially prevents Al3+ from entering root cells. Conversely, in the xylem and leaves, Al3+ is absorbed and transported by forming complexes with citric acid [18,19,20]. Additionally, several genes related to Al accumulation have been identified. Some ABC transporter genes, including the multidrug and toxic compound extrusion (MATE) gene and the NRAMP aluminum transporter, are highly expressed in the roots and leaves of H. macrophylla under Al stress [21,22]. These transporters mediate the transport of organic acids, participate in the absorption, transport, and storage of Al3+, and play a crucial role in signaling, thereby enhancing plant resistance to Al stress [21,22]. Moreover, two aquaporin genes, H. macrophylla vacuolar Al transporter (HmVALT) and H. macrophylla plasma membrane Al transporter 1 (HmPALT1), have been associated with Al transport. HmPALT1 is responsible for transporting Al3+ from outside the cell into the cytoplasm, while HmVALT transports Al3+ to the vacuole [23]. However, the precise mechanisms of Al3⁺ absorption, transport, and storage in H. macrophylla remain unclear.
In this study, transcriptome data from the sepals of the CK group (plants without Al treatment) and the Tr group (plants with Al treatment) in two cultivars were analyzed. The identification of key candidate genes involved in Al accumulation in sepals was examined. These results lay the foundation for elucidating the Al accumulation mechanism in H. macrophylla and provide a significant theoretical reference for studying the mechanism of Al accumulation in Al-accumulating plants.
2. Materials and Methods
2.1. Plant Materials
Two cultivars of H. macrophylla, ‘Bailmer’ (Endless Summer™) and ‘Duro’, were used in this study and were cultivated in a greenhouse at the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences. Following flower bud formation, the plants of both cultivars were subjected to a low temperature (2 °C~6 °C) for approximately six weeks. Subsequently, the plants were returned to the greenhouse for flowering.
Half of the plants from each cultivar (Tr group) were grown in a mixed substrate (peat: perlite = 7:3, v/v). A 400 mL solution of aluminum sulfate (Al2(SO4)3 · 18H2O dissolved in water) was applied to the substrate at the concentration of 6 g/L every seven to ten days, starting from the flower bud formation stage until flowering. While the remaining half, without Al treatment, cultivated in the same mixed substrate served as the control (CK group). Under Al sulfate treatment, the sepal color changed from pink to blue in ‘Bailmer’, while in ‘Duro’, it changed only slightly and did not turn blue [16]. Sepals at three different developmental stages (S1, S2, S3) from the Tr and CK groups of ‘Bailmer’ and ‘Duro’ (Figure 1) [16] were sampled on April 28, 2020 and May 19, 2020, respectively, and used in each experiment.

Figure 1.
Sepals at three different developmental stages. S1, Stage 1; S2, Stage 2; S3, Stage 3. Reprinted with permission from Ref. [16]. 2022, Suxia Yuan.
2.2. Al Content Measurement
Initially, all samples were dried to a constant weight. Each 0.2–0.4 g sample was digested and soaked in 5 mL of nitric acid (≥99.8%) overnight. The treated samples were placed in a constant temperature drying oven for 2 h at 80 °C, 2 h at 120 °C, and 4 h at 160 °C, followed by cooling to room temperature. The cooled solution was heated to evaporate the acid, after which the digestive solution was transferred into a 25 mL volumetric flask. The inner tank and cap were rinsed three times with a small amount of 1% nitric acid solution. The rinsed solution was combined in the volumetric flask, diluted to the mark with 1% nitric acid, and thoroughly mixed to determine the Al content. The Al content was determined using inductively coupled plasma mass spectrometry (ICP-MS) following the methods for the determination of multiple elements in food (National Food Safety Standards, GB 5009.268-2016, China) [24]. The samples were characterized by element-specific mass numbers (mass-to-charge ratio, m/z). The detection wavelength was set at 396.152 nm.
2.3. Transcriptome Sequencing, Annotation
In prior research, a full-length transcriptome database was constructed using the PacBio Sequel II platform, incorporating the flowers, pedicels, buds, leaves, stems, and roots of ‘Bailmer’ [25]. The database contained 72,848 high-quality isoforms, of which 67,941 were annotated using the NCBI Non-Redundant (Nr) database (http://www.ncbi.nlm.nih.gov, accessed on 2 December 2020), Swiss-Prot protein database (http://www.expasy.ch/sprot, accessed on 4 December 2020), Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg, accessed on 7 December 2020), and COG/KOG database (http://www.ncbi.nlm.nih.gov/COG, accessed on 9 December 2020), with an E-value threshold of 1e-5. These PacBio Iso-Seq data served as the reference transcriptome for H. macrophylla. cDNA libraries from 36 samples, including three biological replicates, were constructed from sepals at the S1, S2, and S3 stages of CK and Tr groups in ‘Bailmer’ and ‘Duro’ (Figure 1). These 36 cDNA libraries were sequenced using the Illumina HiSeq™ 4000 [16]. Between 89.45% and 92.36% of the high-quality clean reads from ‘Bailmer’, and 83.06% to 85.97% from ‘Duro’, were mapped to the isoforms from the reference transcriptome. Gene abundances were calculated and normalized to reads per kilobase per million mapped reads (RPKM). The mapped isoforms were annotated using the NR, Swiss-Prot, KEGG, and COG/KOG databases, as previously described.
2.4. Analysis of Differentially Expressed Genes (DEIs)
Differentially expressed genes (DEIs) between the CK and Tr groups were identified. The threshold values for screening DEIs were a false discovery rate (FDR) ≤0.05 and |log2 (foldchange)| ≥ 1. DEIs were annotated using the NR, KEGG, COG, and GO databases, as described for isoform annotation.
2.5. Quantitative Real-Time Fluorescent PCR (qRT-PCR) Analysis
The RNA isolation kit is a quick RNA isolation kit (Hua Yue Yang, Beijing, China). First-strand cDNA was synthesized using HiScript III RT SuperMix for qPCR (+gDNA wiper) according to the manufacturer’s instructions. Primer pairs for qRT-PCR were designed using Primer Premier v5.0 software (Premier Biosoft Int., Palo Alto, CA, USA) (Table S1). The specificity and efficiency of the primers were confirmed using 1.5% agarose gel electrophoresis. All reactions were conducted in 96-well plates using the CFX96 Real-Time System (Bio-Rad, Hercules, CA, USA). qRT-PCR analyses were conducted using Taq Pro Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China). Each PCR reaction (20 μL) contained 5 μL of cDNA (100–200 ng/μL), 0.4 μL of each forward and reverse primer (10 pmol/μL), 10 μL of 2 × Taq Pro Universal SYBR qPCR Master Mix, and 4.2 μL of ddH2O. Amplification reactions were performed under the following conditions: 95 °C for 30 s, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s. A melting curve was generated for each PCR to examine the amplification specificity of primers and the presence of reaction contaminants. The melting curve was gained by heating the amplification products under the following conditions: 65–95 °C with a temperature increment of 0.5 °C every 5 s. The HmActin gene was used as the internal control for normalization. The primer sequences were HmActin-F (5′-GCCTGCCATGTATGTTGCCATC-3′) and HmActin-R (5′-CGGAATCCAGCACAATACCAGTTG-3′) [26]. The relative expression levels of the target genes were calculated using the 2−ΔΔCT method [27] relative to the internal control.
2.6. Protein Interaction Analysis
Protein interactions were predicted using the online software STRING (functional protein association networks; https://string-db.org/, accessed on 24 March 2024).
2.7. Data Analysis
Data are expressed as means ± standard deviations (SDs) of three replicates. Data analysis was conducted using Microsoft Office Excel 2010 and IBM SPSS Statistics 24.0. Bar graphs were generated using Origin 9.0.
3. Results
3.1. Al Contents in the Sepals
The Al content in sepals from the Tr groups was higher than that in sepals from the CK groups at all three stages (S1, S2, and S3) in both ‘Bailmer’ and ‘Duro’. Under Al treatment, sepals accumulated a substantial amount of Al, particularly at S1. The Al content in sepals was 2260.67 μg/g dry weight (DW) for ‘Bailmer’ and 1204.67 μg/g DW for ‘Duro’, which were approximately 2.90 and 5.47 times higher than that of the CK group (779.67 and 220.33 μg/g DW), respectively (Figure 2).
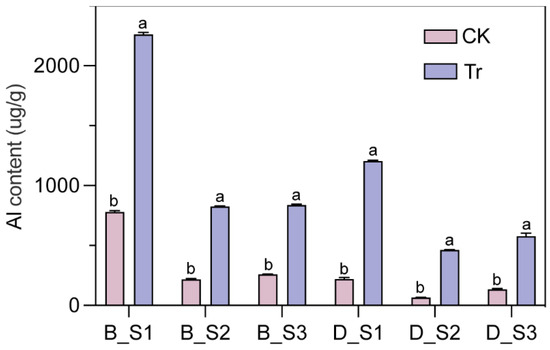
Figure 2.
Al content in sepals at three different developmental stages (S1, S2, S3) in ‘Bailmer’ and ‘Duro’. B and D represent ‘Bailmer’ and ‘Duro’, respectively. Different lowercase letters indicate significant differences between two treatments at p < 0.05 according to Duncan’s multiple range test.
3.2. Analysis of Illumina Transcriptome Data
In our previous study, 72,848 high-quality, full-length isoforms were obtained through full-length transcriptome sequencing. Of these, 71,617 isoforms were expressed in the Illumina-sequenced samples, and a total of 67,932 isoforms were annotated in at least one database. Of these, 44,208 (65.08%), 66,896 (98.47%), 32,980 (48.54%), and 55,845 (82.21%) isoforms were annotated according to the KOG, NR, KEGG, and Swiss-Prot databases, respectively. Across the four databases, 27,077 isoforms were annotated in all of them (Figure 3).
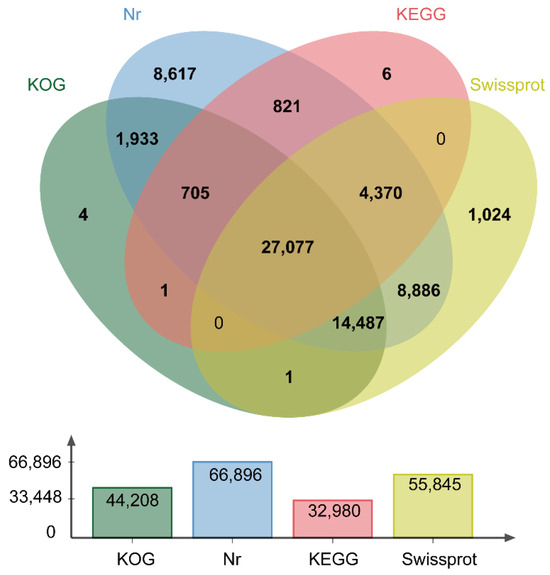
Figure 3.
Venn diagram of NR, KOG, KEGG, and Swiss-Prot function annotations.
3.3. Identification of Key Candidate Genes Mediating Aluminum Accumulation in Sepals
Differentially expressed isoforms (DEIs) between the CK group and the Tr group were identified at each of the three stages (S1–S3) in every cultivar (Figure 4). Considering all three developmental stages (S1–S3) of sepals, a total of 6575 DEIs were obtained in ‘Bailmer’, of which 2972 isoforms were upregulated and 3603 were downregulated. In total, 788 DEIs were identified in ‘Duro’, of which 426 were upregulated and 362 were downregulated. Among these DEIs, 84 upregulated isoforms and 70 downregulated isoforms were identified in both cultivars (Figure 5 and Figure S1).
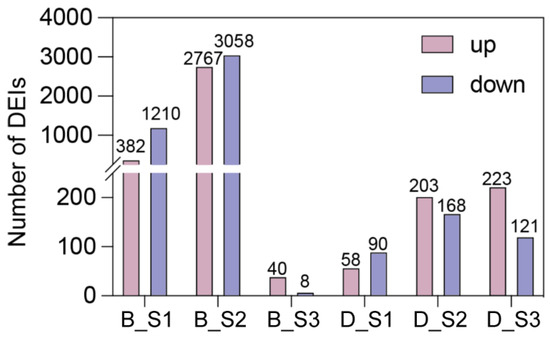
Figure 4.
The number of differentially expressed isoforms in the comparison of treatment versus control and three different stages (S1, S2, and S3). B and D represent ‘Bailmer’ and ‘Duro’, respectively.
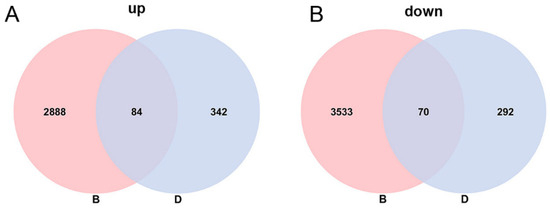
Figure 5.
Venn diagram of DEIs between two cultivars. (A) Upregulated expression genes under Al treatment in ‘Bailmer’ and ‘Duro’. (B) Downregulated expression genes under Al treatment in ‘Bailmer’ and ‘Duro’. B and D represent ‘Bailmer’ and ‘Duro’, respectively.
According to GO annotation, the 154 DEIs (84 upregulated isoforms and 70 downregulated isoforms) were classified into three categories: biological process, cellular component, and molecular function (Figure 6). Among these, 55 upregulated isoforms and 39 downregulated isoforms were significantly enriched (p ≤ 0.05). Isoforms with over 99% similarity in the CDS region were identified as the same gene, resulting in 30 upregulated genes and 13 downregulated genes (Table S2). Of these, seven upregulated and two downregulated genes were related to stress resistance, thirteen upregulated and six downregulated genes were involved in metabolism, six upregulated and two downregulated genes were involved in both metabolism and ion transport, and the remaining (four upregulated and three downregulated genes) were transporter genes.
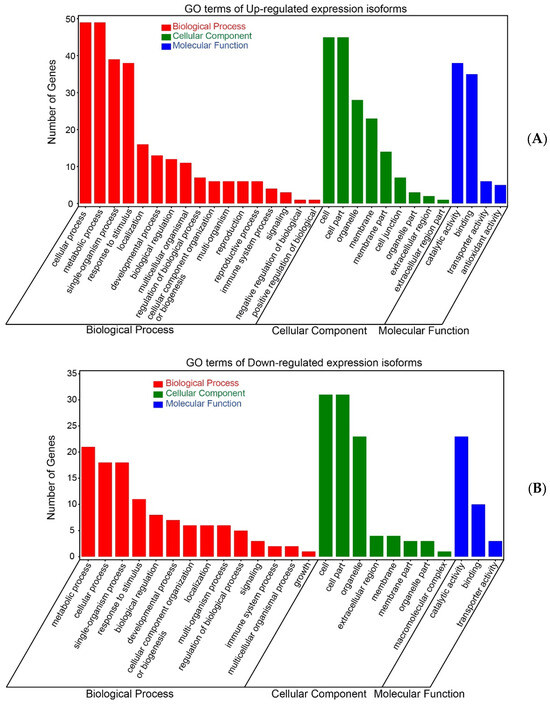
Figure 6.
GO enrichment analysis of DEIs. (A) GO terms of upregulated expression isoforms. (B) GO terms of downregulated expression isoforms.
3.4. Expression Verification of Key Candidate Genes Using qRT-PCR
The relative expression of 43 differentially expressed genes (30 upregulated and 13 downregulated) was verified using qRT-PCR. Seven genes, including HmDDS (Dammarenediol II synthase-like), HmDHK (1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 1), HmALS3.1 (protein Aluminum sensitive 3), HmABAH4 (Abscisic acid 8′-hydroxylase), HmMSMO1-1 (Methylsterol monooxygenase 1-1), HmPAP8 (Purple acid phosphatase 8-like), and HmSULTR3;1 (Sulfate transporter 3.1-like), were upregulated across all three stages of sepals in both cultivars under Al treatment (Figure 7). Additionally, the specificity and efficiency of PCR amplification were confirmed. PCR amplifications for each primer pair showed a single and desired amplicon of the expected size on a 1.5% agarose gel (Figure 8A), and a single peak in melt curves (Figure 8B). This pattern was consistent with the Al content in the sepals and the gene expression profile based on transcriptome data for the two cultivars (Figure 2 and Figure 5). These seven genes were identified as being related to Al accumulation in sepals.
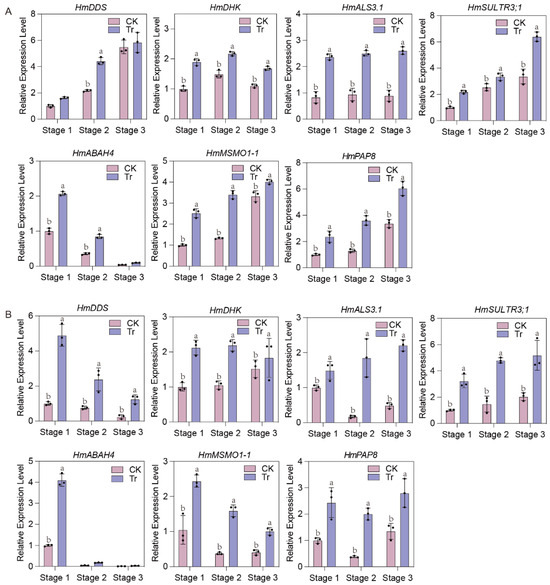
Figure 7.
Expression of seven genes between two treatments in ‘Bailmer’ (A) and ‘Duro’ (B). Different lowercase letters indicate significant differences between two treatments at p ≤ 0.05 according to Duncan’s multiple range test.
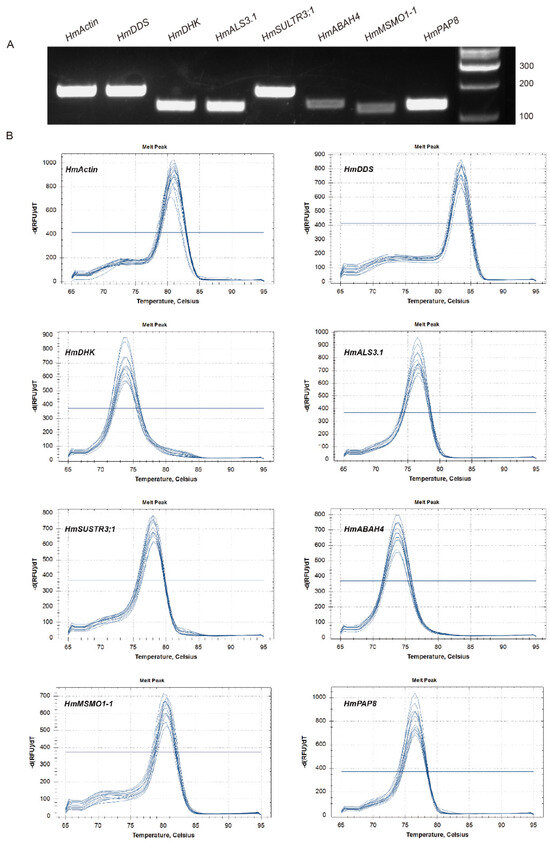
Figure 8.
Specificity of qRT-PCR amplification for HmAction and seven candidate genes. (A) Amplification of a specific PCR product for each gene through agarose gel. (B) Melting curves for each gene.
According to gene functions (Table S2), HmDDS and HmMSMO1-1 are associated with organic substance biosynthesis; HmDHK, HmABAH4, and HmPAP8 are primarily involved in macromolecule metabolism and transition metal ion binding; and HmALS3.1 and HmSULTR3;1 are transporters responsible for metal ion and sulfate transport, respectively.
3.5. Protein Interaction Analysis
Among the seven genes, only the HmALS3.1 gene was found to have an interaction network with several proteins (Figure 9). HmALS3.1, as the central protein, was closely linked to the ATP-binding cassette (ABC) transporter protein family. The only experimentally verified interaction with the HmALS3.1 protein is ABCI17, which belongs to the ABC transporter superfamily. Among the predicted interactions, ABCB27, ABCI17, and LPR2 are identified as neighborhood proteins to HmALS3.1. Through automated and unsupervised text mining, nine proteins (ABCB27, ABCI1, ABCI17, ALMT1, Q9LJC4_ARATH, DTX42, MRS2-10, STOP1, and STOP2) were frequently mentioned and were the first shell of interactors with HmALS3.1.
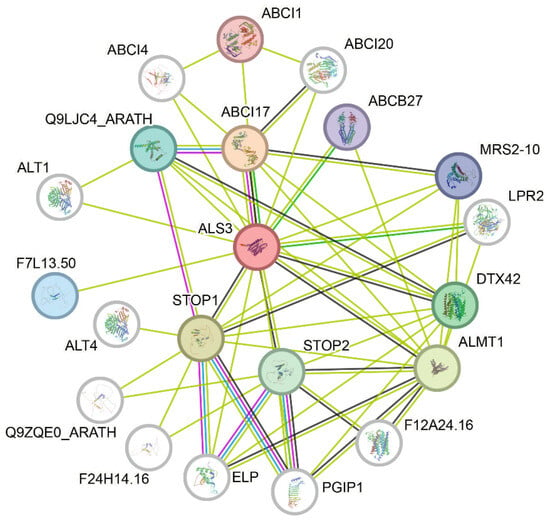
Figure 9.
Protein interaction network diagram of HmALS3.1. The blue line indicates interactions known from curated databases; the pink line indicates experimentally determined interactions; the green line represents text mining-based interactions; and the black line denotes co-expression interactions.
4. Discussion
H. macrophylla is famous for its sepal color changes under different cultivation conditions. The sepal color is pink-red when the plant is grown in neutral to alkaline soils and blue when grown in acidic soils. The main reason for sepals bluing is that Al3+ is absorbed by roots in acidic soil and transported to sepals to participate in the formation of blue complex [28,29]. In fact, not every cultivar with a pink-red sepal color has blue sepals in acidic soils with Al3+ because the bluing capability of the sepal color was positively correlated with the proportion of delphinidin-3-glucoside among all the anthocyanins in sepals [16,30]. In this experiment, two cultivars with different sepal color bluing capabilities were utilized. The sepal color of ‘Bailmer’ varied significantly from pink to blue under Al treatment, whereas ‘Duro’ exhibited slightly and did not turn blue from pink [16]. However, both cultivars had higher Al content in sepals under Al treatment. Consequently, we analyzed the transcriptome data from the two cultivars to elucidate how H. macrophylla responded to Al.
To identify genes associated with Al accumulation, transcriptome data from two cultivars of H. macrophylla were used to screen 43 differentially expressed genes. Additionally, seven genes associated with Al accumulation were identified using the qRT-PCR technique: HmDDS, HmDHK, HmALS3.1, HmABAH4, HmMSMO1-1, HmPAP8, and HmSULTR3;1 (Figure 7). In the research, we failed to obtain hydroponically cultivated flowering plants, so all the plants were grown in the mixed substrate containing peat and perlite (7:3, by volume). The lower Al3+ level in peat might affect gene expression. The seven genes screened exhibited significant differences in the expression level at all three developmental stages between the CK and Tr groups in both cultivars.
Except for HmALS3.1, the other six genes have not been reported in the literature as being associated with Al stress. In the experiments, Al3+ in the form of Al2(SO4)3 was applied to the substrate; thus, the addition of sulfate ions may have significantly increased the expression of HmSULTR3;1 [31].
Aluminum sensitive 3 (ALS3) encodes a half-size ABC protein, which belongs to the large family of ATP-binding cassette (ABC) proteins [32]. Studies have shown that the ALS3 gene functions in Al resistance across various plant species. In Arabidopsis thaliana, the expression of AtALS3 was regulated by Al signal in roots and shoots [32,33]. Homologs of ALS3 genes required for Al resistance have also been identified in Rhododendron yunnanense [34], soybean [35], and buckwheat [36]. The potential role of this protein is to transfer Al3+ from sensitive root tissues to less sensitive shoots, thereby reducing Al toxicity [32,37]. Plant roots can also tolerate Al3+ internally by sequestering it into vacuoles [38,39].
Simulations using the Arabidopsis database (Figure 9) revealed that some proteins interacting with HmALS3.1 are also involved in plant Al resistance. Among them, two organic acid transporters were mentioned, ALMT1 and DTX42. Aluminum-activated malate transporter 1 (ALMT1) was identified as an Al-activated malate efflux transporter, secreting malate to chelate Al3+ in the rhizosphere, thus preventing Al from sensitive root cells [40,41,42]. The expression of ALMT1 plays an important role in improving Al resistance in wheat [43], Arabidopsis [44], and apple [45]. Although DTX42 belonging to the MATE efflux family has not yet been verified as related to Al response, such as DTX30 from the same family, it indirectly modulates citrate exudation to promote Al tolerance by modulating auxin levels in root [46]. The HmALS3.1 gene was also closely associated with the other ATP-binding cassette (ABC) transporter proteins. The ABC transporter family is divided into eight subfamilies based on the evolutionary relationships of conserved regions, with ABCB transporter proteins being the second-largest subfamily [47]. ABCB transporter proteins mediate the transport of heavy metal ions, including cadmium, lead, and Al, and enhance plant tolerance to heavy metals [32,48]. Among them, ABCB27 (ALS1) may play a role in Al intracellular movement [32,49]. Additionally, some ABCI transporter proteins also respond to Al stress. In soybeans, GsABCI1 transgenic plants exhibited resistance to Al treatment through ion translocation or alterations in root components [50]. The ABCI17 (sensitive to aluminum rhizotoxicity 1 (STAR1)) can interact with STAR2 to form a complex as a bacterial-type ABC transporter, which transports UDP-Glc and is required for Al tolerance in rice [39]. HmABCI17 from H. macrophylla has been shown to enhance Al tolerance in yeast [51]. Q9LJC4_ARATH also belongs to the ABC transporter family, but it has not been verified as being related to the Al response yet. Research has shown that the transcription factor (TF) sensitive to proton rhizotoxicity1 (STOP1) regulates ALS3, AtALMT1, and AtMATE expression under Al stress [52,53]. Additionally, the STOP1-independent NO signaling pathway and STOP1-dependent regulation in the phosphoinositide (PI) signaling pathway are involved in the regulation of PGIP1 expression under Al stress [54]. To some extent, STOP2 can substitute for STOP1 in activating the transcription of genes regulated by STOP1, including ALS3 and AtMATE [55,56]. In Arabidopsis, ALS3 and STAR1 form the ATP-binding cassette (ABC) transporter complex located in the vacuolar membrane. ALS3/STAR1 promotes the degradation of the Al resistance transcription factor STOP1 to regulate ALMT1 expression [57]. Two metal transporters are also mentioned in the protein interaction network, magnesium transporter 1 (MRS2-10) and low phosphate root 2 (LPR2). MRS2-10 has shown the activity to transport Al in Arabidopsis [58]. These proteins mentioned by the protein network may become the key points in the future study on Al accumulation in H. macrophylla.
Additionally, other Al resistance genes have been identified in various plants, including those from the MATE family first described in sorghum [59]. Subsequently, VrMATE19 and VrMATE30 were found to play a potential role in Al tolerance in mung beans [60]. In soybeans, the GmMATE13 and GmMATE75 genes enhanced Al tolerance in the plant through citrate secretion [61]. Moreover, the nbatural resistance-associated macrophage protein 1 (Nrat1) gene may be a useful tool for enhancing Al tolerance in rice and Arabidopsis [62]. A knockout mutation in the Nrat1 gene resulted in decreased Al uptake and increased binding of Al3+ to the cell wall [63]. In H. macrophylla, two aquaporin genes, HmVALT and HmPALT1, were found to be associated with Al transport.
In our study, HmALS3.1 was identified as being closely related to Al accumulation in sepals of H. macrophylla, while the functions of the remaining six genes require further investigation. At present, the specific functions and regulatory mechanisms of HmALS3.1 participating in Al accumulation are being studied in our laboratory.
5. Conclusions
This study first reports the identification of HmALS3.1 as a key gene related to Al accumulation in the sepals of H. macrophylla. By analyzing the transcriptome data from the CK group and the Al treatment group of the two cultivars of H. macrophylla, 43 differentially expressed genes (30 upregulated and 13 downregulated under the Al treatment) in sepals were identified. Further research indicated that seven genes upregulated across all three stages under Al treatment in both cultivars, including HmDDS, HmDHK, HmALS3.1, HmABAH4, HmMSMO1-1, HmPAP8, and HmSUSTR3;1. Among the seven, the gene HmALS3.1 was identified as a potential key player in Al transport within the sepals of H. macrophylla. This study lays the groundwork for further exploration into the mechanisms by which HmALS3.1 regulates Al accumulation in H. macrophylla.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/horticulturae10111180/s1: Figure S1: Heat maps of expression of 84 upregulated and 70 downregulated isoforms based on transcriptome data; Table S1: Primers used for qRT-PCR in this study; and Table S2: Annotation of 96 DEIs related to aluminum accumulation in sepals of hydrangea.
Author Contributions
Conceptualization, S.Y.; methodology, S.Y.; validation, Y.F.; formal analysis, Y.L.; investigation, Y.L.; resources, Y.L. and Y.W.; data curation, S.L.; writing—original draft preparation, S.L.; writing—review and editing, S.Y.; visualization, S.L.; supervision, C.L.; project administration, S.Y.; funding acquisition, C.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Central Public-interest Scientific Institution Basal Research Fund (grant number: IVF-BRF2022017 and IVF-BRF2023009), the Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (grant number: CAAS-ASTIP-2022-IVFCAAS and CAAS-ASTIP-2023-IVFCAAS), the National Flower Improvement Center, and the Key Laboratory of Biology and Genetic Improvement of Flower Crops (North China), Ministry of Agriculture and Rural Affairs, China.
Data Availability Statement
The RNA-Seq data generated in this study are available at the SRA Archive (https://submit.ncbi.nlm.nih.gov/subs/sra/, accessed on 10 May 2020), with accession number PRJNA1125644.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ma, J.F. Role of organic acids in detoxification of aluminum in higher plants. Plant Cell Physiol. 2000, 41, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Mortvedt, J.J.; Asher, C.J. Beneficial Elements, Functional Nutrients, and Possible New Essential Elements. Micronutr. Agric. 1991, 4, 703–723. [Google Scholar]
- Famoso, A.N.; Clark, R.T.; Shaff, J.E.; Craft, E.; McCouch, S.R.; Kochian, L.V. Development of a Novel Aluminum Tolerance Phenotyping Platform Used for Comparisons of Cereal Aluminum Tolerance and Investigations into Rice Aluminum Tolerance Mechanisms. Plant Physiol. 2010, 153, 1678–1691. [Google Scholar] [CrossRef]
- Xu, Q.S.; Wang, Y.; Ding, Z.T.; Song, L.B.; Li, Y.S.; Ma, D.X.; Wang, Y.; Shen, J.Z.; Jia, S.S.; Sun, H.W.; et al. Aluminum induced metabolic responses in two tea cultivars. Plant Physiol. Biochem. 2016, 101, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Bojórquez-Quintal, E.; Escalante-Magaña, C.; Echevarría-Machado, I.; Martínez-Estévez, M. Aluminum, a Friend or Foe of Higher Plants in Acid Soils. Front. Plant Sci. 2017, 8, 1767. [Google Scholar] [CrossRef]
- Muhammad, N.; Zvobgo, G.; Zhang, G.P. A review: The beneficial effects and possible mechanisms of aluminum on plant growth in acidic soil. J. Integr. Agric. 2019, 18, 1518–1528. [Google Scholar] [CrossRef]
- Chauhan, D.K.; Yadav, V.; Vaculik, M.; Gassmann, W.; Pike, S.; Arif, N.; Singh, V.P.; Deshmukh, R.; Sahi, S.; Tripathi, D.K. Aluminum toxicity and aluminum stress-induced physiological tolerance responses in higher plants. Crit. Rev. Biotechnol. 2021, 41, 715–730. [Google Scholar] [CrossRef]
- Schmitt, M.; Watanabe, T.; Jansen, S. The effects of aluminium on plant growth in a temperate and deciduous aluminium accumulating species. AoB Plants 2016, 8, plw065. [Google Scholar] [CrossRef] [PubMed]
- Jansen, S.; Broadley, M.R.; Robbrecht, E.; Smets, E. Aluminum hyperaccumulation in angiosperms: A review of its phylogenetic significance. Bot. Rev. 2002, 68, 235–269. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, M.; Liu, X.; Mao, Q.; Shi, C.; Kochian, L.V.; Liao, H. Aluminium is essential for root growth and development of tea plants (Camellia sinensis). J. Integr. Plant Biol. 2020, 62, 984–997. [Google Scholar] [CrossRef]
- Ma, J.F.; Hiradate, S.; Nomoto, K.; Iwashita, T.; Matsumoto, H. Internal detoxification mechanism of Al in hydrangea—Identification of Al form in the leaves. Plant Physiol. 1997, 113, 1033–1039. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, X.; Xu, L.F. Identification and Bioinformatics Analysis of ABC Transporter Gene Family in Hydrangea under Aluminum Stress. Mol. Plant Breed. 2022, 13, 1–15. [Google Scholar] [CrossRef]
- Ito, D.; Shinkai, Y.; Kato, Y.; Kondo, T.; Yoshida, K. Chemical Studies on Different Color Development in Blue- and Red-Colored Sepal Cells of. Biosci. Biotechnol. Biochem. 2009, 73, 1054–1059. [Google Scholar] [CrossRef] [PubMed]
- Osaki, M.; Watanabe, T.; Tadano, T. Beneficial effect of aluminum on growth of plants adapted to low pH soils. Soil Sci. Plant Nutr. 1997, 43, 551–563. [Google Scholar] [CrossRef]
- Peng, J.; Dong, X.; Xue, C.; Liu, Z.; Cao, F. Exploring the Molecular Mechanism of Blue Flower Color Formation in Hydrangea macrophylla cv. “Forever Summer”. Front. Plant Sci. 2021, 12, 585665. [Google Scholar] [CrossRef]
- Yuan, S.X.; Qi, H.; Yang, S.N.; Chu, Z.Y.; Zhang, G.T.; Liu, C. Role of delphinidin-3-glucoside in the sepal blue color change among Hydrangea macrophylla cultivars. Sci. Hortic. 2023, 313, 111902. [Google Scholar] [CrossRef]
- Liang, C.Y.; Rengasamy, K.P.; Huang, L.M.; Hsu, C.C.; Jeng, M.F.; Chen, W.H.; Chen, H.H. Assessment of violet-blue color formation in Phalaenopsis orchids. BMC Plant Biol. 2020, 20, 212. [Google Scholar] [CrossRef]
- Naumann, A.; Horst, W.J. Effect of aluminium supply on aluminium uptake, translocation and blueing of Hydrangea macrophylla (Thunb.) Ser. cultivars in a peatclay substrate. J. Hortic. Sci. Biotechnol. 2003, 78, 463–469. [Google Scholar] [CrossRef]
- Hotta, H.; Wang, Q.; Fukuda, M.; Aizawa, S.; Umemura, T.; Sekizawa, K.; Tsunoda, K.I. Identification of aluminum species in an aluminum-accumulating plant, hydrangea (Hydrangea macrophylla), by electrospray ionization mass spectrometry. Anal. Sci. 2008, 24, 795–798. [Google Scholar] [CrossRef][Green Version]
- Dong, B.; Meng, D.; Song, Z.; Cao, H.; Du, T.; Qi, M.; Wang, S.; Xue, J.; Yang, Q.; Fu, Y. CcNFYB3-CcMATE35 and LncRNA CcLTCS-CcCS modules jointly regulate the efflux and synthesis of citrate to enhance aluminium tolerance in pigeon pea. Plant Biotechnol. J. 2024, 22, 181–199. [Google Scholar] [CrossRef]
- Wilkens, S. Structure and mechanism of ABC transporters. F1000Prime Rep. 2015, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Aryal, B.; Laurent, C.; Geisler, M. Learning from each other: ABC transporter regulation by protein phosphorylation in plant and mammalian systems. Biochem. Soc. Trans. 2015, 43, 966–974, Erratum in Biochem. Soc. Trans. 2016, 44, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Negishi, T.; Oshima, K.; Hattori, M.; Kanai, M.; Mano, S.; Nishimura, M.; Yoshida, K. Tonoplast- and plasma membrane-localized aquaporin-family transporters in blue hydrangea sepals of aluminum hyperaccumulating plant. PLoS ONE 2012, 7, e43189. [Google Scholar] [CrossRef] [PubMed]
- GB 5009.268–2016; National Food Safety Standard. Determination of Multi-elements in Food. General Administration of Quality Supervision, Inspection and Quarantine of the People’s Republic of China: Beijing, China, 2016. (In Chinese)
- Qi, H.; Zhang, G.T.; Chu, Z.Y.; Liu, C.; Yuan, S.X. Identification of Seven Key Structural Genes in the Anthocyanin Biosynthesis Pathway in Sepals of Hydrangea macrophylla. Curr. Issues Mol. Biol. 2022, 44, 4167–4180. [Google Scholar] [CrossRef]
- Zhang, G.T.; Yuan, S.X.; Qi, H.; Chu, Z.Y.; Liu, C. Identification of Reliable Reference Genes for the Expression of Hydrangea macrophylla ‘Bailmer’ and ‘Duro’ Sepal Color. Horticulturae 2022, 8, 835. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Rahmati, R.; Hamid, R.; Ghorbanzadeh, Z.; Jacob, F.; Azadi, P.; Zeinalabedini, M.; Karimi Farsad, L.; Kazemi, M.; Ebrahimi, M.A.; Shahinnia, F.; et al. Comparative Transcriptome Analysis Unveils the Molecular Mechanism Underlying Sepal Colour Changes under Acidic pH Substratum in Hydrangea macrophylla. Int. J. Mol. Sci. 2022, 23, 15428. [Google Scholar] [CrossRef]
- Ito, T.; Oyama, K.I.; Yoshida, K. Direct Observation of Hydrangea Blue-Complex Composed of 3-O-Glucosyldelphinidin, Al3+ and 5-O-Acylquinic Acid by ESI-Mass Spectrometry. Molecules 2018, 23, 1424. [Google Scholar] [CrossRef]
- Yoshida, K.; Ito, D.; Miki, N.; Kondo, T. Single-cell analysis clarifies mosaic color development in purple hydrangea sepal. New Phytol. 2021, 229, 3549–3557. [Google Scholar] [CrossRef]
- Zhang, H.J.; Hao, X.Y.; Zhang, J.J.; Wang, L.; Wang, Y.C.; Li, N.N.; Guo, L.N.; Ren, H.Z.; Zeng, J.M. Genome-wide identification of SULTR genes in tea plant and analysis of their expression in response to sulfur and selenium. Protoplasma 2022, 259, 127–140. [Google Scholar] [CrossRef]
- Larsen, P.B.; Cancel, J.; Rounds, M.; Ochoa, V. Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 2007, 225, 1447–1458. [Google Scholar] [CrossRef] [PubMed]
- Sadhukhan, A.; Agrahari, R.K.; Wu, L.J.; Watanabe, T.; Nakano, Y.; Panda, S.K.; Koyama, H.; Kobayashi, Y. Expression genome-wide association study identifies that phosphatidylinositol-derived signalling regulates ALUMINIUM SENSITIVE3 expression under aluminium stress in the shoots of Arabidopsis thaliana. Plant Sci. 2021, 302, 110711. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.X.; Lei, Y.S.; Huang, S.X.; Zhang, J.; Wan, Z.Y.; Zhu, X.T.; Jin, S.H. Combined de novo transcriptomic and physiological analyses reveal RyALS3-mediated aluminum tolerance in Rhododendron yunnanense Franch. Front. Plant Sci. 2022, 13, 951003. [Google Scholar] [CrossRef]
- Agrahari, R.K.; Kobayashi, Y.; Borgohain, P.; Panda, S.K.; Koyama, H. Aluminum-Specific Upregulation of GmALS3 in the Shoots of Soybeans: A Potential Biomarker for Managing Soybean Production in Acidic Soil Regions. Agronomy 2020, 10, 1228. [Google Scholar] [CrossRef]
- Reyna-Llorens, I.; Corrales, I.; Poschenrieder, C.; Barcelo, J.; Cruz-Ortega, R. Both aluminum and ABA induce the expression of an ABC-like transporter gene (FeALS3) in the Al-tolerant species. Environ. Exp. Bot. 2015, 111, 74–82. [Google Scholar] [CrossRef]
- Larsen, P.B.; Tai, C.Y.; Kochian, L.V.; Howell, S.H. Arabidopsis mutants with increased sensitivity to aluminum. Plant Physiol. 1996, 110, 743–751. [Google Scholar] [CrossRef]
- Huang, C.F.; Yamaji, N.; Ma, J.F. Knockout of a Bacterial-Type ATP-Binding Cassette Transporter Gene, AtSTAR1, Results in Increased Aluminum Sensitivity in Arabidopsis. Plant Physiol. 2010, 153, 1669–1677. [Google Scholar] [CrossRef]
- Huang, C.F.; Yamaji, N.; Mitani, N.; Yano, M.; Nagamura, Y.; Ma, J.F. A Bacterial-Type ABC Transporter Is Involved in Aluminum Tolerance in Rice. Plant Cell 2009, 21, 655–667. [Google Scholar] [CrossRef]
- Kochian, L.V.; Piñeros, M.A.; Liu, J.P.; Magalhaes, J.V. Plant Adaptation to Acid Soils: The Molecular Basis for Crop Aluminum Resistance. Annu. Rev. Plant Biol. 2015, 66, 571–598. [Google Scholar] [CrossRef]
- Ma, J.F.; Chen, Z.C.; Shen, R.F. Molecular mechanisms of Al tolerance in gramineous plants. Plant Soil 2014, 381, 1–12. [Google Scholar] [CrossRef]
- Peng, F.C.; Yuan, M.; Zhou, L.; Zheng, B.Q.; Wang, Y. Identification and Analysis of Aluminum-Activated Malate Transporter Gene Family Reveals Functional Diversification in Orchidaceae and the Expression Patterns of Dendrobium catenatum Aluminum-Activated Malate Transporters. Int. J. Mol. Sci. 2024, 25, 9662. [Google Scholar] [CrossRef] [PubMed]
- Ligaba, A.; Dreyer, I.; Margaryan, A.; Schneider, D.J.; Kochian, L.; Piñeros, M. Functional, structural and phylogenetic analysis of domains underlying the Al sensitivity of the aluminum-activated malate/anion transporter, TaALMT1. Plant J. 2013, 76, 766–780. [Google Scholar] [CrossRef]
- Wang, J.Q.; Yu, X.F.; Ding, Z.J.; Zhang, X.K.; Luo, Y.P.; Xu, X.M.; Xie, Y.; Li, X.X.; Yuan, T.; Zheng, S.J.; et al. Structural basis of ALMT1-mediated aluminum resistance in Arabidopsis. Cell Res. 2022, 32, 89–98. [Google Scholar] [CrossRef]
- Lu, J.; Liu, X.; Ma, Q.J.; Kang, H.; Liu, Y.J.; Hao, Y.J.; You, C.X. Molecular cloning and functional characterization of the Aluminum-activated malate transporter gene. Sci. Hortic. 2019, 244, 208–217. [Google Scholar] [CrossRef]
- Upadhyay, N.; Kar, D.; Datta, S. A multidrug and toxic compound extrusion (MATE) transporter modulates auxin levels in root to regulate root development and promotes aluminium tolerance. Plant Cell Environ. 2020, 43, 745–759. [Google Scholar] [CrossRef]
- Lee, M.; Choi, Y.; Burla, B.; Kim, Y.Y.; Jeon, B.; Maeshima, M.; Yoo, J.Y.; Martinoia, E.; Lee, Y. The ABC transporter AtABCB14 is a malate importer and modulates stomatal response to CO2. Nat. Cell Biol. 2008, 10, 1217–1223. [Google Scholar] [CrossRef]
- Devi, R.; Goyal, P.; Verma, B.; Hussain, S.; Chowdhary, F.; Arora, P.; Gupta, S. A transcriptome-wide identification of ATP-binding cassette (ABC) transporters revealed participation of ABCB subfamily in abiotic stress management of Glycyrrhiza glabra L. BMC Genomics 2024, 25, 315. [Google Scholar] [CrossRef]
- Fang, C.; Wu, J.; Liang, W. Systematic Investigation of Aluminum Stress-Related Genes and Their Critical Roles in Plants. Int. J. Mol. Sci. 2024, 25, 9045. [Google Scholar] [CrossRef]
- Wen, K.; Pan, H.T.; Li, X.A.; Huang, R.; Ma, Q.B.; Nian, H. Identification of an ATP-Binding Cassette Transporter Implicated in Aluminum Tolerance in Wild Soybean (Glycine soja). Int. J. Mol. Sci. 2021, 22, 13264. [Google Scholar] [CrossRef]
- Chen, S.S.; Qi, X.Y.; Feng, J.; Chen, H.J.; Qin, Z.Y.; Wang, H.D.; Deng, Y.M. Biochemistry and transcriptome analyses reveal key genes and pathways involved in high-aluminum stress response and tolerance in hydrangea sepals. Plant Physiol. Biochem. 2022, 185, 268–278. [Google Scholar] [CrossRef]
- Sawaki, K.; Sawaki, Y.; Zhao, C.R.; Kobayashi, Y.; Koyama, H. Specific transcriptomic response in the shoots of Arabidopsis thaliana after exposure to Al rhizotoxicity: - Potential gene expression biomarkers for evaluating Al toxicity in soils. Plant Soil 2016, 409, 131–142. [Google Scholar] [CrossRef]
- Sawaki, Y.; Iuchi, S.; Kobayashi, Y.; Kobayashi, Y.; Ikka, T.; Sakurai, N.; Fujita, M.; Shinozaki, K.; Shibata, D.; Kobayashi, M.; et al. STOP1 Regulates Multiple Genes That Protect Arabidopsis from Proton and Aluminum Toxicities. Plant Physiol. 2009, 150, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Agrahari, R.K.; Enomoto, T.; Ito, H.; Nakano, Y.; Yanase, E.; Watanabe, T.; Sadhukhan, A.; Iuchi, S.; Kobayashi, M.; Panda, S.K.; et al. Expression GWAS of PGIP1 Identifies STOP1-Dependent and STOP1-Independent Regulation of PGIP1 in Aluminum Stress Signaling in Arabidopsis. Front. Plant Sci. 2021, 12, 774687. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Ohyama, Y.; Kobayashi, Y.; Ito, H.; Iuchi, S.; Fujita, M.; Zhao, C.R.; Tanveer, T.; Ganesan, M.; Kobayashi, M.; et al. STOP2 Activates Transcription of Several Genes for Al- and Low pH-Tolerance that Are Regulated by STOP1 in. Mol. Plant 2014, 7, 311–322. [Google Scholar] [CrossRef]
- Sadhukhan, A.; Kobayashi, Y.; Iuchi, S.; Koyama, H. Synergistic and antagonistic pleiotropy of STOP1 in stress tolerance. Trends Plant Sci. 2021, 26, 1014–1022. [Google Scholar] [CrossRef]
- Fan, N.; Li, X.B.; Xie, W.X.; Wei, X.; Fang, Q.; Xu, J.Y.; Huang, C.F. Modulation of external and internal aluminum resistance by ALS3-dependent STAR1-mediated promotion of STOP1 degradation. New Phytol. 2024, 244, 511–527. [Google Scholar] [CrossRef]
- Ishijima, S.; Manabe, Y.; Shinkawa, Y.; Hotta, A.; Tokumasu, A.; Ida, M.; Sagami, I. The homologous Arabidopsis MRS2/MGT/CorA-type Mg channels, AtMRS2-10 and AtMRS2-1 exhibit different aluminum transport activity. Biochim. Biophys. Acta BBA-Biomembr. 2018, 1860, 2184–2191. [Google Scholar] [CrossRef]
- Magalhaes, J.V.; Liu, J.; Guimaraes, C.T.; Lana, U.G.P.; Alves, V.M.C.; Wang, Y.H.; Schaffert, R.E.; Hoekenga, O.A.; Piñeros, M.A.; Shaff, J.E.; et al. A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat. Genet. 2007, 39, 1156–1161. [Google Scholar] [CrossRef]
- Singh, D.; Tripathi, A.; Mitra, R.; Bhati, J.; Rani, V.; Taunk, J.; Singh, D.; Yadav, R.K.; Siddiqui, M.H.; Pal, M. Genome-wide identification of MATE and ALMT genes and their expression profiling in mungbean (Vigna radiata L.) under aluminium stress. Ecotoxicol. Environ. Saf. 2024, 280, 116558. [Google Scholar] [CrossRef]
- Gao, P.X.; Han, R.R.; Xu, H.; Wei, Y.M.; Yu, Y.X. Identification of MATE Family and Characterization of GmMATE1 and GmMATE75 in Soybean’s Response to Stress. Int. J. Mol. Sci. 2024, 25, 3711. [Google Scholar] [CrossRef]
- Li, J.Y.; Liu, J.P.; Dong, D.K.; Jia, X.M.; McCouch, S.R.; Kochian, L.V. Natural variation underlies alterations in Nramp aluminum transporter (NRAT1) expression and function that play a key role in rice aluminum tolerance. Proc. Natl. Acad. Sci. USA 2014, 111, 6503–6508. [Google Scholar] [CrossRef]
- Ofoe, R.; Thomas, R.H.; Asiedu, S.K.; Wang-Pruski, G.; Fofana, B.; Abbey, L. Aluminum in plant: Benefits, toxicity and tolerance mechanisms. Front. Plant Sci. 2023, 13, 1085998. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).