Cucumber Auxin Response Factor CsARF10a Regulates Leaf Morphogenesis and Parthenocarpic Fruit Set in Tomato
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatment Methods
2.2. Bioinformatics Analysis of CsARF10a
2.3. RNA Extraction and RT-qPCR Analysis
2.4. Transgenic Tomato Construction
2.5. Phenotypical and Physiological Characterizations of Transgenic Tomato Plants
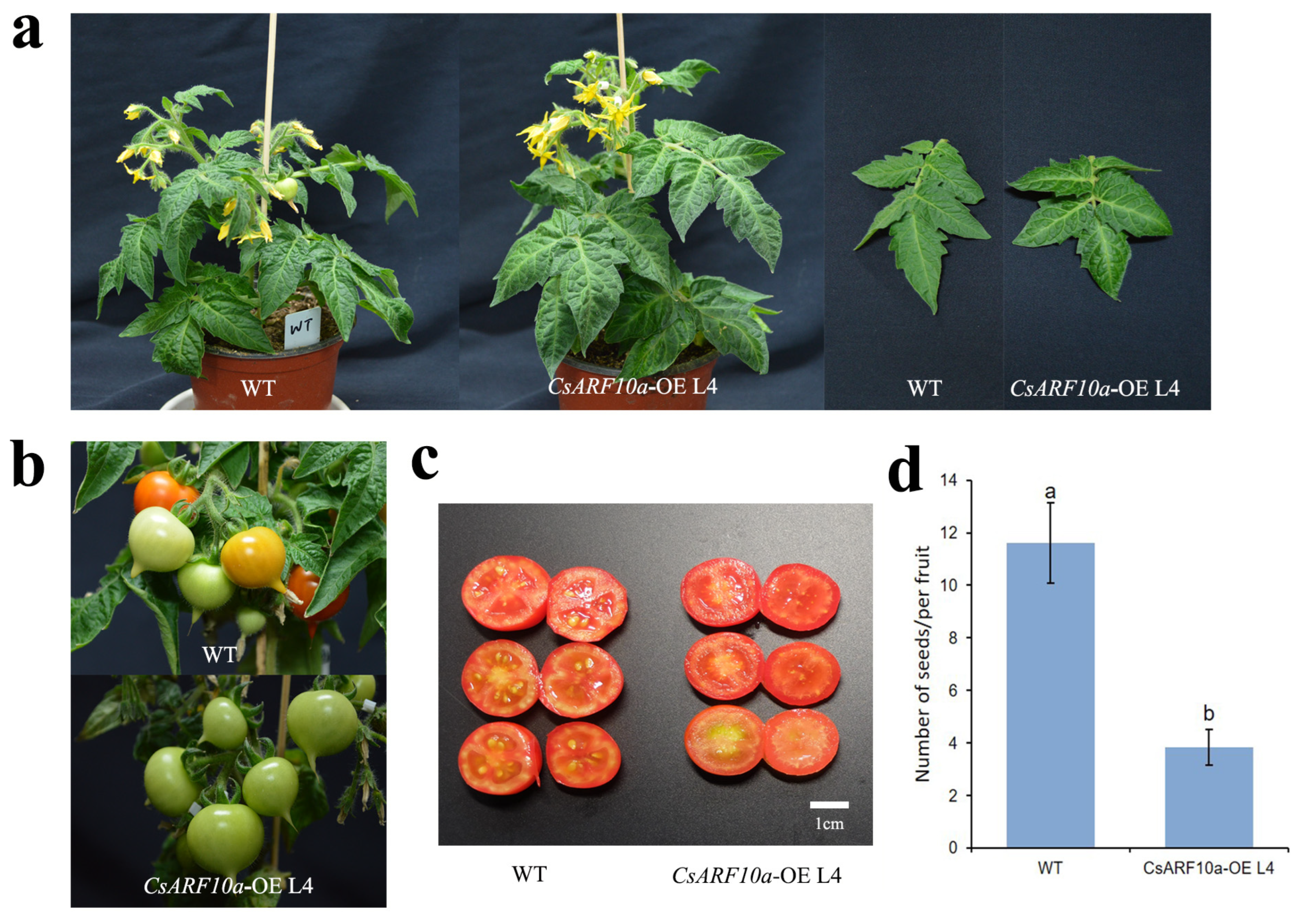
3. Results
3.1. CsARF10a Belongs to the Clade III Family of ARF
3.2. Expression Analysis of CsARF10a Gene in Cucumber
3.3. Functional Analysis of CsARF10a Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- McAtee, P.; Karim, S.; Schaffer, R.; David, K. A dynamic interplay between phytohormones is required for fruit development, maturation, and ripening. Front. Plant Sci. 2013, 4, 79. [Google Scholar] [CrossRef]
- Mapelli, S.; Frova, C.; Torti, G.; Soressi, G.P. Relationship between set, development and activities of growth-regulators in tomato fruits. Plant Cell Physiol. 1978, 19, 1281–1288. [Google Scholar]
- Friml, J. Auxin transport—Shaping the plant. Curr. Opin. Plant Biol. 2003, 6, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Sundberg, E.; Ostergaard, L. Distinct and dynamic auxin activities during reproductive development. Cold Spring Harb. Perspect. Biol. 2009, 1, a001628. [Google Scholar] [CrossRef] [PubMed]
- Pattison, R.J.; Csukasi, F.; Catalá, C. Mechanisms regulating auxin action during fruit development. Physiol. Plant. 2014, 151, 62–72. [Google Scholar] [CrossRef]
- Godoy, F.; Kühn, N.; Muñoz, M.; Marchandon, G.; Gouthu, S.; Deluc, L.; Delrot, S.; Lauvergeat, V.; Arce-Johnson, P. The role of auxin during early berry development in grapevine as revealed by transcript profiling from pollination to fruit set. Hortic. Res. 2021, 8, 140. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, J.; Xu, X.; Wang, R.; Liu, Y.; Huang, S.; Wei, H.; Wei, Z. Molecular mechanisms of diverse auxin responses during plant growth and development. Int. J. Mol. Sci. 2022, 23, 12495. [Google Scholar] [CrossRef]
- Hagen, G.; Guilfoyle, T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef]
- Szemenyei, H.; Hannon, M.; Long, J.A. TOPLESS mediates auxin-dependent transcriptional repression during embryogenesis. Science 2008, 319, 1384–1386. [Google Scholar] [CrossRef]
- Gray, W.M.; Kepinski, S.; Rouse, D.; Leyser, O.; Estelle, M. Auxin regulates SCF-dependent degradation of AUX/IAA proteins. Nature 2001, 414, 271–276. [Google Scholar] [CrossRef]
- Rogg, L.E.; Bartel, B. Auxin signaling: Derepression through regulated proteolysis. Dev. Cell 2001, 1, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Kepinski, S.; Leyser, O. Ubiquitination and auxin signaling: A degrading story. Plant Cell 2002, 14, S81–S95. [Google Scholar] [CrossRef] [PubMed]
- Dharmasiri, N.; Dharmasiri, S.; Estelle, M. The F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 441–445. [Google Scholar] [CrossRef]
- Kepinski, S.; Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 446–451. [Google Scholar] [CrossRef]
- Weijers, D.; Friml, J. SnapShot: Auxin signaling and transport. Cell 2009, 136, 1172–1172.e1. [Google Scholar] [CrossRef]
- Li, S.B.; Xie, Z.Z.; Hu, C.G.; Zhang, J.Z. A review of auxin response factors (ARFs) in plants. Front. Plant Sci. 2016, 7, 47. [Google Scholar] [CrossRef]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. J. Plant Growth Regul. 2001, 20, 281–291. [Google Scholar] [CrossRef]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 2003, 15, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.K.; Pei, K.M.; Fu, Y.P.; Sun, Z.X.; Li, S.J.; Liu, H.Q.; Tang, K.; Han, B.; Tao, Y.Z. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 2007, 394, 13–24. [Google Scholar] [CrossRef]
- Remington, D.L.; Vision, T.J.; Guilfoyle, T.J.; Reed, J.W. Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 2004, 135, 1738–1752. [Google Scholar] [CrossRef]
- Wang, L.; Hua, D.P.; He, J.N.; Duan, Y.; Chen, Z.Z.; Hong, X.H.; Gong, Z.Z. Auxin response factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis. PLoS Genet. 2011, 7, e1002172. [Google Scholar] [CrossRef] [PubMed]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef]
- Nemhauser, J.L.; Feldman, L.J.; Zambryski, P.C. Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 2000, 127, 3877–3888. [Google Scholar] [CrossRef]
- Zhou, J.H.; Sittmann, J.; Guo, L.; Xiao, Y.W.; Huang, X.L.; Pulapaka, A.; Liu, Z.C. Gibberellin and auxin signaling genes and repress accessory fruit initiation in diploid strawberry. Plant Physiol. 2021, 185, 1059–1075. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hua, W.; Hu, Z.Y.; Yang, H.L.; Zhang, L.; Li, R.J.; Deng, L.B.; Sun, X.C.; Wang, X.F.; Wang, H.Z. Natural variation in gene simultaneously affects seed weight and silique length in polyploid rapeseed. Proc. Natl. Acad. Sci. USA 2015, 112, E5123–E5132. [Google Scholar] [CrossRef]
- Sagar, M.; Chervin, C.; Mila, I.; Hao, Y.W.; Roustan, J.P.; Benichou, M.; Gibon, Y.; Biais, B.; Maury, P.; Latché, A.; et al. SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiol. 2013, 161, 1362–1374. [Google Scholar] [CrossRef]
- Dong, X.X.; Li, Y.J.; Guan, Y.H.; Wang, S.X.; Luo, H.; Li, X.M.; Li, H.; Zhang, Z.H. Auxin-induced AUXIN RESPONSE FACTOR4 activates APETALA1 and FRUITFULL to promote flowering in woodland strawberry. Hortic. Res. 2021, 8, 115. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Y.; Zhang, Y.W.; Feng, Q.S.; Qin, L.; Pan, C.T.; Lamin-Samu, A.T.; Lu, G. Tomato AUXIN RESPONSE FACTOR 5 regulates fruit set and development via the mediation of auxin and gibberellin signaling. Sci. Rep. 2018, 8, 2971. [Google Scholar] [CrossRef]
- Liu, Z.N.; Miao, L.M.; Huo, R.X.; Song, X.Y.; Johnson, C.; Kong, L.J.; Sundaresan, V.; Yu, X.L. ARF2–ARF4 and ARF5 are essential for female and male gametophyte development in Arabidopsis. Plant Cell Physiol. 2018, 59, 179–189. [Google Scholar] [CrossRef]
- Nagpal, P.; Ellis, C.M.; Weber, H.; Ploense, S.E.; Barkawi, L.S.; Guilfoyle, T.J.; Hagen, G.; Alonso, J.M.; Cohen, J.D.; Farmer, E.E.; et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 2005, 132, 4107–4118. [Google Scholar] [CrossRef]
- Yuan, Y.J.; Xu, X.; Gong, Z.H.; Tang, Y.W.; Wu, M.B.; Yan, F.; Zhang, X.L.; Zhang, Q.; Yang, F.Q.; Hu, X.W.; et al. Auxin response factor 6A regulates photosynthesis, sugar accumulation, and fruit development in tomato. Hortic. Res. 2019, 6, 85. [Google Scholar] [CrossRef] [PubMed]
- Goetz, M.; Hooper, L.C.; Johnson, S.D.; Rodrigues, J.C.M.; Vivian-Smith, A.; Koltunow, A.M. Expression of aberrant forms of AUXIN RESPONSE FACTOR8 stimulates parthenocarpy in Arabidopsis and tomato. Plant Physiol. 2007, 145, 351–366. [Google Scholar] [CrossRef]
- de Jong, M.; Wolters-Arts, M.; Feron, R.; Mariani, C.; Vriezen, W.H. The auxin response factor 7 (ARF7) regulates auxin signaling during tomato fruit set and development. Plant J. 2009, 57, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Wang, L.J.; Mao, Y.B.; Cai, W.J.; Xue, H.W.; Chen, X.Y. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 2005, 17, 2204–2216. [Google Scholar] [CrossRef]
- Liu, X.D.; Huang, J.; Wang, Y.; Khanna, K.; Xie, Z.X.; Owen, H.A.; Zhao, D.Z. The role of floral organs in carpels, an Arabidopsis loss-of-function mutation in MicroRNA160a, in organogenesis and the mechanism regulating its expression. Plant J. 2010, 62, 416–428. [Google Scholar] [CrossRef]
- Hendelman, A.; Buxdorf, K.; Stav, R.; Kravchik, M.; Arazi, T. Inhibition of lamina outgrowth following Solanum lycopersicum AUXIN RESPONSE FACTOR 10 (SlARF10) derepression. Plant Mol. Biol. 2012, 78, 561–576. [Google Scholar] [CrossRef]
- Liu, X.; Dong, X.F.; Liu, Z.H.; Shi, Z.H.; Jiang, Y.; Qi, M.F.; Xu, T.; Li, T.L. Repression of ARF10 by microRNA160 plays an important role in the mediation of leaf water loss. Plant Mol. Biol. 2016, 92, 313–336. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.J.; Mei, L.H.; Wu, M.B.; Wei, W.; Shan, W.; Gong, Z.H.; Zhang, Q.; Yang, F.Q.; Yan, F.; Zhang, Q.; et al. SIARF10, an auxin response factor, is involved in chlorophyll and sugar accumulation during tomato fruit development. J. Exp. Bot. 2018, 69, 5507–5518. [Google Scholar] [CrossRef]
- Guo, Q.W.; Li, J.; Cui, L.; Zhang, T.L.; Kere, G.M.; Chen, J.F. Cloning and expression analysis of cucumber CsARF10 genes subfamily. Hortic. Plant J. 2013, 40, 1071–1080. [Google Scholar]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Xu, J.; Li, J.; Cui, L.; Zhang, T.; Wu, Z.; Zhu, P.Y.; Meng, Y.J.; Zhang, K.J.; Yu, X.Q.; Lou, Q.F.; et al. New insights into the roles of cucumber TIR1 homologs and miR393 in regulating fruit/seed set development and leaf morphogenesis. BMC Plant Biol. 2017, 17, 130. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.X.; Li, Z.G.; Miao, Q.; Yang, Y.W.; Deng, W.; Hao, Y.W. The auxin receptor homologue in Solanum lycopersicum stimulates tomato fruit set and leaf morphogenesis. J. Exp. Bot. 2011, 62, 2815–2826. [Google Scholar] [CrossRef]
- Finet, C.; Berne-Dedieu, A.; Scutt, C.P.; Marlétaz, F. Evolution of the ARF gene family in land plants: Old domains, new tricks. Mol. Biol. Evol. 2013, 30, 45–56. [Google Scholar] [CrossRef]
- Rademacher, E.H.; Lokerse, A.S.; Schlereth, A.; Llavata-Peris, C.I.; Bayer, M.; Kientz, M.; Rios, A.F.; Borst, J.W.; Lukowitz, W.; Jürgens, G.; et al. Different auxin response machineries control distinct cell fates in the early plant embryo. Dev. Cell 2012, 22, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Okushima, Y.; Overvoorde, P.J.; Arima, K.; Alonso, J.M.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Lui, A.; Nguyen, D.; et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 2005, 17, 444–463. [Google Scholar] [CrossRef] [PubMed]
- Bustos-Sanmamed, P.; Bazin, J.; Hartmann, C.; Crespi, M.; Lelandais-Brière, C. Small RNA pathways and diversity in model legumes: Lessons from genomics. Front. Plant Sci. 2013, 4, 236. [Google Scholar] [CrossRef]
- Turner, M.; Nizampatnam, N.R.; Baron, M.; Coppin, S.; Damodaran, S.; Adhikari, S.; Arunachalam, S.P.; Yu, O.; Subramanian, S. Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiol. 2013, 162, 2042–2055. [Google Scholar] [CrossRef]
- Liu, P.P.; Montgomery, T.A.; Fahlgren, N.; Kasschau, K.D.; Nonogaki, H.; Carrington, J.C. Repression of AUXIN response FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007, 52, 133–146. [Google Scholar] [CrossRef]
- Damodharan, S.; Zhao, D.Z.; Arazi, T. A common miRNA160-based mechanism regulates ovary patterning, floral organ abscission and lamina outgrowth in tomato. Plant J. 2016, 86, 458–471. [Google Scholar] [CrossRef]
- Ben-Gera, H.; Dafna, A.; Alvarez, J.P.; Bar, M.; Mauerer, M.; Ori, N. Auxin-mediated lamina growth in tomato leaves is restricted by two parallel mechanisms. Plant J. 2016, 86, 443–457. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.X.; Chen, J.H.; Li, B.L.; Zhang, D.Q. Association genetics in Populus reveals the interactions between Pto-miR160a and its target Pto-ARF16. Mol. Genet. Genom. 2016, 291, 1069–1082. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.L.; Lai, Z.X.; Tian, Q.L.; Lin, L.X.; Lai, R.L.; Yang, M.M.; Zhang, D.M.; Chen, Y.K.; Zhang, Z.H. Endogenous target mimics down-regulate miR160 mediation of ARF10, -16, and -17 cleavage during somatic embryogenesis in Dimocarpus Longan Lour. Front. Plant Sci. 2015, 6, 956. [Google Scholar] [CrossRef] [PubMed]
- Damodharan, S.; Corem, S.; Gupta, S.K.; Arazi, T. Tuning of SlARF10A dosage by sly-miR160a is critical for auxin-mediated compound leaf and flower development. Plant J. 2018, 96, 855–868. [Google Scholar] [CrossRef]
- Dai, X.H.; Lu, Q.; Wang, J.; Wang, L.L.; Xiang, F.N.; Liu, Z.H. MiR160 and its target genes ARF10, ARF16 and ARF17 modulate hypocotyl elongation in a light, BRZ, or PAC-dependent manner in Arabidopsis: miR160 promotes hypocotyl elongation. Plant Sci. 2021, 303, 110686. [Google Scholar] [CrossRef]




| Primer Name | Forward | Reverse |
|---|---|---|
| CsARF10a-CDS | 5′-GGGTTTATTTTACATTTGGG-3′ | 5′-ACATTTCTTGGGTTCATTTT-3′ |
| CsActin | 5′-TTCTGGTGATGGTGTGAGTC-3′ | 5′-GGCAGTGGTGGTGAACATG-3′ |
| SlActin | 5′-TGTCCCTATTTACGAGGGTTATGC-3′ | 5′-CAGTTAAATCACGACCAGCAAGAT-3′ |
| CsARF10a-RT-qPCR | 5′-CAATTCCCACTGTCGTCATC-3′ | 5′-GTATGCCTGGCTCCCTGTAT-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Zhu, P.; Yao, X.; Meng, Y.; Lou, L.; Zhang, M.; Liu, G.; Yang, X.; Liu, J.; Zhu, L.; et al. Cucumber Auxin Response Factor CsARF10a Regulates Leaf Morphogenesis and Parthenocarpic Fruit Set in Tomato. Horticulturae 2024, 10, 79. https://doi.org/10.3390/horticulturae10010079
Xu J, Zhu P, Yao X, Meng Y, Lou L, Zhang M, Liu G, Yang X, Liu J, Zhu L, et al. Cucumber Auxin Response Factor CsARF10a Regulates Leaf Morphogenesis and Parthenocarpic Fruit Set in Tomato. Horticulturae. 2024; 10(1):79. https://doi.org/10.3390/horticulturae10010079
Chicago/Turabian StyleXu, Jian, Pinyu Zhu, Xiefeng Yao, Yongjiao Meng, Lina Lou, Man Zhang, Guang Liu, Xingping Yang, Jinqiu Liu, Lingli Zhu, and et al. 2024. "Cucumber Auxin Response Factor CsARF10a Regulates Leaf Morphogenesis and Parthenocarpic Fruit Set in Tomato" Horticulturae 10, no. 1: 79. https://doi.org/10.3390/horticulturae10010079
APA StyleXu, J., Zhu, P., Yao, X., Meng, Y., Lou, L., Zhang, M., Liu, G., Yang, X., Liu, J., Zhu, L., Hou, Q., Li, J., & Xu, J. (2024). Cucumber Auxin Response Factor CsARF10a Regulates Leaf Morphogenesis and Parthenocarpic Fruit Set in Tomato. Horticulturae, 10(1), 79. https://doi.org/10.3390/horticulturae10010079






