Petroleum Hydrocarbon Catabolic Pathways as Targets for Metabolic Engineering Strategies for Enhanced Bioremediation of Crude-Oil-Contaminated Environments
Abstract
1. Introduction
2. Microbe-Assisted Remediation of Crude-Oil-Contaminated Environment
2.1. Regulatory Factors Involve in Microbial Degradation of Crude Oil
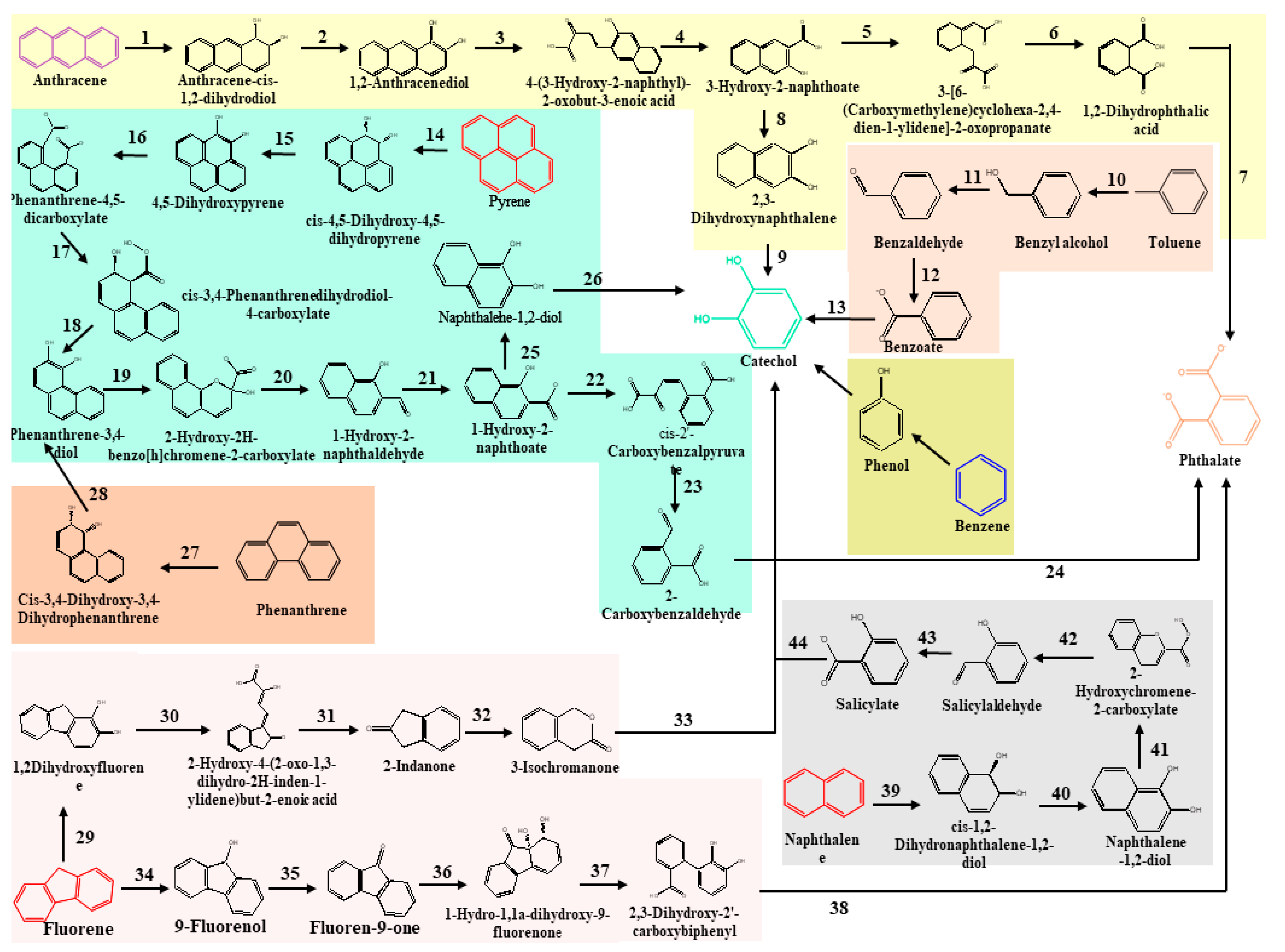
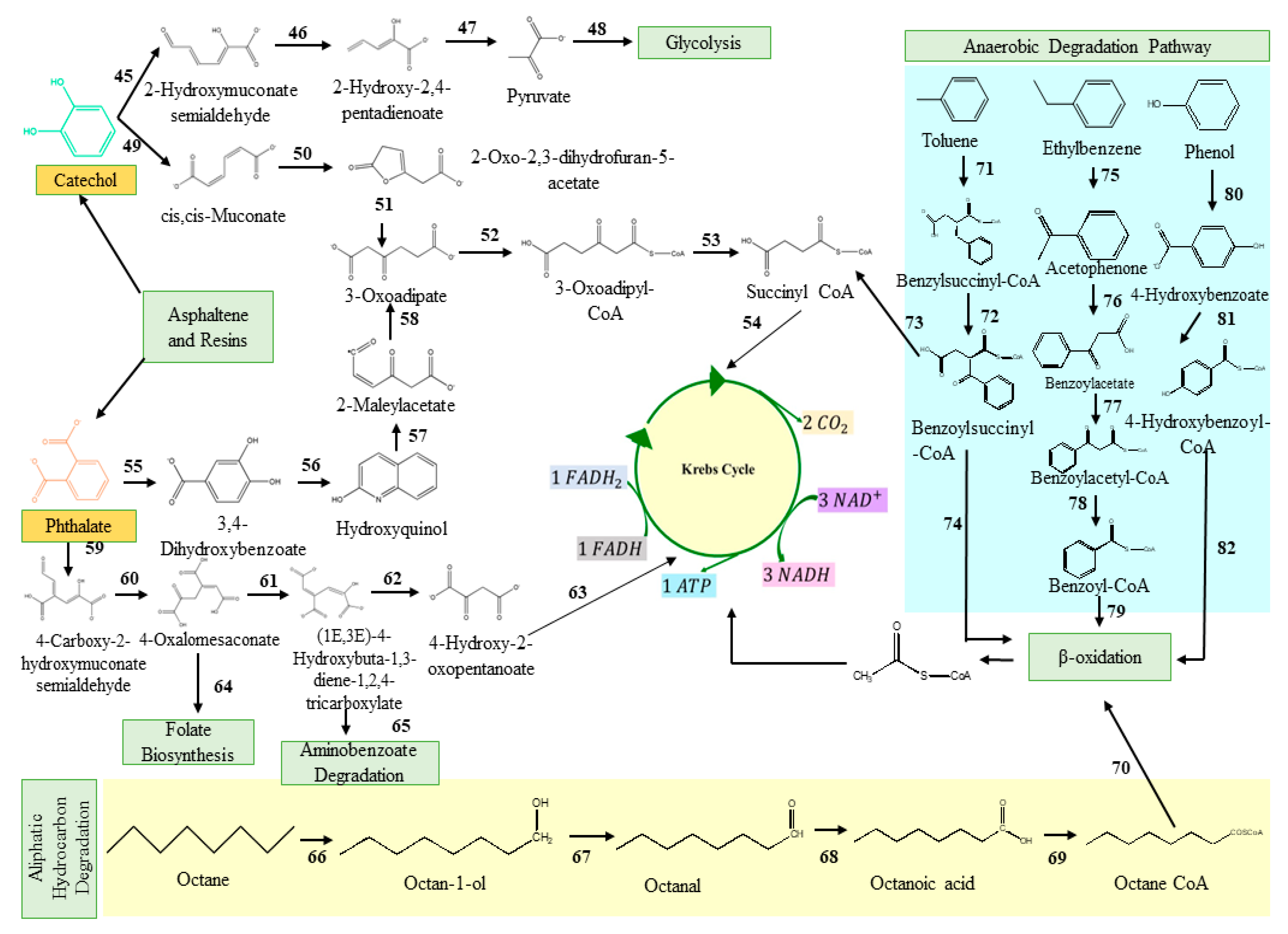
2.2. Classical Metabolic Pathways Involved in Degradation of Total Petroleum Hydrocarbons
3. Metabolic Pathway Engineering for Bioremediation of Crude Oil Contamination
3.1. Contaminant’s Characteristics and Selection of Suitable Microbial Host
3.2. Preference for Utilization of Substrate
3.3. Computational Application for Metabolic Pathway Prediction
3.4. Toxicity Determination of Metabolic Pathways
3.5. Transcriptional Modification of Regulatory Factors
3.6. Prediction and Engineering of Metabolic Building Blocks
4. Metabolic Engineering Studies Integrating Systems Biology
4.1. Multi-Omics Approaches for Metabolic Pathway Engineering
4.2. Computational Analytical Software Used in System Biology
4.3. Limitations in System Biology Approach
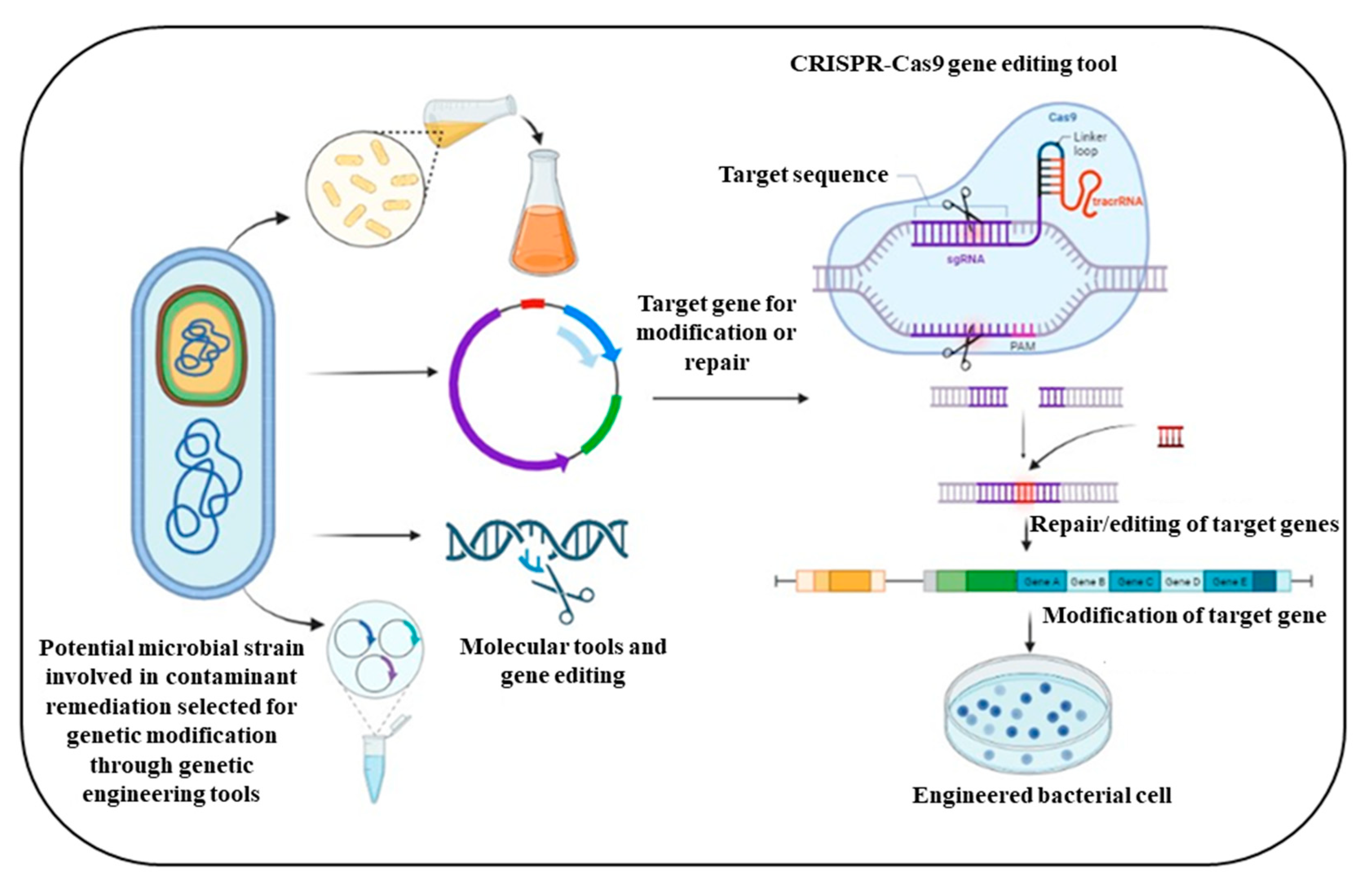
5. Synthetic Biology Approach in the Field of Metabolic Engineering
5.1. Construction of Synthetic Consortia to Enhance Biodegradation
5.2. Risk Assessment of Synthetic Consortium
5.3. Synthetic Orthogonal Approach for Bioremediation Enhancement
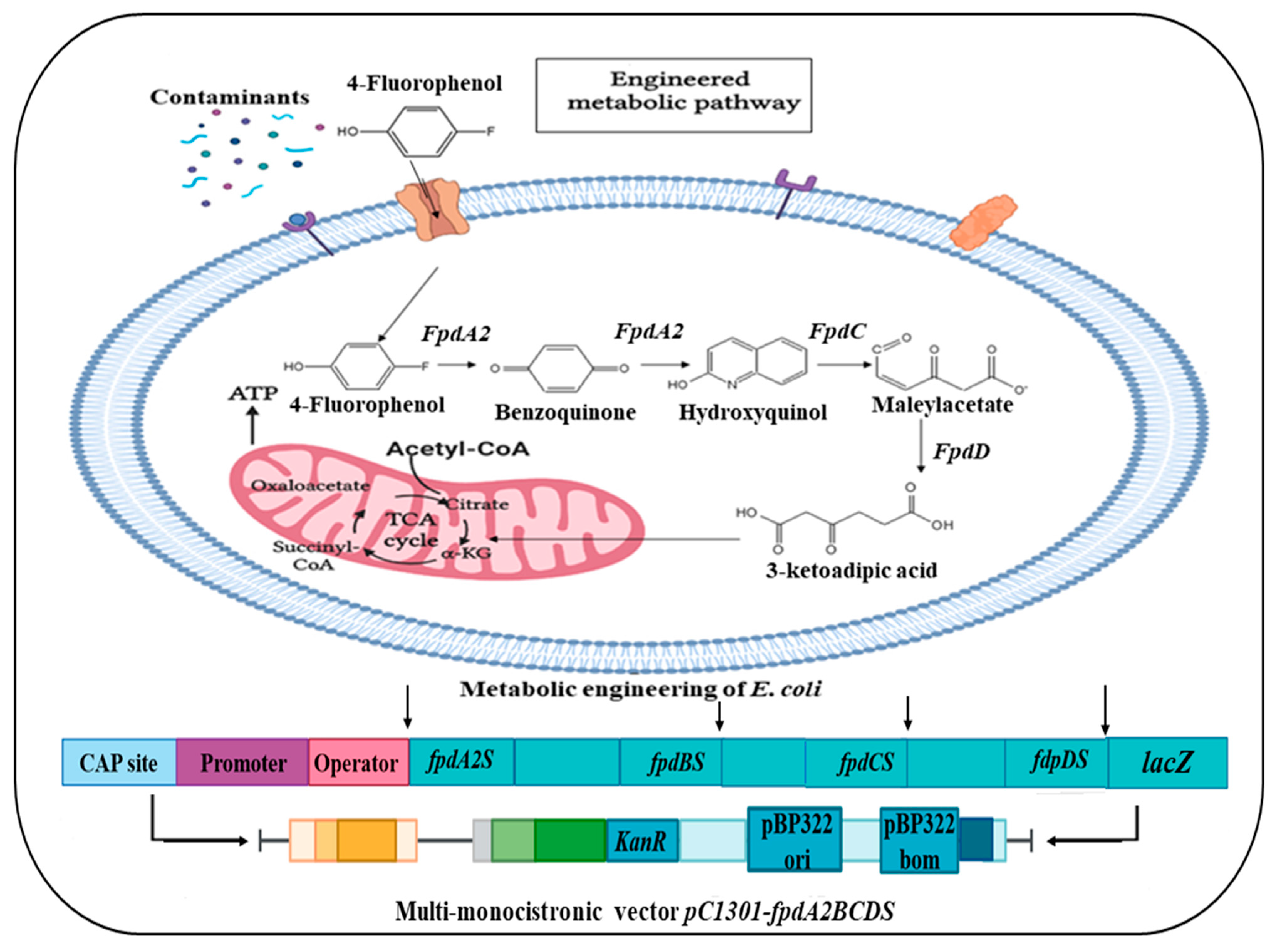
5.4. Experimental Strategies for Metabolic Pathway Engineering
6. Conclusions and Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GMO | Genetically modified organisms |
| GEMs | Genetically engineered microorganisms |
| HMW | High molecular weight |
| LMW | Low molecular weight |
| PAH | Polycyclic aromatic hydrocarbons |
| NSO | Nitrogen, sulphur, and oxygen |
| DEGs | Differentially expressed genes |
References
- Singh, H.; Bhardwaj, N.; Arya, S.K.; Khatri, M. Environmental impacts of oil spills and their remediation by magnetic nanomaterials. Environ. Nanotechnol. Monit. Manag. 2020, 14, 100305. [Google Scholar] [CrossRef]
- Brkić, D.; Praks, P. Probability analysis and prevention of offshore oil and gas accidents: Fire as a cause and a consequence. Fire 2021, 4, 71. [Google Scholar] [CrossRef]
- Atlas, R.M.; Hazen, T.C. Oil biodegradation and bioremediation: A tale of the two worst spills in U.S. history. Environ. Sci. Technol. 2011, 45, 6709–6715. [Google Scholar] [CrossRef]
- Patel, A.B.; Shaikh, S.; Jain, K.R.; Desai, C.; Madamwar, D. Polycyclic Aromatic Hydrocarbons: Sources, Toxicity, and Remediation Approaches. Front. Microbiol. 2020, 11, 562813. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Feng, Z.H.; Fang, W.; Huo, Q.L.; Zhang, K.; Li, J.K.; Zeng, H.S.; Zhang, B.W. Crude-oil hydro-carbon composition characteristics and oil viscosity prediction in the northern Songliao Basin. Sci. China Earth Sci. 2014, 57, 297–312. [Google Scholar] [CrossRef]
- Speight, J.G. The Chemistry and Technology of Petroleum; CRC Press: Boca Raton, FL, USA, 2006. [Google Scholar] [CrossRef]
- Speight, J.G. Organic Chemistry. In Environmental Organic Chemistry for Engineers; Butterworth-Heinemann: Oxford, UK, 2017; pp. 43–86. [Google Scholar] [CrossRef]
- Gailiūtė, I.; Račkauskienė, G.; Grigiškis, S. Changes in crude oil hydrocarbon composition during biodegradation by Arthrobacter sp. M1 and Acinetobacter sp. Pr82 in selected optimal conditions. Biologija 2014, 60, 134–141. [Google Scholar] [CrossRef]
- Klamerus-Iwan, A.; Błońska, E.; Lasota, J.; Kalandyk, A.; Waligórski, P. Influence of Oil Contamination on Physical and Biological Properties of Forest Soil after Chainsaw Use. Water Air Soil Pollut. 2015, 226, 389. [Google Scholar] [CrossRef]
- Galazka, A.; Grzadziel, J.; Galazka, R.; Ukalska-Jaruga, A.; Strzelecka, J.; Smreczak, B.; Tarafdar, A.; Sinha, A.; Horel, A.; Bernard, R.J.; et al. Microbial diversity and hydrocarbon degrading gene capacity of a crude oil field soil as determined by met-agenomics analysis. Front. Microbiol. 2018, 9, 638–648. [Google Scholar] [CrossRef]
- Englande, A.J., Jr.; Krenkel, P.; Shamas, J. Wastewater Treatment &Water Reclamation. Ref. Modul. Earth Syst. Environ. Sci. 2015, 1–32. [Google Scholar] [CrossRef]
- Pichtel, J. Oil and gas production wastewater: Soil contamination and pollution prevention. Appl. Environ. Soil Sci. 2016, 2016, 2707989. [Google Scholar] [CrossRef]
- National Oceanic and Atmospheric Administration (NOAA). Climate Change Impacts. Available online: https://www.noaa.gov/ (accessed on 13 August 2021).
- McDonnell, T. California’s Oil Spill Is Proof That Climate Action Is Worth the Cost. Available online: https://qz.com/2068783/who-pays-to-clean-up-californias-oil-spill (accessed on 4 October 2021).
- Cronin, M.A.; Bickham, J.W. A population genetic analysis of the potential for a crude oil spill to induce heritable mutations and impact natural populations. Ecotoxicology 1998, 7, 259–278. [Google Scholar] [CrossRef]
- Alzahrani, A.M.; Rajendran, P. Petroleum hydrocarbon and living organisms. In Hydrocarbon Pollution and Its Effect on the Environment; IntechOpen: London, UK, 2019; pp. 71–82. [Google Scholar]
- Gupta, S.K.; Sriwastav, A.; Ansari, F.A.; Nasr, M.; Nema, A.K. Phycoremediation: An ecofriendly algal technology for bioremediation and bioenergy production. In Phytoremediation Potential of Bioenergy Plants; Springer: Berlin/Heidelberg, Germany, 2017; pp. 431–456. [Google Scholar]
- Kotoky, R.; Pandey, P. Difference in the rhizosphere microbiome of Melia azedarach during removal of benzo (a) pyrene from cadmium co-contaminated soil. Chemosphere 2020, 258, 127175. [Google Scholar] [CrossRef]
- Wang, Y.; Oyaizu, H. Enhanced remediation of dioxins-spiked soil by a plant-microbe system using a dibenzofuran-degrading Comamonas sp. and Trifolium repens L. Chemosphere 2011, 85, 1109–1114. [Google Scholar] [CrossRef]
- Gaskin, S.E.; Bentham, R.H. Rhizoremediation of hydrocarbon contaminated soil using Australian native grasses. Sci. Total Environ. 2010, 408, 3683–3688. [Google Scholar] [CrossRef] [PubMed]
- Rajkumari, J.; Choudhury, Y.; Bhattacharjee, K.; Pandey, P. Rhizodegradation of pyrene by a non-pathogenic Klebsiella pneumoniae isolate applied with Tagetes erecta l. and changes in the rhizobacterial community. Front. Microbiol. 2021, 12, 593023. [Google Scholar] [CrossRef] [PubMed]
- Raymond, R.L.; Jamison, V.W.; Hudson, J.O. Beneficial stimulation of bacterial activity in groundwaters containing petroleum products. AIChE Symp. Ser. 1976, 73, 390. [Google Scholar]
- Sharma, I. Bioremediation Techniques for Polluted Environment: Concept, Advantages, Limitations, and Prospects. In Trace Metals in the Environment—New Approaches and Recent Advances; IntechOpen: London, UK, 2021. [Google Scholar] [CrossRef]
- Chuah, L.F.; Chew, K.W.; Bokhari, A.; Mubashir, M.; Show, P.L. Biodegradation of crude oil in seawater by using a consortium of symbiotic bacteria. Environ. Res. 2022, 213, 113721. [Google Scholar] [CrossRef]
- Al-Wasify, R.S.; Hamed, S.R. Bacterial biodegradation of crude oil using local isolates. Int. J. Bacteriol. 2014, 2014, 863272. [Google Scholar] [CrossRef]
- Aditiawati, P.; Kamarisima, K. Isolation of asphaltene-degrading bacteria from sludge oil. Makara J. Sci. 2015, 19, 13–20. [Google Scholar] [CrossRef]
- Ali, H.R.; El-Gendy, N.S.; Moustafa, Y.M.; Roushdy, M.I.; Hashem, A.I. Degradation of asphaltenic fraction by locally isolated halotolerant bacterial strains. Int. Sch. Res. Not. 2012, 2012, 435485. [Google Scholar] [CrossRef]
- Abatenh, E.; Gizaw, B.; Tsegaye, Z.; Wassie, M. The Role of Microorganisms in Bioremediation—A Review. Open J. Environ. Biol. 2017, 2, 38–46. [Google Scholar] [CrossRef]
- Ławniczak, Ł.; Woźniak-Karczewska, M.; Loibner, A.P.; Heipieper, H.J.; Chrzanowski, Ł. Microbial degradation of hydrocarbons—Basic principles for bioremediation: A review. Molecules 2020, 25, 856. [Google Scholar] [CrossRef] [PubMed]
- Bala, S.; Garg, D.; Thirumalesh, B.V.; Sharma, M. Recent Strategies for Bioremediation of Emerging Pollutants: A Review for a Green and Sustainable Environment. Toxics 2022, 10, 484. [Google Scholar] [CrossRef]
- Tribedi, P.; Goswami, M.; Chakraborty, P.; Mukherjee, K.; Mitra, G.; Bhattacharyya, P.; Dey, S. Bioaugmentation and biostimulation: A potential strategy for environmental remediation. J. Microbiol. Exp. 2018, 6, 223–231. [Google Scholar] [CrossRef]
- Das, N.; Chandran, P. Microbial Degradation of Petroleum Hydrocarbon Contaminants: An Overview. Biotechnol. Res. Int. 2011, 2011, 941810. [Google Scholar] [CrossRef]
- Jaiswal, S.; Singh, D.K.; Shukla, P. Gene editing and systems biology tools for pesticide bioremediation: A review. Front. Microbiol. 2019, 10, 87. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.B.; Kim, D.Y.; Ko, J.H.; Kim, Y.S. Recent advances in the CRISPR genome editing tool set. Exp. Mol. Med. 2019, 51, 1–11. [Google Scholar] [CrossRef]
- Kumar, R.R.; Prasad, S. Metabolic Engineering of Bacteria. Indian J. Microbiol. 2011, 51, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Megharaj, M.; Venkateswarlu, K.; Naidu, R. Bioremediation. In Encyclopedia of Toxicology, 3rd ed.; Academic Press: Cambridge, MA, USA, 2014; Volume 1, pp. 485–489. [Google Scholar] [CrossRef]
- Tyagi, A.; Kumar, A.; Aparna, S.V.; Mallappa, R.H.; Grover, S.; Batish, V.K. Synthetic biology: Applications in the food sector. Crit. Rev. Food Sci. Nutr. 2016, 56, 1777–1789. [Google Scholar] [CrossRef] [PubMed]
- Kotoky, R.; Rajkumari, J.; Pandey, P. The rhizosphere microbiome: Significance in rhizoremediation of polyaromatic hydrocarbon contaminated soil. J. Environ. Manag. 2018, 217, 858–870. [Google Scholar] [CrossRef]
- Azad, M.A.K.; Amin, L.; Sidik, N.M. Genetically engineered organisms for bioremediation of pollutants in contaminated sites. Chin. Sci. Bull. 2014, 59, 703–714. [Google Scholar] [CrossRef]
- Koshlaf, E.; Ball, A.S. Soil bioremediation approaches for petroleum hydrocarbon polluted environments. AIMS Microbiol. 2017, 3, 25–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, D.; Yu, Z.; Dong, X.; Liu, S.; Cui, H.; Sun, B. Advances in research on petroleum biodegradability in soil. Environ. Sci. Process. Impacts 2021, 23, 9–27. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, Z.W.; Liu, R.X.; Ju, H.Y.; Bian, X.K.; Zhang, W.Z.; Zhang, C.B.; Yang, T.; Guo, B.; Xiao, C.L.; et al. Research progress in bioremediation of petroleum pollution. Environ. Sci. Pollut. Res. 2021, 28, 46877–46893. [Google Scholar] [CrossRef]
- Rahman, K.S.M.; Thahira-Rahman, J.; Lakshmanaperumalsamy, P.; Banat, I.M. Towards efficient crude oil degradation by a mixed bacterial consortium. Bioresour. Technol. 2002, 85, 257–261. [Google Scholar] [CrossRef]
- Das, K.; Mukherjee, A.K. Crude petroleum-oil biodegradation efficiency of Bacillus subtilis and Pseudomonas aeruginosa strains isolated from a petroleum-oil contaminated soil from North-East India. Bioresour. Technol. 2007, 98, 1339–1345. [Google Scholar] [CrossRef] [PubMed]
- Zucchi, M.; Angiolini, L.; Borin, S.; Brusetti, L.; Dietrich, N.; Gigliotti, C.; Barbieri, P.; Sorlini, C.; Daffonchio, D. Response of bacterial community during bioremediation of an oil-polluted soil. J. Appl. Microbiol. 2003, 94, 248–257. [Google Scholar] [CrossRef]
- Abbasian, F.; Lockington, R.; Mallavarapu, M.; Naidu, R. A Comprehensive Review of Aliphatic Hydrocarbon Biodegradation by Bacteria. Appl. Biochem. Biotechnol. 2015, 176, 670–699. [Google Scholar] [CrossRef]
- Megharaj, M.; Ramakrishnan, B.; Venkateswarlu, K.; Sethunathan, N.; Naidu, R. Bioremediation approaches for organic pollutants: A critical perspective. Environ. Int. 2011, 37, 1362–1375. [Google Scholar] [CrossRef]
- Gargouri, B.; Karray, F.; Mhiri, N.; Aloui, F.; Sayadi, S. Bioremediation of petroleum hydrocarbons-contaminated soil by bacterial consortium isolated from an industrial wastewater treatment plant. J. Chem. Technol. Biotechnol. 2014, 89, 978–987. [Google Scholar] [CrossRef]
- Roy, A.S.; Baruah, R.; Borah, M.; Singh, A.K.; Boruah, H.P.D.; Saikia, N.; Deka, M.; Dutta, N.; Bora, T.C. Bioremediation potential of native hydrocarbon degrading bacterial strains in crude oil contaminated soil under microcosm study. Int. Biodeterior. Biodegrad. 2014, 94, 79–89. [Google Scholar] [CrossRef]
- Lee, H.; Yun, S.Y.; Jang, S.; Kim, G.H.; Kim, J.J. Bioremediation of polycyclic aromatic hydrocarbons in creosote-contaminated soil by Peniophora incarnata KUC8836. Bioremed. J. 2015, 19, 1–8. [Google Scholar] [CrossRef]
- Hesnawi, R.M.; Adbeib, M.M. Effect of Nutrient Source on Indigenous Biodegradation of Diesel Fuel Contaminated Soil. APCBEE Procedia 2013, 5, 557–561. [Google Scholar] [CrossRef]
- Gurav, R.; Lyu, H.; Ma, J.; Tang, J.; Liu, Q. Degradation of n-alkanes and PAHs from the heavy crude oil using salt-tolerant bacterial consortia and analysis of their catabolic genes. Environ. Sci. Pollut. Res. 2017, 24, 11392–11403. [Google Scholar] [CrossRef] [PubMed]
- El-Liethy, M.A.; El-Noubi, M.M.; Abia, A.L.K.; El-Malky, M.G.; Hashem, A.I.; El-Taweel, G.E. Eco-friendly bioremediation approach for crude oil-polluted soils using a novel and biostimulated Enterobacter hormaechei ODB H32 strain. Int. J. Environ. Sci. Technol. 2022, 19, 10577–10588. [Google Scholar] [CrossRef]
- Hajieghrari, M.; Hejazi, P. Enhanced biodegradation of n-Hexadecane in solid-phase of soil by employing immobilized Pseudomonas Aeruginosa on size-optimized coconut fibers. J. Hazard. Mater. 2020, 389, 122134. [Google Scholar] [CrossRef] [PubMed]
- Throne-Holst, M.; Wentzel, A.; Ellingsen, T.E.; Kotlar, H.K.; Zotchev, S.B. Identification of novel genes involved in long-chain n-alkane degradation by Acinetobacter sp. strain DSM 17874. Appl. Environ. Microbiol. 2007, 73, 3327–3332. [Google Scholar] [CrossRef]
- Okoye, A.U.; Chikere, C.B.; Okpokwasili, G.C. Characterization of potential paraffin wax removing bacteria for sustainable biotechnological application. In Proceedings of the Society of Petroleum Engineers—SPE Nigeria Annual International Conference and Exhibition 2019, NAIC 2019, Lagos, Nigeria, 6 August 2019. [Google Scholar] [CrossRef]
- Rahman, R.N.Z.R.A.; Latip, W.; Adlan, N.A.; Sabri, S.; Ali, M.S.M. Bacteria consortia enhanced hydrocarbon degradation of waxy crude oil. Arch. Microbiol. 2022, 204, 701. [Google Scholar] [CrossRef]
- Ali, N.; Khanafer, M.; Al-Awadhi, H. Indigenous oil-degrading bacteria more efficient in soil bioremediation than microbial consortium and active even in super oil-saturated soils. Front. Microbiol. 2022, 13, 2840. [Google Scholar] [CrossRef]
- Gangola, S.; Kumar, R.; Sharma, A.; Singh, H. Bioremediation of petrol engine oil polluted soil using microbial consortium and wheat crop. J. Pure Appl. Microbiol. 2017, 11, 1583–1588. [Google Scholar]
- Xu, X.; Liu, W.; Tian, S.; Wang, W.; Qi, Q.; Jiang, P.; Gao, X.; Li, F.; Li, H.; Yu, H. Petroleum hydrocarbon-degrading bacteria for the remediation of oil pollution under aerobic conditions: A perspective analysis. Front. Microbiol. 2018, 9, 2885. [Google Scholar] [CrossRef] [PubMed]
- Lakshmi, G.; Kishore Babu, A. Isolation and Study of Biodegradation Capability of Hydrocarbonoclastic Bacteria from Industrial Waste Lubrication Oil Contaminated Sites. Int. J. Curr. Microbiol. Appl. Sci. 2021, 10, 163–172. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, Q.; Li, D.; Du, J.; Wang, C.; Qin, J. Rapid degradation of long-chain crude oil in soil by indigenous bacteria using fermented food waste supernatant. Waste Manag. 2019, 85, 361–373. [Google Scholar] [CrossRef]
- Mohanty, G.; Mukherji, S. Biodegradation rate of diesel range n-alkanes by bacterial cultures Exiguobacterium aurantiacum and Burkholderia cepacia. Int. Biodeterior. Biodegrad. 2008, 61, 240–250. [Google Scholar] [CrossRef]
- Brito, E.M.S.; Guyoneaud, R.; Goñi-Urriza, M.; Ranchou-Peyruse, A.; Verbaere, A.; Crapez, M.A.; Wasserman, J.C.A.; Duran, R. Characterization of hydrocarbonoclastic bacterial communities from mangrove sediments in Guanabara Bay, Brazil. Res. Microbiol. 2006, 157, 752–762. [Google Scholar] [CrossRef] [PubMed]
- Thangarajan, R.; Adetutu, E.M.; Moore, R.B.; Ogunbanwo, S.T.; Ball, A.S. Comparison between different bio-treatments of a hydrocarbon contaminated soil from a landfill site. Afr. J. Biotechnol. 2011, 10, 15151–15162. [Google Scholar] [CrossRef]
- Bociu, I.; Shin, B.; Wells, W.; Kostka, J.E.; Konstantinidis, K.T.; Huettel, M. Decomposition of sediment-oil-agglomerates in a Gulf of Mexico sandy beach. Sci. Rep. 2019, 9, 10071. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, S.; Méndez, V.; Aguila, P.; Seeger, M. Bioremediation of petroleum hydrocarbons: Catabolic genes, microbial communities, and applications. Appl. Microbiol. Biotechnol. 2014, 98, 4781–4794. [Google Scholar] [CrossRef]
- Zhou, L.; Li, H.; Zhang, Y.; Han, S.; Xu, H. Sphingomonas from petroleum-contaminated soils in Shenfu, China and their PAHs degradation abilities. Braz. J. Microbiol. 2016, 47, 271–278. [Google Scholar] [CrossRef]
- Ganesh Kumar, A.; Mathew, N.C.; Sujitha, K.; Kirubagaran, R.; Dharani, G. Genome analysis of deep sea piezotolerant Nesiotobacter exalbescens COD22 and toluene degradation studies under high pressure condition. Sci. Rep. 2019, 9, 18724. [Google Scholar] [CrossRef]
- Zhang, X.; Kong, D.; Liu, X.; Xie, H.; Lou, X.; Zeng, C. Combined microbial degradation of crude oil under alkaline conditions by Acinetobacter baumannii and Talaromyces sp. Chemosphere 2021, 273, 129666. [Google Scholar] [CrossRef] [PubMed]
- Miglani, R.; Parveen, N.; Kumar, A.; Ansari, M.A.; Khanna, S.; Rawat, G.; Panda, A.K.; Bisht, S.S.; Upadhyay, J.; Ansari, M.N. Degradation of xenobiotic pollutants: An environmentally sustainable approach. Metabolites 2022, 12, 818. [Google Scholar] [CrossRef]
- Turner, J.T. Zooplankton faecal pellets, marine snow, phytodetritus and the ocean’s biological pump. Prog. Oceanogr. 2015, 130, 205–248. [Google Scholar] [CrossRef]
- Hazen, T.C.; Prince, R.C.; Mahmoudi, N. Marine Oil Biodegradation. Environ. Sci. Technol. 2016, 50, 2121–2129. [Google Scholar] [CrossRef] [PubMed]
- Dyksterhouse, S.E.; Gray, J.P.; Herwig, R.P.; Lara, J.C.; Staley, J.T. Cycloclasticus pugetii gen-nov, sp-nov, an aromatic hydrocarbon-degrading bacterium from marine-sediments. Int. J. Syst. Bacteriol. 1995, 45, 116−123. [Google Scholar] [CrossRef]
- Bruns, A.; Berthe-Corti, L. Fundibacter jadensis gen. nov., sp. nov., a new slightly halophilic bacterium, isolated from intertidal sediment. Int. J. Syst. Bacteriol. 1999, 49, 441−448. [Google Scholar] [CrossRef]
- Yakimov, M.M.; Giuliano, L.; Gentile, G.; Crisafi, E.; Chernikova, T.N.; Abraham, W.R.; Lunsdorf, H.; Timmis, K.N.; Golyshin, P.N. Oleispira antarctica gen. nov., sp nov., a novel hydrocarbonoclastic marine bacterium isolated from Antarctic coastal sea water. Int. J. Syst. Evol. Microbiol. 2003, 53, 779−785. [Google Scholar] [CrossRef]
- Wang, B.; Lai, Q.; Cui, Z.; Tan, T.; Shao, Z. A pyrene-degrading consortium from deep-sea sediment of the West Pacific and its key member Cycloclasticus sp. P1. Environ. Microbiol. 2008, 10, 1948–1963. [Google Scholar] [CrossRef]
- Chen, Q.; Li, J.; Liu, M.; Sun, H.; Bao, M. Study on the biodegradation of crude oil by free and immobilized bacterial consortium in marine environment. PLoS ONE 2017, 12, e0174445. [Google Scholar] [CrossRef]
- Farag, S.; Soliman, N.A.; Abdel-Fattah, Y.R. Statistical optimization of crude oil bio-degradation by a local marine bacterium isolate Pseudomonas sp. sp48. J. Genet. Eng. Biotechnol. 2018, 16, 409–420. [Google Scholar] [CrossRef]
- Li, M.; Yin, H.; Zhu, M.; Yu, Y.; Lu, G.; Dang, Z. Co-metabolic and biochar-promoted biodegradation of mixed PAHs by highly efficient microbial consortium QY1. J. Environ. Sci. 2021, 107, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hu, H.; Zanaroli, G.; Xu, P.; Tang, H. A Pseudomonas sp. strain uniquely degrades PAHs and heterocyclic derivatives via lateral dioxygenation pathways. J. Hazard. Mater. 2021, 403, 123956. [Google Scholar] [CrossRef] [PubMed]
- Medić, A.; Lješević, M.; Inui, H.; Beškoski, V.; Kojić, I.; Stojanović, K.; Karadžić, I. Efficient biodegradation of petroleum n-alkanes and polycyclic aromatic hydrocarbons by polyextremophilic Pseudomonas aeruginosa san ai with multidegradative capacity. RSC Adv. 2020, 10, 14060–14070. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Rathour, R.; Singh, R.; Kumari, T.; Thakur, I.S. Degradation and detoxification of phenanthrene by actinobacterium Zhihengliuella sp. ISTPL4. Environ. Sci. Pollut. Res. 2020, 27, 27256–27267. [Google Scholar] [CrossRef]
- Rabodonirina, S.; Rasolomampianina, R.; Krier, F.; Drider, D.; Merhaby, D.; Net, S.; Ouddane, B. Degradation of fluorene and phenanthrene in PAHs-contaminated soil using Pseudomonas and Bacillus strains isolated from oil spill sites. J. Environ. Manag. 2019, 232, 1–7. [Google Scholar] [CrossRef]
- Zeng, J.; Zhu, Q.; Li, Y.; Dai, Y.; Wu, Y.; Sun, Y.; Miu, L.; Chen, H.; Lin, X. Isolation of diverse pyrene-degrading bacteria via introducing readily utilized phenanthrene. Chemosphere 2019, 222, 534–540. [Google Scholar] [CrossRef]
- Qin, W.; Fan, F.; Zhu, Y.; Huang, X.; Ding, A.; Liu, X.; Dou, J. Anaerobic biodegradation of benzo (a) pyrene by a novel Cellulosimicrobium cellulans CWS2 isolated from polycyclic aromatic hydrocarbon-contaminated soil. Braz. J. Microbiol. 2018, 49, 258–268. [Google Scholar] [CrossRef]
- Zhu, X.; Ni, X.; Waigi, M.G.; Liu, J.; Sun, K.; Gao, Y. Biodegradation of mixed PAHs by PAH-degrading endophytic bacteria. Int. J. Environ. Res. Public Health 2016, 13, 805. [Google Scholar] [CrossRef]
- Haritash, A.K.; Kaushik, C.P. Degradation of low molecular weight polycyclic aromatic hydrocarbons by microorganisms isolated from contaminated soil. Int. J. Environ. Sci. 2016, 6, 808–819. [Google Scholar]
- Zhu, X.; Wang, W.; Crowley, D.E.; Sun, K.; Hao, S.; Waigi, M.G.; Gao, Y. The endophytic bacterium Serratia sp. PW7 degrades pyrene in wheat. Environ. Sci. Pollut. Res. 2017, 24, 6648–6656. [Google Scholar] [CrossRef]
- Valsala, H.; Prakash, P.; Elavarasi, V.; Pugazhendhi, A.; Thamaraiselvi, K. Isolation of Staphylococcus nepalensis for Degradation of pyrene from Diesel Contaminated Site. Int. J. Comput. Appl. 2014, 1, 21–24. [Google Scholar]
- Hesham, A.E.L.; Mawad, A.M.; Mostafa, Y.M.; Shoreit, A. Biodegradation ability and catabolic genes of petroleum-degrading Sphingomonas koreensis strain ASU-06 isolated from Egyptian oily soil. BioMed Res. Int. 2014, 2014, 127674. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, P.; Sharma, R.; Singh, S.B.; Nain, L. Bioremediation of PAH by Streptomyces sp. Bull. Environ. Contam. Toxicol. 2011, 86, 268–271. [Google Scholar] [CrossRef] [PubMed]
- Arulazhagan, P.; Vasudevan, N. Biodegradation of polycyclic aromatic hydrocarbons by a halotolerant bacterial strain Ochrobactrum sp. VA1. Mar. Pollut. Bull. 2011, 62, 388–394. [Google Scholar] [CrossRef]
- Das, N.; Bhuyan, B.; Pandey, P. Correlation of soil microbiome with crude oil contamination drives detection of hydrocarbon degrading genes which are independent to quantity and type of contaminants. Environ. Res. 2022, 215 Pt 1, 114185. [Google Scholar] [CrossRef]
- Meckenstock, R.U.; Safinowski, M.; Griebler, C. Anaerobic degradation of polycyclic aromatic hydrocarbons. FEMS Microbiol. Ecol. 2004, 49, 27–36. [Google Scholar] [CrossRef]
- Varjani, S.; Thaker, M.; Upasani, V. Optimization of growth conditions of native hydrocarbon utilizing bacterial consortium “HUBC” obtained from petroleum pollutant contaminated sites. Indian J. Appl. Res. 2014, 4, 474–476. [Google Scholar]
- Shafiee, P.; SHOJA, A.S.; Charkhabi, A.H. Biodegradation of polycyclic aromatic hydrocarbons by aerobic mixed bacterial culture isolated from hydrocarbon polluted soils. Iran. J. Chem. Chem. Eng. 2006, 25, 73–78. [Google Scholar]
- Edet, U.O.; Anika, O.C.; Umoren, E. Toxicity Profile of Crude Oil on Degrading Bacterial Isolates. Int. J. Sci. Eng. Res. 2019, 10, 1131–1135. [Google Scholar] [CrossRef]
- Obi, L.U.; Atagana, H.I.; Adeleke, R.A. Isolation and characterisation of crude oil sludge degrading bacteria. SpringerPlus 2016, 5, 1946. [Google Scholar] [CrossRef]
- Ribicic, D.; McFarlin, K.M.; Netzer, R.; Brakstad, O.G.; Winkler, A.; Throne-Holst, M.; Størseth, T.R. Oil type and temperature dependent biodegradation dynamics—Combining chemical and microbial community data through multivariate analysis. BMC Microbiol. 2018, 18, 83. [Google Scholar] [CrossRef]
- Devatha, C.P.; Vishnu Vishal, A.; Purna Chandra Rao, J. Investigation of physical and chemical characteristics on soil due to crude oil contamination and its remediation. Appl. Water Sci. 2019, 9, 89. [Google Scholar] [CrossRef]
- Al-Hawash, A.B.; Dragh, M.A.; Li, S.; Alhujaily, A.; Abbood, H.A.; Zhang, X.; Ma, F. Principles of microbial degradation of petroleum hydrocarbons in the environment. Egypt. J. Aquat. Res. 2018, 44, 71–76. [Google Scholar] [CrossRef]
- Wang, D.; Lin, J.; Lin, J.; Wang, W.; Li, S. Biodegradation of petroleum hydrocarbons by bacillus subtilis BL-27, a strain with weak hydrophobicity. Molecules 2019, 24, 3021. [Google Scholar] [CrossRef] [PubMed]
- Dubinsky, E.A.; Conrad, M.E.; Chakraborty, R.; Bill, M.; Borglin, S.E.; Hollibaugh, J.T.; Mason, O.U.; Piceno, Y.M.; Reid, F.C.; Stringfellow, W.T.; et al. Succession of hydrocarbon-degrading bacteria in the aftermath of the deepwater horizon oil spill in the Gulf of Mexico. Environ. Sci. Technol. 2013, 47, 10860–10867. [Google Scholar] [CrossRef] [PubMed]
- Atlas, R.M.; Stoeckel, D.M.; Faith, S.A.; Minard-Smith, A.; Thorn, J.R.; Benotti, M.J. Oil Biodegradation and oil-degrading microbial populations in marsh sediments impacted by oil from the Deepwater Horizon well blowout. Environ. Sci. Technol. 2015, 49, 8356–8366. [Google Scholar] [CrossRef] [PubMed]
- Alegbeleye, O.O.; Opeolu, B.O.; Jackson, V.A. Polycyclic aromatic hydrocarbons: A critical review of environmental occurrence and bioremediation. Environ. Manag. 2017, 60, 758–783. [Google Scholar] [CrossRef]
- Kliem, S.; Kreutzbruck, M.; Bonten, C. Review on the biological degradation of polymers in various environments. Materials 2020, 13, 4586. [Google Scholar] [CrossRef] [PubMed]
- Fragkou, E.; Antoniou, E.; Daliakopoulos, I.; Manios, T.; Theodorakopoulou, M.; Kalogerakis, N. In situ aerobic bioremediation of sediments polluted with petroleum hydrocarbons: A critical review. J. Mar. Sci. Eng. 2021, 9, 1003. [Google Scholar] [CrossRef]
- Hamamura, N.; Olson, S.H.; Ward, D.M.; Inskeep, W.P. Microbial population dynamics associated with crude-oil biodegradation in diverse soils. Appl. Environ. Microbiol. 2006, 72, 6316–6324. [Google Scholar] [CrossRef]
- Kumari, S.; Regar, R.K.; Manickam, N. Improved polycyclic aromatic hydrocarbon degradation in a crude oil by individual and a consortium of bacteria. Bioresour. Technol. 2018, 254, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Baboshin, M.; Golovleva, L. Aerobic bacterial degradation of polycyclic aromatic hydrocarbons (PAHs) and its kinetic aspects. Microbiology 2012, 81, 639–650. [Google Scholar] [CrossRef]
- Sun, S.; Wang, H.; Chen, Y.; Lou, J.; Wu, L.; Xu, J. Salicylate and phthalate pathways contributed differently on phenanthrene and pyrene degradations in Mycobacterium sp. WY10. J. Hazard. Mater. 2019, 364, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, S.; Jain, K.; Madamwar, D. Metabolism of pyrene through phthalic acid pathway by enriched bacterial consortium composed of Pseudomonas, Burkholderia, and Rhodococcus (PBR). 3 Biotech 2017, 7, 29. [Google Scholar] [CrossRef]
- Moody, J.D.; Freeman, J.P.; Doerge, D.R.; Cerniglia, C.E. Degradation of phenanthrene and anthracene by cell suspensions of Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 2001, 67, 1476–1483. [Google Scholar] [CrossRef]
- Zhao, H.-P.; Wu, Q.-S.; Wang, L.; Zhao, X.-T.; Gao, H.-W. Degradation of phenanthrene by bacterial strain isolated from soil in oil refinery fields in Shanghai China. J. Hazard. Mater. 2009, 164, 863–869. [Google Scholar] [CrossRef]
- Peng, R.H.; Xiong, A.S.; Xue, Y.; Fu, X.Y.; Gao, F.; Zhao, W.; Tian, Y.S.; Yao, Q.H. Microbial biodegradation of polyaromatic hydrocarbons. FEMS Microbiol. Rev. 2008, 32, 927–955. [Google Scholar] [CrossRef] [PubMed]
- Barnsley, E.A. Naphthalene metabolism by pseudomonads: The oxidation of 1, 2-dihydroxynaphthalene to 2-hydroxychromene-2-carboxylic acid and the formation of 2′-hydroxybenzalpyruvate. Biochem. Biophys. Res. Commun. 1976, 72, 1116–1121. [Google Scholar] [CrossRef]
- Anokhina, T.; Esikova, T.; Gafarov, A.; Polivtseva, V.; Baskunov, B.; Solyanikova, I. Alternative naphthalene metabolic pathway includes formation of ortho-phthalic acid and cinnamic acid derivatives in the Rhodococcus opacus strain 3D. Biochemistry 2020, 85, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.M.; Kamal, M.S.; Al-Harthi, M. Polymeric and low molecular weight shale inhibitors: A review. Fuel 2019, 251, 187–217. [Google Scholar] [CrossRef]
- Rojo, F. Degradation of alkanes by bacteria. Environ. Microbiol. 2009, 11, 2477–2490. [Google Scholar] [CrossRef]
- Ji, Y.; Mao, G.; Wang, Y.; Bartlam, M. Structural insights into diversity and n-alkane biodegradation mechanisms of alkane hydroxylases. Front. Microbiol. 2013, 4, 58. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Shao, Z. Enzymes and genes involved in aerobic alkane degradation. Front. Microbiol. 2013, 4, 116. [Google Scholar] [CrossRef] [PubMed]
- Zuo, P.; Qu, S.; Shen, W. Asphaltenes: Separations, structural analysis and applications. J. Energy Chem. 2019, 34, 186–207. [Google Scholar] [CrossRef]
- Parra-Barraza, H.; Hernández-Montiel, D.; Lizardi, J.; Hernández, J.; Urbina, R.H.; Valdez, M.A. The zeta potential and surface properties of asphaltenes obtained with different crude oil/n-heptane proportions. Fuel 2003, 82, 869–874. [Google Scholar] [CrossRef]
- Foght, J. Anaerobic biodegradation of aromatic hydrocarbons: Pathways and prospects. Microb. Physiol. 2008, 15, 93–120. [Google Scholar] [CrossRef]
- Malla, M.A.; Dubey, A.; Yadav, S.; Kumar, A.; Hashem, A.; Abd-Allah, E.F. Understanding and designing the strategies for the microbe-mediated remediation of environmental contaminants using omics approaches. Front. Microbiol. 2018, 9, 1132. [Google Scholar] [CrossRef]
- Copley, S.D. Evolution of efficient pathways for degradation of anthropogenic chemicals. Nat. Chem. Biol. 2009, 5, 559–566. [Google Scholar] [CrossRef]
- Baoune, H.; Aparicio, J.D.; Acuña, A.; El Hadj-khelil, A.O.; Sanchez, L.; Polti, M.A.; Alvarez, A. Effectiveness of the Zea mays-Streptomyces association for the phytoremediation of petroleum hydrocarbons impacted soils. Ecotoxicol. Environ. Saf. 2019, 184, 109591. [Google Scholar] [CrossRef]
- Chen, W.; Kong, Y.; Li, J.; Sun, Y.; Min, J.; Hu, X. Enhanced biodegradation of crude oil by constructed bacterial consortium comprising salt-tolerant petroleum degraders and biosurfactant producers. Int. Biodeterior. Biodegrad. 2020, 154, 105047. [Google Scholar] [CrossRef]
- Nevoigt, E. Progress in metabolic engineering of Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2008, 72, 379–412. [Google Scholar] [CrossRef]
- Bailey, J. Toward a science of metabolic engineering. Science 1991, 252, 1668–1675. [Google Scholar] [CrossRef] [PubMed]
- Jawed, K.; Yazdani, S.S.; Koffas, M.A. Advances in the development and application of microbial consortia for metabolic engineering. Metab. Eng. Commun. 2019, 9, e00095. [Google Scholar] [CrossRef]
- Weeks, A.M.; Chang, M.C. Constructing de novo biosynthetic pathways for chemical synthesis inside living cells. Biochemistry 2011, 50, 5404–5418. [Google Scholar] [CrossRef] [PubMed]
- Markina, N.M.; Kotlobay, A.A.; Tsarkova, A.S. Heterologous Metabolic Pathways: Strategies for Optimal Expression in Eukaryotic Hosts. Acta Nat. 2020, 12, 28–39. [Google Scholar] [CrossRef]
- Khanijou, J.K.; Kulyk, H.; Bergès, C.; Khoo, L.W.; Ng, P.; Yeo, H.C.; Helmy, M.; Bellvert, F.; Chew, W.; Selvarajoo, K. Metabolomics and modelling approaches for systems metabolic engineering. Metab. Eng. Commun. 2022, 15, e00209. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yan, J.; Wang, L.; Zhang, Y.; Liu, D.; Geng, H.; Xiong, L. Characterization of the phthalate acid catabolic gene cluster in phthalate acid esters transforming bacterium-Gordonia sp. strain HS-NH1. Int. Biodeterior. Biodegrad. 2016, 106, 34–40. [Google Scholar] [CrossRef]
- Flannagan, R.S.; Linn, T.; Valvano, M.A. A system for the construction of targeted unmarked gene deletions in the genus Burkholderia. Environ. Microbiol. 2008, 10, 1652–1660. [Google Scholar] [CrossRef]
- Parellada, E.A.; Igarza, M.; Isacc, P.; Bardón, A.; Ferrero, M.; Ameta, K.L.; Neske, A. Squamocin, an annonaceous acetogenin, enhances naphthalene degradation mediated by Bacillus atrophaeus CN4. Rev. Argent. Microbiol. 2017, 49, 282–288. [Google Scholar] [CrossRef]
- Nakamura, C.E.; Whited, G.M. Metabolic engineering for the microbial production of 1, 3-propanediol. Curr. Opin. Biotechnol. 2003, 14, 454–459. [Google Scholar] [CrossRef]
- Devers, M.; Rouard, N.; Martin-Laurent, F. Fitness drift of an atrazine-degrading population under atrazine selection pressure. Environ. Microbiol. 2008, 10, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Dellamatrice, P.M.; Silva-Stenico, M.E.; Moraes, L.A.B.D.; Fiore, M.F.; Monteiro, R.T.R. Degradation of textile dyes by cyanobacteria. Braz. J. Microbiol. 2017, 48, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Ghoshal, A.K. Bioremediation of petroleum wastewater by hyper-phenol tolerant Bacillus cereus: Preliminary studies with laboratory-scale batch process. Bioengineered 2017, 8, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Yang, J.W.; Cho, W.; Kwak, S.; Park, S.; Lim, Y.; Choe, J.W.; Kim, H.S. Oxidative biodegradation of 4-chlorophenol by using recombinant monooxygenase cloned and overexpressed from Arthrobacter chlorophenolicus A6. Bioresour. Technol. 2017, 240, 123–129. [Google Scholar] [CrossRef]
- Mishra, S.; Lin, Z.; Pang, S.; Zhang, W.; Bhatt, P.; Chen, S. Recent Advanced Technologies for the Characterization of Xenobiotic-Degrading Microorganisms and Microbial Communities. Front. Bioeng. Biotechnol. 2021, 9, 632059. [Google Scholar] [CrossRef]
- Singha, L.P.; Pandey, P. Rhizosphere assisted bioengineering approaches for the mitigation of petroleum hydrocarbons contamination in soil. Crit. Rev. Biotechnol. 2021, 41, 749–766. [Google Scholar] [CrossRef]
- Azubuike, C.C.; Chikere, C.B.; Okpokwasili, G.C. Bioremediation techniques—Classification based on site of application: Principles, advantages, limitations and prospects. World J. Microbiol. Biotechnol. 2016, 32, 180. [Google Scholar] [CrossRef]
- Popoola, L.T.; Yusuff, A.S. Optimization and characterization of crude oil contaminated soil bioremediation using bacteria isolates: Plant growth effect. S. Afr. J. Chem. Eng. 2021, 37, 206–213. [Google Scholar]
- Calero, P.; Nikel, P.I. Chasing bacterial chassis for metabolic engineering: A perspective review from classical to non-traditional microorganisms. Microb. Biotechnol. 2019, 12, 98–124. [Google Scholar] [CrossRef]
- Zhang, W.; Lin, Z.; Pang, S.; Bhatt, P.; Chen, S. Insights Into the Biodegradation of Lindane (γ-Hexachlorocyclohexane) Using a Microbial System. Front. Microbiol. 2020, 11, 522. [Google Scholar] [CrossRef]
- Singh, B.K.; Walker, A. Microbial degradation of organophosphorus compounds. FEMS Microbiol. Rev. 2006, 30, 428–471. [Google Scholar] [CrossRef] [PubMed]
- Trögl, J.; Chauhan, A.; Ripp, S.; Layton, A.C.; Kuncová, G.; Sayler, G.S. Pseudomonas fluorescens HK44: Lessons learned from a model whole-cell bioreporter with a broad application history. Sensors 2012, 12, 1544–1571. [Google Scholar] [CrossRef] [PubMed]
- Lawson, C.E.; Wu, S.; Bhattacharjee, A.S.; Hamilton, J.J.; McMahon, K.D.; Goel, R.; Noguera, D.R. Metabolic network analysis reveals microbial community interactions in anammox granules. Nat. Commun. 2017, 8, 15416. [Google Scholar] [CrossRef]
- Gupta, A.; Gupta, R.; Singh, R.L. Microbes and environment. In Principles and Applications of Environmental Biotechnology for a Sustainable Future; Springer: Berlin/Heidelberg, Germany, 2017; pp. 43–84. [Google Scholar]
- Koussa, J.; Chaiboonchoe, A.; Salehi-Ashtiani, K. Computational approaches for microalgal biofuel optimization: A review. BioMed Res. Int. 2014, 2014, 649453. [Google Scholar] [CrossRef] [PubMed]
- Massie, C.E.; Mills, I.G. Mapping protein–DNA interactions using ChIP-sequencing. In Transcriptional Regulation: Methods and Protocols; Humana Press: Totowa, NJ, USA, 2012; pp. 157–173. [Google Scholar]
- Norsigian, C.J.; Pusarla, N.; McConn, J.L.; Yurkovich, J.T.; Dräger, A.; Palsson, B.O.; King, Z. BiGG Models 2020: Multi-strain genome-scale models and expansion across the phylogenetic tree. Nucleic Acids Res. 2020, 48, D402–D406. [Google Scholar] [CrossRef] [PubMed]
- Hafner, J.; MohammadiPeyhani, H.; Sveshnikova, A.; Scheidegger, A.; Hatzimanikatis, V. Updated ATLAS of biochemistry with new metabolites and improved enzyme prediction power. ACS Synth. Biol. 2020, 9, 1479–1482. [Google Scholar] [CrossRef]
- Shuikan, A.M.; Hozzein, W.N.; Alshuwaykan, R.M.; Arif, I.A. Metabolomics and Genetic Engineering for Secondary Metabolites Discovery. In Secondary Metabolites: Trends and Reviews; IntechOpen: London, UK, 2022; p. 71. [Google Scholar]
- Kanehisa, M. Enzyme annotation and metabolic reconstruction using KEGG. In Protein Function Prediction: Methods and Protocols; Humana Press: Totowa, NJ, USA, 2017; pp. 135–145. [Google Scholar]
- Caspi, R.; Foerster, H.; Fulcher, C.A.; Hopkinson, R.; Ingraham, J.; Kaipa, P.; Krummenacker, M.; Paley, S.; Pick, J.; Rhee, S.Y.; et al. MetaCyc: A multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 2006, 34 (Suppl. S1), D511–D516. [Google Scholar] [CrossRef] [PubMed]
- Schomburg, I.; Chang, A.; Placzek, S.; Söhngen, C.; Rother, M.; Lang, M.; Munaretto, C.; Ulas, S.; Stelzer, M.; Grote, A.; et al. BRENDA in 2013: Integrated reactions, kinetic data, enzyme function data, improved disease classification: New options and contents in BRENDA. Nucleic Acids Res. 2012, 41, D764–D772. [Google Scholar] [CrossRef]
- Wang, L.; Dash, S.; Ng, C.Y.; Maranas, C.D. A review of computational tools for design and reconstruction of metabolic pathways. Synth. Syst. Biotechnol. 2017, 2, 243–252. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, Z.; Wei, D. Progress in the applications of flux analysis of metabolic networks. Chin. Sci. Bull. 2010, 55, 2315–2322. [Google Scholar] [CrossRef]
- Dangi, A.K.; Sharma, B.; Hill, R.T.; Shukla, P. Bioremediation through microbes: Systems biology and metabolic engineering approach. Crit. Rev. Biotechnol. 2019, 39, 79–98. [Google Scholar] [CrossRef]
- Jhariya, U.; Srivastava, S.; Das, S.; Bombaywala, S.; Mahallea, S.; Dafale, N.A. Understanding the role of genetic and protein networking involved in microbial bioremediation. In Bioremediation of Environmental Pollutants: Emerging Trends and Strategies; Springer: Berlin/Heidelberg, Germany, 2022; pp. 187–219. [Google Scholar]
- Moriya, Y.; Shigemizu, D.; Hattori, M.; Tokimatsu, T.; Kotera, M.; Goto, S.; Kanehisa, M. PathPred: An enzyme-catalyzed metabolic pathway prediction server. Nucleic Acids Res. 2010, 38 (Suppl. S2), W138–W143. [Google Scholar] [CrossRef] [PubMed]
- Finley, S.D.; Broadbelt, L.J.; Hatzimanikatis, V. Computational framework for predictive biodegradation. Bio-Technol. Bioeng. 2009, 104, 1086–1097. [Google Scholar] [CrossRef] [PubMed]
- Hadadi, N. Computational Studies on Cellular Metabolism: From Biochemical Pathways to Complex Metabolic Networks; EPFL: Lausanne, Switzerland, 2015; p. 6667. [Google Scholar]
- Pazos, F.; Guijas, D.; Valencia, A.; De Lorenzo, V. MetaRouter: Bioinformatics for bioremediation. Nucleic Acids Res. 2005, 33, D588–D592. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Van Raaphorst, R.; Takano, E.; Breitling, R. Computational tools for the synthetic design of biochemical pathways. Nat. Rev. Microbiol. 2012, 10, 191–202. [Google Scholar] [CrossRef]
- Seressiotis, A.; Bailey, J.E. MPS: An artificially intelligent software system for the analysis and synthesis of metabolic pathways. Biotechnol. Bioeng. 1988, 31, 587–602. [Google Scholar] [CrossRef]
- Spurgeon, D.; Lahive, E.; Robinson, A.; Short, S.; Kille, P. Species Sensitivity to Toxic Substances: Evolution, Ecology and Applications. Front. Environ. Sci. 2020, 8, 588380. [Google Scholar] [CrossRef]
- Belfield, S.J.; Firman, J.W.; Enoch, S.J.; Madden, J.C.; Tollefsen, K.E.; Cronin, M.T. A Review of Quantitative Structure-Activity Relationship Modelling Approaches to Predict the Toxicity of Mixtures. Comput. Toxicol. 2022, 25, 100251. [Google Scholar] [CrossRef]
- Schmidt, U.; Struck, S.; Gruening, B.; Hossbach, J.; Jaeger, I.S.; Parol, R.; Lindequist, U.; Teuscher, E.; Preissner, R. SuperToxic: A comprehensive database of toxic compounds. Nucleic Acids Res. 2009, 37 (Suppl. S1), 295–299. [Google Scholar] [CrossRef]
- Judson, R.; Richard, A.; Dix, D.J.; Houck, K.; Martin, M.; Kavlock, R.; Dellarco, V.; Henry, T.; Holderman, T.; Sayre, P.; et al. The toxicity data landscape for environmental chemicals. Environ. Health Perspect. 2009, 117, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Planson, A.G.; Carbonell, P.; Paillard, E.; Pollet, N.; Faulon, J.L. Compound toxicity screening and structure-activity relationship modeling in Escherichia coli. Biotechnol. Bioeng. 2012, 109, 846–850. [Google Scholar] [CrossRef] [PubMed]
- Petsas, A.S.; Vagi, M.C. Trends in the bioremediation of pharmaceuticals and other organic contaminants using native or genetically modified microbial strains: A review. Curr. Pharm. Biotechnol. 2019, 20, 787–824. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jiang, Y.; Guengerich, X.F.P.; Ma, L.; Li, S.; Zhang, W. Engineering cytochrome P450 enzyme systems for biomedical and biotechnological applications. J. Biol. Chem. 2020, 295, 833–849. [Google Scholar] [CrossRef] [PubMed]
- Pieper, D.H.; Reineke, W. Engineering bacteria for bioremediation. Curr. Opin. Biotechnol. 2000, 11, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, L.M.; Pfaender, F.K. The effect of inorganic and organic supplements on the microbial degradation of phenanthrene and pyrene in soils. Biodegradation 1997, 8, 1–13. [Google Scholar] [CrossRef]
- Kowalko, J.E.; Ma, L.; Jeffery, W.R. Genome editing in Astyanax mexicanus using transcription activator-like effector nucleases (TALENs). JoVE J. Vis. Exp. 2016, 112, e54113. [Google Scholar]
- Wu, C.; Li, F.; Yi, S.; Ge, F. Genetically engineered microbial remediation of soils co-contaminated by heavy metals and polycyclic aromatic hydrocarbons: Advances and ecological risk assessment. J. Environ. Manag. 2021, 296, 113185. [Google Scholar] [CrossRef]
- Kumari, S.; Mangwani, N.; Das, S. Synergistic effect of quorum sensing genes in biofilm development and PAHs degradation by a marine bacterium. Bioengineered 2016, 7, 205–211. [Google Scholar] [CrossRef]
- Cases, I.; De Lorenzo, V. The black cat/white cat principle of signal integration in bacterial promoters. EMBO J. 2001, 20, 1–11. [Google Scholar] [CrossRef]
- Wang, L.; Peng, R.; Tian, Y.; Xu, J.; Wang, B.; Han, H.; Fu, X.; Gao, J.; Yao, Q. Metabolic engineering of Escherichia coli for efficient degradation of 4-fluorophenol. AMB Express 2022, 12, 55. [Google Scholar] [CrossRef]
- Wicker, J.; Fenner, K.; Ellis, L.; Wackett, L.; Kramer, S. Predicting biodegradation products and pathways: A hybrid knowledge- and machine learning-based approach. Bioinformatics 2010, 26, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Park, J.M.; Kim, T.Y. Application of metabolic flux analysis in metabolic engineering. In Methods in Enzymology, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2011; Volume 498. [Google Scholar] [CrossRef]
- Burgard, A.P.; Pharkya, P.; Maranas, C.D. OptKnock: A Bilevel Programming Framework for Identifying Gene Knockout Strategies for Microbial Strain Optimization. Biotechnol. Bioeng. 2003, 84, 647–657. [Google Scholar] [CrossRef]
- Wang, X.; Yu, L.; Chen, S. UP Finder: A COBRA toolbox extension for identifying gene overexpression strategies for targeted overproduction. Metab. Eng. Commun. 2017, 5, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, S.; Suthers, P.F.; Maranas, C.D. OptForce: An optimization procedure for identifying all genetic manipulations leading to targeted overproductions. PLoS Comput. Biol. 2010, 6, e1000744. [Google Scholar] [CrossRef]
- Hoops, S.; Gauges, R.; Lee, C.; Pahle, J.; Simus, N.; Singhal, M.; Xu, L.; Mendes, P.; Kummer, U. COPASI—A Complex Pathway Simulator. Bioinformatics 2006, 22, 3067–3074. [Google Scholar] [CrossRef] [PubMed]
- Minkiewicz, P.; Darewicz, M.; Iwaniak, A.; Bucholska, J.; Starowicz, P.; Czyrko, E. Internet databases of the properties, enzymatic reactions, and metabolism of small molecules—Search options and applications in food science. Int. J. Mol. Sci. 2016, 17, 2039. [Google Scholar] [CrossRef]
- Lange, V.; Picotti, P.; Domon, B.; Aebersold, R. Selected reaction monitoring for quantitative proteomics: A tutorial. Mol. Syst. Biol. 2008, 4, 222. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, R.; Wu, C.H.; Hazen, T.C. Systems biology approach to bioremediation. Curr. Opin. Bio-Technol. 2012, 23, 483–490. [Google Scholar] [CrossRef]
- Wanichthanarak, K.; Fahrmann, J.F.; Grapov, D. Genomic, proteomic, and metabolomic data integration strategies. Biomark. Insights 2015, 10, 1–6. [Google Scholar] [CrossRef]
- Manzoni, C.; Kia, D.A.; Vandrovcova, J.; Hardy, J.; Wood, N.W.; Lewis, P.A.; Ferrari, R. Genome, transcriptome and proteome: The rise of omics data and their integration in biomedical sciences. Brief. Bioinform. 2018, 19, 286–302. [Google Scholar] [CrossRef]
- Handelsman, J. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 2004, 68, 669–685. [Google Scholar] [CrossRef]
- Desai, C.; Pathak, H.; Madamwar, D. Advances in molecular and “omics” technologies to gauge microbial communities and bioremediation at xenobiotic/anthropogen contaminated sites. Bioresour. Technol. 2010, 101, 1558–1569. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Kundu, A.; Banerjee, T.D.; Mohapatra, B.; Roy, A.; Manna, R.; Sar, P.; Kazy, S.K. Genome analysis of crude oil degrading Franconibacter pulveris strain DJ34 revealed its genetic basis for hydrocarbon degradation and survival in oil contaminated environment. Genomics 2017, 109, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Ikrema, H.; Eastman, A.W.; Brian, W.; Eltayeb, M.; Yanful, E.K.; Ze-Chun, Y. Complete Genome Sequence of Arthrobacter sp. Strain LS16, Isolated from Agricultural Soils with Potential for Applications in Bioremediation and Bioproducts. Microbiol. Resour. Announc. 2016, 4, e01586-15. [Google Scholar]
- Neifar, M.; Chouchane, H.; Najjari, A.; El Hidri, D.; Mahjoubi, M.; Ghedira, K.; Naili, F.; Soufi, L.; Raddadi, N.; Sghaier, H.; et al. Genome analysis provides insights into crude oil degradation and biosurfactant production by extremely halotolerant Halomonas desertis G11 isolated from Chott El-Djerid salt-lake in Tunisian desert. Genomics 2019, 111, 1802–1814. [Google Scholar] [CrossRef] [PubMed]
- Kube, M.; Beck, A.; Meyerdierks, A.; Amann, R.; Reinhardt, R.; Rabus, R. A catabolic gene cluster for anaerobic benzoate degradation in methanotrophic microbial Black Sea mats. Syst. Appl. Microbiol. 2005, 28, 287–294. [Google Scholar] [CrossRef]
- Lamendella, R.; Strutt, S.; Borglin, S.; Chakraborty, R.; Tas, N.; Mason, O.U.; Hultman, J.; Prestat, E.; Hazen, T.C.; Jansson, J.K. Assessment of the Deepwater Horizon oil spill impact on Gulf coast microbial communities. Front. Microbiol. 2014, 5, 130. [Google Scholar] [CrossRef]
- Xiong, W.; Abraham, P.E.; Li, Z.; Pan, C.; Hettich, R.L. Microbial metaproteomics for characterizing the range of metabolic functions and activities of human gut microbiota. Proteomics 2015, 15, 3424–3438. [Google Scholar] [CrossRef]
- Gillan, D.C. Metal resistance systems in cultivated bacteria: Are they found in complex communities? Curr. Opin. Biotechnol. 2016, 38, 123–130. [Google Scholar] [CrossRef]
- Kim, S.J.; Kweon, O.; Sutherland, J.B.; Kim, H.L.; Jones, R.C.; Burback, B.L.; Graves, S.W.; Psurny, E.; Cerniglia, C.E. Dynamic response of Mycobacterium vanbaalenii PYR-1 to BP Deepwater Horizon crude oil. Appl. Environ. Microbiol. 2015, 81, 4263–4276. [Google Scholar] [CrossRef]
- Luo, F.; Gitiafroz, R.; Devine, C.E.; Gong, Y.; Hug, L.A.; Raskin, L.; Edwards, E.A. Metatranscriptome of an an-aerobic benzene-degrading, nitrate-reducing enrichment culture reveals involvement of carboxylation in benzene ring activation. Appl. Environ. Microbiol. 2014, 80, 4095–4107. [Google Scholar] [CrossRef]
- Cao, B.; Geng, A.; Loh, K.C. Induction of ortho-and meta-cleavage pathways in Pseudomonas in biodegradation of high benzoate concentration: MS identification of catabolic enzymes. Appl. Microbiol. Biotechnol. 2008, 81, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Armengaud, J.; Trapp, J.; Pible, O.; Geffard, O.; Chaumot, A.; Hartmann, E.M. Non-model organisms, a species endangered by proteogenomics. J. Proteom. 2014, 105, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics technologies. PLoS Comput. Biol. 2017, 13, e1005457. [Google Scholar] [CrossRef] [PubMed]
- Jennings, L.K.; Chartrand, M.M.; Lacrampe-Couloume, G.; Lollar, B.S.; Spain, J.C.; Gossett, J.M. Proteomic and transcriptomic analyses reveal genes upregulated by cis-dichloroethene in Polaromonas sp. strain JS666. Appl. Environ. Microbiol. 2009, 75, 3733–3744. [Google Scholar] [CrossRef] [PubMed]
- Das, D.; Mawlong, G.T.; Sarki, Y.N.; Singh, A.K.; Chikkaputtaiah, C.; Boruah, H.P.D. Transcriptome analysis of crude oil degrading Pseudomonas aeruginosa strains for identification of potential genes involved in crude oil degradation. Gene 2020, 755, 144909. [Google Scholar] [CrossRef] [PubMed]
- Tribelli, P.M.; Rossi, L.; Ricardi, M.M.; Gomez-Lozano, M.; Molin, S.; Raiger Iustman, L.J.; Lopez, N.I. Microaer-ophilic alkane degradation in Pseudomonas extremaustralis: A transcriptomic and physiological approach. J. Ind. Microbiol. Biotechnol. 2018, 45, 15–23. [Google Scholar] [CrossRef]
- Hong, Y.H.; Ye, C.C.; Zhou, Q.Z.; Wu, X.Y.; Yuan, J.P.; Peng, J.; Deng, H.; Wang, J.H. Genome sequencing reveals the potential of Achromobacter sp. HZ01 for bioremediation. Front. Microbiol. 2017, 8, 1507. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Xu, H.; Qiu, S.; Wang, X. Cell metabolomics. Omics J. Integr. Biol. 2013, 17, 495–501. [Google Scholar] [CrossRef]
- Mani, I. Metagenomics approach for bioremediation: Challenges and perspectives. In Bioremediation of Pollutants; Elsevier: Amsterdam, The Netherlands, 2020; pp. 275–285. [Google Scholar]
- Davey, M.E.; O’toole, G.A. Microbial biofilms: From ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 2000, 64, 847–867. [Google Scholar] [CrossRef]
- Furbank, R.T.; Tester, M. Phenomics-technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 2011, 16, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Salt, D.E.; Baxter, I.; Lahner, B. Ionomics and the study of the plant ionome. Annu. Rev. Plant Biol. 2008, 59, 709–733. [Google Scholar] [CrossRef]
- Zhang, W.; Li, F.; Nie, L. Integrating multiple ‘omics’ analysis for microbial biology: Application and methodologies. Microbiology 2010, 156, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, V.M.; Ivanova, N.N.; Szeto, E.; Palaniappan, K.; Chu, K.; Dalevi, D.; Chen, I.M.A.; Grechkin, Y.; Dubchak, I.; Anderson, I.; et al. IMG/M: A data management and analysis system for metagenomes. Nucleic Acids Res. 2007, 36 (Suppl. S1), D534–D538. [Google Scholar] [CrossRef]
- Chen, I.M.A.; Markowitz, V.M.; Chu, K.; Palaniappan, K.; Szeto, E.; Pillay, M.; Ratner, A.; Huang, J.; Andersen, E.; Huntemann, M.; et al. IMG/M: Integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 2016, 45, gkw929. [Google Scholar] [CrossRef]
- Sharma, V.K.; Kumar, N.; Prakash, T.; Taylor, T.D. MetaBioME: A database to explore commercially useful enzymes in metagenomic datasets. Nucleic Acids Res. 2010, 38 (Suppl. S1), D468–D472. [Google Scholar] [CrossRef]
- Hoff, K.J.; Lingner, T.; Meinicke, P.; Tech, M. Orphelia: Predicting genes in metagenomic sequencing reads. Nucleic Acids Res. 2009, 37 (Suppl. S2), W101–W105. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Bilal, M.; Iqbal, H.M.; Raj, A. Trends in predictive biodegradation for sustainable mitigation of envi-ronmental pollutants: Recent progress and future outlook. Sci. Total Environ. 2021, 770, 144561. [Google Scholar] [CrossRef]
- Raman, K.; Chandra, N. Flux balance analysis of biological systems: Applications and challenges. Brief. Bio-Inform. 2009, 10, 435–449. [Google Scholar] [CrossRef]
- Antoniewicz, M.R. Methods and advances in metabolic flux analysis: A mini-review. J. Ind. Microbiol. Biotechnol. 2015, 42, 317–325. [Google Scholar] [CrossRef]
- Sturm, M.; Bertsch, A.; Gröpl, C.; Hildebrandt, A.; Hussong, R.; Lange, E.; Pfeifer, N.; Schulz-Trieglaff, O.; Zerck, A.; Reinert, K.; et al. OpenMS—An open-source software framework for mass spectrometry. BMC Bioinform. 2008, 9, 163. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef] [PubMed]
- Haug, K.; Cochrane, K.; Nainala, V.C.; Williams, M.; Chang, J.; Jayaseelan, K.V.; O’Donovan, C. MetaboLights: A resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020, 48, D440–D444. [Google Scholar] [CrossRef]
- Weber, T.; Kim, H.U. The secondary metabolite bioinformatics portal: Computational tools to facilitate synthetic biology of secondary metabolite production. Synth. Syst. Biotechnol. 2016, 1, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Pande, V.; Pandey, S.C.; Sati, D.; Pande, V.; Samant, M. Bioremediation: An emerging effective approach towards environment restoration. Environ. Sustain. 2020, 3, 91–103. [Google Scholar] [CrossRef]
- Cheng, A.A.; Lu, T.K. Synthetic biology: An emerging engineering discipline. Annu. Rev. Biomed. Eng. 2012, 14, 155–178. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, K.; Loor, J.J. Application of top-down and bottom-up systems approaches in ruminant physiology and metabolism. Curr. Genom. 2012, 13, 379–394. [Google Scholar] [CrossRef]
- Bhatt, P.; Gangola, S.; Bhandari, G.; Zhang, W.; Maithani, D.; Mishra, S.; Chen, S. New insights into the degradation of synthetic pollutants in contaminated environments. Chemosphere 2021, 268, 128827. [Google Scholar] [CrossRef]
- Wang, S.; Tang, H.; Peng, F.; Yu, X.; Su, H.; Xu, P.; Tan, T. Metabolite-based mutualism enhances hydrogen production in a two-species microbial consortium. Commun. Biol. 2019, 2, 82. [Google Scholar] [CrossRef]
- Patowary, K.; Patowary, R.; Kalita, M.C.; Deka, S. Development of an efficient bacterial consortium for the potential remediation of hydrocarbons from contaminated sites. Front. Microbiol. 2016, 7, 1092. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, H.C.; Carlson, R.P. Microbial consortia engineering for cellular factories: In vitro to in silico systems. Comput. Struct. Biotechnol. J. 2012, 3, e201210017. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Díaz, V.; Pedroza-Rodríguez, A.M.; Ramos-Monroy, O.; Castillo-Carvajal, L.C. Synthetic Biology: A New Era in Hydrocarbon Bioremediation. Processes 2022, 10, 712. [Google Scholar] [CrossRef]
- Kim, D.; Lee, J.W. Genetic biocontainment systems for the safe use of engineered microorganisms. Biotechnol. Bioprocess Eng. 2020, 25, 974–984. [Google Scholar] [CrossRef]
- Liang, Y.; Ma, A.; Zhuang, G. Construction of environmental synthetic microbial consortia: Based on engineering and ecological principles. Front. Microbiol. 2022, 13, 437. [Google Scholar] [CrossRef] [PubMed]
- Esvelt, K.M.; Wang, H.H. Genome-scale engineering for systems and synthetic biology. Mol. Syst. Biol. 2013, 9, 641. [Google Scholar] [CrossRef] [PubMed]
- Dvořák, P.; Nikel, P.I.; Damborský, J.; de Lorenzo, V. Bioremediation 3.0: Engineering pollutant-removing bacteria in the times of systemic biology. Biotechnol. Adv. 2017, 35, 845–866. [Google Scholar] [CrossRef]
- Budisa, N. Engineering the Genetic Code: Expanding the Amino Acid Repertoire for the Design of Novel Proteins; John Wiley Sons: Hoboken, NJ, USA, 2006. [Google Scholar]
- Holland, J.H. Adaptation in Natural and Artificial Systems: An Introductory Analysis with Applications to Biology, Control, and Artificial Intelligence; MIT Press: Cambridge, MA, USA, 1992. [Google Scholar]
- Isaacs, F.J.; Carr, P.A.; Wang, H.H.; Lajoie, M.J.; Sterling, B.; Kraal, L.; Tolonen, A.C.; Gianoulis, T.A.; Goodman, D.B.; Reppas, N.B.; et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science 2011, 333, 348–353. [Google Scholar] [CrossRef]
- Wang, B.; Kitney, R.I.; Joly, N.; Buck, M. Engineering modular and orthogonal genetic logic gates for robust digi-tal-like synthetic biology. Nat. Commun. 2011, 2, 508. [Google Scholar] [CrossRef]
- Benedetti, I.; de Lorenzo, V.; Nikel, P.I. Genetic programming of catalytic Pseudomonas putida biofilms for boosting biodegradation of haloalkanes. Metab. Eng. 2016, 33, 109–118. [Google Scholar] [CrossRef]
- Benedetti, I. Design of Standardized Molecular Tools to Analyze Regulatory Properties and Biotechnological Applications of the Soil Bacterium Pseudomonas Putida. Doctoral Dissertation, Universidad Autónoma de Madrid, Madrid, Spain, 2014. [Google Scholar]
- Chung, T.H.; Meshref, M.N.; Dhar, B.R. A review and roadmap for developing microbial electrochemical cell-based biosensors for recalcitrant environmental contaminants, emphasis on aromatic compounds. Chem. Eng. J. 2021, 424, 130245. [Google Scholar] [CrossRef]
- Meeske, A.J.; Nakandakari-Higa, S.; Marraffini, L.A. Cas13-induced cellular dormancy prevents the rise of CRISPR-resistant bacteriophage. Nature 2019, 570, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Scheben, A.; Wolter, F.; Batley, J.; Puchta, H.; Edwards, D. Towards CRISPR/Cas crops-bringing together genomics and genome editing. New Phytol. 2017, 216, 682–698. [Google Scholar] [CrossRef] [PubMed]
- Durai, S.; Mani, M.; Kandavelou, K.; Wu, J.; Porteus, M.H.; Chandrasegaran, S. Zinc finger nucleases: Custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005, 33, 5978–5990. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Lee, H.J.; Kim, H.; Cho, S.W.; Kim, J.S. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009, 19, 1279–1288. [Google Scholar] [CrossRef]
- Sprink, T.; Metje, J.; Hartung, F. Plant genome editing by novel tools: TALEN and other sequence specific nucleases. Curr. Opin. Biotechnol. 2015, 32, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Yaashikaa, P.R.; Devi, M.K.; Kumar, P.S. Engineering microbes for enhancing the degradation of environmental pollutants: A detailed review on synthetic biology. Environ. Res. 2022, 214, 113868. [Google Scholar] [CrossRef]
- Panigrahy, N.; Priyadarshini, A.; Sahoo, M.M.; Verma, A.K.; Daverey, A.; Sahoo, N.K. A comprehensive review on eco-toxicity and biodegradation of phenolics: Recent progress and future outlook. Environ. Technol. Innov. 2022, 27, 102423. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Q.; Guo, S.H.; Cheng, M.G.; Zhao, M.J.; Hong, Q.; Huang, X. Cloning, expression and mutation of a triazophos hydrolase gene from Burkholderia sp. SZL-1. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef]

| Sl. No. | Microorganism | Specific Polyaromatic Hydrocarbon (PAHs) | Degradation Percentage (%) | Concentration (mg/L) | Environment from Which Bacteria Are Isolated | References |
|---|---|---|---|---|---|---|
| 1 | Methylobacterium, Burkholderia and Stenotrophomonas. | Phenanthrene | 94.5 | 500 | Heavy-metal- and PAH-contaminated sites | [80] |
| Pyrene | 17.8 | 10 | ||||
| 2 | Pseudomonas brassicacearum strain MPDS | Naphthalene | 50 | PAH-contaminated sites | [81] | |
| Fluorene | 40.3 | 5 | ||||
| Dibenzofuran | 65.7 | 5 | ||||
| Dibenzothiophene | 32.1 | 5 | ||||
| 3 | Pseudomonas aeruginosa | Fluorene | 96 | 20 | Hydrocarbon-contaminated sites | [82] |
| Phenanthrene | 50 | 20 | ||||
| Pyrene | 41 | 20 | ||||
| 4 | Zhihengliuella sp. ISTPL4 | Phenanthrene | 87 | 250 | Contaminated frozen sites | [83] |
| 5 | Bacillus pumilus | Fluoranthene | 76.03 | 500 | Oil-spill sites | [84] |
| Phenanthrene | 87.98 | 500 | ||||
| 6 | Bacillus simplex | Fluoranthene | 86.89 | 500 | Oil-spill sites | [84] |
| Phenanthrene | 95.13 | 500 | ||||
| 7 | Pseudomonas stutzeri | Fluoranthene | 64.97 | 500 | Oil-spill sites | [84] |
| Phenanthrene | 86.32 | 500 | ||||
| 8 | Bosea, Arthrobacter, Paenibacillus, Bacillus, and Rhodococcus | Pyrene | 100 | Farmland | [85] | |
| Benzo [a]pyrene | 26.9–71.5 | |||||
| 9 | Sphingobium sp. NS7 | Pyrene | 5.6 | Farmland | [85] | |
| Benzo[a]pyrene | 8.6 | |||||
| 10 | Cellulosimicrobium cellulans CWS2 | Benzo[a]pyrene | 78.8 | 10 | PAH-contaminated soil | [86] |
| 11 | Pseudomonas sp. | Naphthalene | 95.3 | 100 | Plants from PAH-contaminated site | [87] |
| Fluoranthene | 87.9 | 100 | ||||
| Phenanthrene | 90.4 | 100 | ||||
| Pyrene | 6.9 | 100 | ||||
| 12 | Stenotrophomonas sp. | Naphthalene | 98.0 | 100 | Plants from PAH-contaminated sites | [87] |
| Fluoranthene | 83.1 | 100 | ||||
| Phenanthrene | 87.8 | 100 | ||||
| Pyrene | 14.4 | 100 | ||||
| Benzo[a]pyrene | 1.6 | 10 | ||||
| 13 | Micrococcus luteus | Naphthalene | 68.7 | 1 | Petroleum-contaminated soil | [88] |
| Fluoranthene | 61.4 | 1 | ||||
| Phenanthrene | 62.9 | 1 | ||||
| Pyrene | 61.3 | 1 | ||||
| 14 | Kocuria rosea | Naphthalene | 59.8 | 1 | Petroleum-contaminated soil | [88] |
| Fluoranthene | 53.8 | 1 | ||||
| Phenanthrene | 54.6 | 1 | ||||
| Pyrene | 53.3 | 1 | ||||
| 15 | Serratia sp. PW7 | Pyrene | 51.2 | 50 | Plant from contaminated sites | [89] |
| 16 | Staphylococcus nepalensis | Pyrene | 93.25 | 50 | Diesel-contaminated soil | [90] |
| 17 | Sphingomonas koreensis ASU-06 | Naphthalene | 100 | 100 | Soil from oil refinery | [91] |
| Phenanthrene | 99 | 100 | ||||
| Pyrene | 92.7 | 100 | ||||
| Anthracene | 98 | 100 | ||||
| 18 | Streptomyces sp. | Fluoranthene | 92 | 100 | Bitumen-contaminated soil | [92] |
| Phenanthrene | 80 | 100 | ||||
| Pyrene | 28 | 100 | ||||
| Anthracene | 78.2 | 100 | ||||
| 19 | Ochrobactrum sp. VA1 | Anthracene | 88 | 3 | Petroleum- and coal-contaminated sites | [93] |
| Phenanthrene | 98 | 3 | ||||
| Naphthalene | 90 | 3 | ||||
| Fluorene | 97 | 3 | ||||
| Pyrene | 84 | 3 | ||||
| Benzo[e]pyrene | 50 | 1 | ||||
| Benzo[k]fluoranthene | 57 | 1 |
| Serial No. | Metabolic Engineering Technology | Types of Organisms Used | Specific Pollutants | Strategies | Result of Process | References |
|---|---|---|---|---|---|---|
| 1 | Introduction of entire gene clusters | A gene cluster from Gordonia sp. responsible for phthalate acid degradation (phtBAabcdCR) was expressed in E. coli BL21 (DE3) | Phthalate acid (PA) and protocatechuate acid (PCA) | Gene cluster containing complete catabolic pathways is introduced into a new host that can neutralize pollutants without inhibition. | Gene cluster encodes 3,4-phthalate dioxygenase, which totally oxidizes phthalate acid and is composed of reductase, ferredoxin, and oxygenase. | [136] |
| 2 | Engineered up-regulation of regulatory networks | Streptomyces coelicolor | Antibiotics | This technique entails manipulating the microorganisms so that they continually produce the activator, which further acts on specific targets. | Continuous and increased production of various secondary metabolites has been observed with continuous expression of Streptomyces antibiotic regulatory protein (SARP)-positive regulators. | [35] |
| 3 | Engineered down-regulation of regulatory networks | Streptomyces griseus | Repressor or inhibitor production is interrupted in bacteria, which is the basic principle of this process. | Chromomycin synthesis rises when pathway-specific repressors are turned off. | [139] | |
| 4 | Insertion and deletion of genes | Burkholderia cenocepacia K56–2 | To obtain a desirable phenotype, it may be required to modify the route by adding or deleting one or more genes. | The modified strain can be employed in environmental bioremediation since it is simpler to genetically modify and less likely to cause severe infections. | [137] | |
| 5 | Stimulation by providing precursors | Bacillus atrophaeus CN4 | Naphthalene | Precursor or inducers have the ability to induce specific catabolic pathways involved in bioremediation. | Squamocin, a kind of acetogenin that can break down naphthalene, was a biofilm-inducing agent in the studied bacteria. | [138] |
| 6 | Gene duplication | Pseudomonas sp. strain ADP | Atrazine | New genetic material is created by replicating the portion of genomic DNA that includes the gene responsible for protein coding. | The atzB gene, which encodes the second enzyme in the atrazine catabolic pathway, was tandem duplicated in this mutant strain of Pseudomonas sp. strain ADP. | [140] |
| 7 | Whole-genome duplication | Phormidium autumnale UTEX1580 | Dyes used in textile industry | Duplicating an organism’s whole genome, which over time leads to speciation and divergence. | The polyploid cells of Cyanobacterium were observed during the process of textile dye degradation. | [141] |
| 8 | Assembly Likelihood Evaluation (ALE) | Bacillus cereus | Wastewater (phenolic compounds) | Effectiveness of enzymes was increased by increasing the exposure time of the microbe to the toxic pollutant. | Enhanced degradation and a significant change in cell membrane was observed after prolonged exposure to xenobiotics. | [142] |
| 9 | Heterologous expression of genes | cphC-I and cphB from Arthrobacter chlorophenolicus, which encodes monooxygenase complex, were expressed in E. coli | Chlorophenolic compounds degradation | The gene or gene cluster is cloned and expressed in other competent bacterium to increase the production of important compounds. | The inducer was produced firmly under the influence of a strong promoter, which further regulates the production of various metabolites. | [143] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Das, N.; Das, A.; Das, S.; Bhatawadekar, V.; Pandey, P.; Choure, K.; Damare, S.; Pandey, P. Petroleum Hydrocarbon Catabolic Pathways as Targets for Metabolic Engineering Strategies for Enhanced Bioremediation of Crude-Oil-Contaminated Environments. Fermentation 2023, 9, 196. https://doi.org/10.3390/fermentation9020196
Das N, Das A, Das S, Bhatawadekar V, Pandey P, Choure K, Damare S, Pandey P. Petroleum Hydrocarbon Catabolic Pathways as Targets for Metabolic Engineering Strategies for Enhanced Bioremediation of Crude-Oil-Contaminated Environments. Fermentation. 2023; 9(2):196. https://doi.org/10.3390/fermentation9020196
Chicago/Turabian StyleDas, Nandita, Ankita Das, Sandeep Das, Vasudha Bhatawadekar, Prisha Pandey, Kamlesh Choure, Samir Damare, and Piyush Pandey. 2023. "Petroleum Hydrocarbon Catabolic Pathways as Targets for Metabolic Engineering Strategies for Enhanced Bioremediation of Crude-Oil-Contaminated Environments" Fermentation 9, no. 2: 196. https://doi.org/10.3390/fermentation9020196
APA StyleDas, N., Das, A., Das, S., Bhatawadekar, V., Pandey, P., Choure, K., Damare, S., & Pandey, P. (2023). Petroleum Hydrocarbon Catabolic Pathways as Targets for Metabolic Engineering Strategies for Enhanced Bioremediation of Crude-Oil-Contaminated Environments. Fermentation, 9(2), 196. https://doi.org/10.3390/fermentation9020196





