Enhancement of 2,3-Butanediol Production by Klebsiella pneumoniae: Emphasis on the Mediation of sRNA-SgrS on the Carbohydrate Utilization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biochemical Reagents
2.2. Strains, Media, and Growth Conditions
2.3. Obtainment of sgrS Gene Knockout and Overexpression Strains
2.4. Sugar Tolerance Test
2.5. Fermentation Kinetics
2.6. Quantitative Real-Time PCR (qRT-PCR) to Measure Gene Expression
2.7. Statistical Analysis
3. Results and Discussion
3.1. Glucose Tolerance of Each Strain
3.2. The Fermentation Results
3.3. Analysis of Differentially Expressed Genes
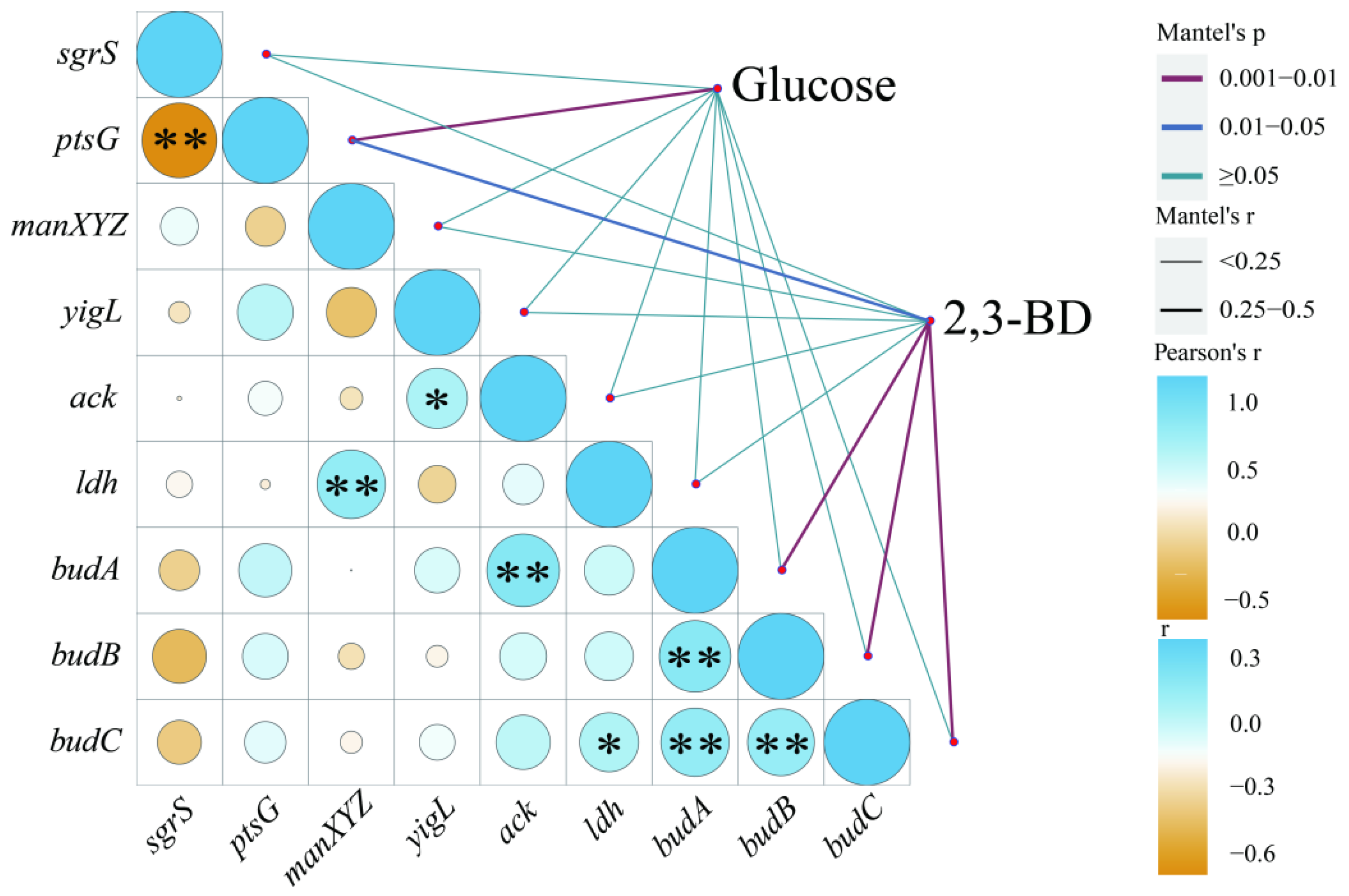
3.4. Effects of sRNA-SgrS on Carbon Metabolic Flux
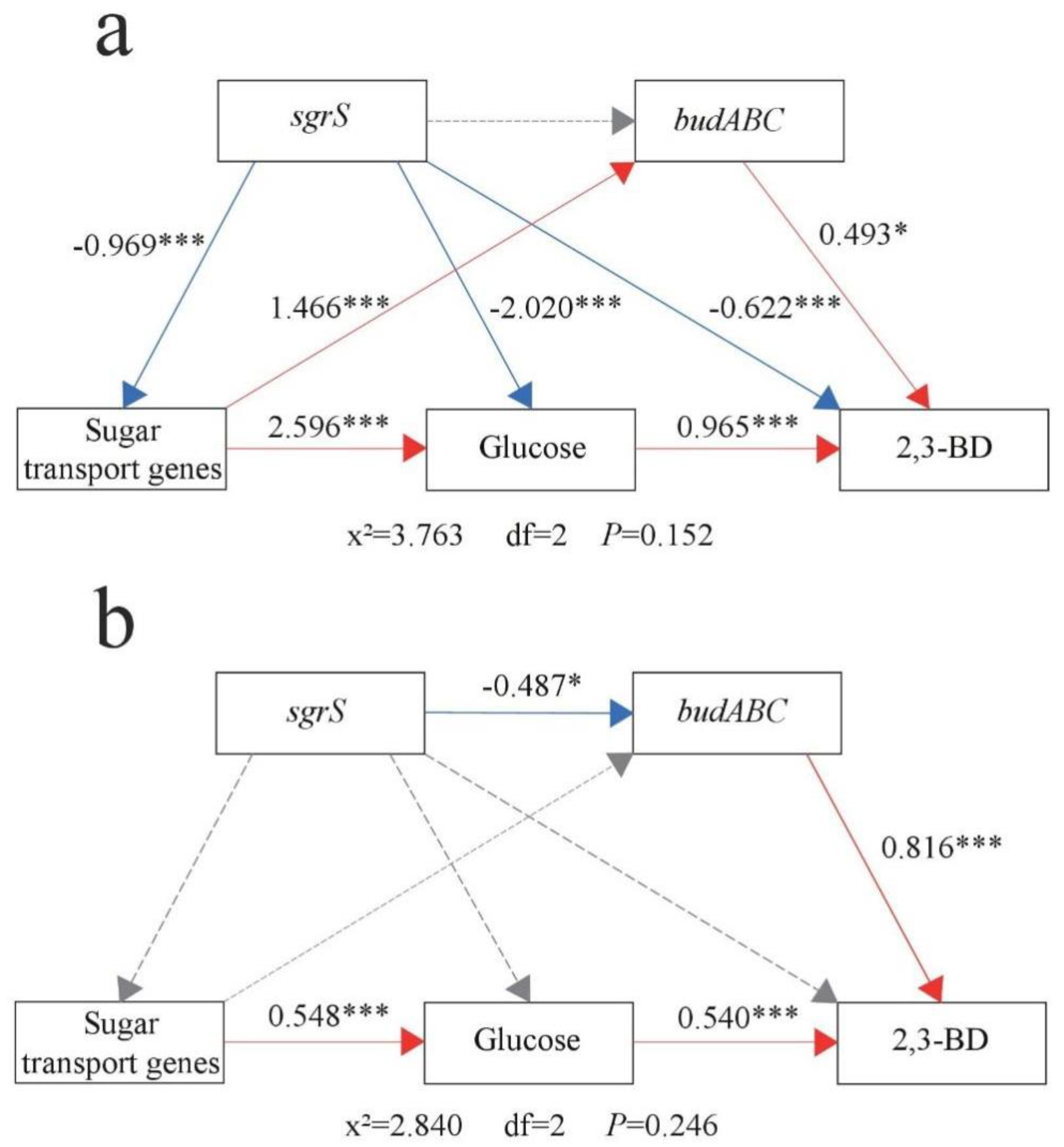
3.5. Relationship of sgrS, Sugar Transport Genes (ptsG, manXYZ), and budABC with Glucose Metabolism and 2,3-BD Production
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Garg, S.K.; Jain, A. Fermentative production of 2,3-butanediol: A review. Bioresour. Technol. 1995, 51, 103–109. [Google Scholar] [CrossRef]
- Tran, A.V.; Chambers, R.P. The dehydration of fermentative 2,3-butanediol into methyl ethyl ketone. Biotechnol. Bioeng. 1987, 29, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Long, S.K.; Patrick, R. Production of 2,3-Butylene Glycol from Citrus Wastes: I. The Aerobacter aerogenes Fermentation. Appl. Microbiol. 1961, 9, 244–248. [Google Scholar] [CrossRef]
- Sun, J.A.; Zhang, L.Y.; Rao, B.; Shen, L.Y.; Wei, Z.D. Enhanced acetoin production by Serratia marcescens H32 with expression of a water forming NADH oxidase. Bioresour. Technol. 2012, 119, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Qin, J.; Gao, C.; Hua, D.L.; Ma, C.Q.; Li, L.X.; Wang, Y.; Xu, P. Production of (2S, 3S)-2,3-butanediol and (3S)-acetoin from glucose using resting cells of Klebsiella pneumonia and Bacillus subtilis. Bioresour. Technol. 2011, 102, 10741–10744. [Google Scholar] [CrossRef]
- Ji, X.J.; Xia, Z.F.; Fu, N.H.; Nie, Z.K.; Shen, M.Q.; Tian, Q.Q.; Huang, H. Cofactor engineering through heterologous expression of an NADH oxidase and its impact on metabolic flux redistribution in Klebsiella pneumoniae. Biotechnol. Biofuels 2013, 6, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Ji, X.J.; Huang, H.; Ouyang, P.K. Microbial 2,3-butanediol production: A state-of-the-art review. Biotechnol. Adv. 2011, 29, 351–364. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, N.; Cheryan, M. Production of 2,3-butanediol by Klebsiella oxytoca. Appl. Microbiol. Biotechnol. 1989, 30, 440–443. [Google Scholar] [CrossRef]
- Celińska, E.; Grajek, W. Biotechnological production of 2,3-butanediol—current state and prospects. Biotechnol. Adv. 2009, 27, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Syu, M.J. Biological production of 2,3-butanediol. Appl. Microbiol. Biotechnol. 2001, 55, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.J.; Huang, H.; Du, J.; Zhu, J.G.; Ren, L.J.; Hu, N.; Li, S. Enhanced 2,3-butanediol production by Klebsiella oxytoca using a two-stage agitation speed control strategy. Bioresour. Technol. 2009, 100, 3410–3414. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.Y.; Jung, M.; Lee, J.; Oh, M.K. Production of 2,3-butanediol in Saccharomyces cerevisiae by in silico aided metabolic engineering. Microb. Cell Factories 2012, 11, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Zhou, X.; Tong, T.; Ge, J. Acetic acid acting as a signaling molecule in the quorum sensing system increases 2,3-butanediol production in Saccharomyces cerevisiae. Prep. Biochem. Biotechnol. 2021, 52, 487–497. [Google Scholar] [CrossRef]
- Cho, S.; Kim, T.; Woo, H.M.; Lee, J.; Kim, Y.; Um, Y. Enhanced 2,3-butanediol production by optimizing fermentation conditions and engineering Klebsiella oxytoca M1 through overexpression of acetoin reductase. PLoS ONE. 2015, 10, e0138109. [Google Scholar] [CrossRef] [Green Version]
- Ji, X.J.; Huang, H.; Li, S.; Du, J.; Lian, M. Enhanced 2,3-butanediol production by altering the mixed acid fermentation pathway in Klebsiella oxytoca. Biotechnol. Lett. 2008, 30, 731–734. [Google Scholar] [CrossRef] [PubMed]
- Jansen, N.B.; Tsao, G.T. Bioconversion of pentoses to 2,3-butanediol by Klebsiella pneumoniae. Pentoses Lignin 1983, 27, 85–99. [Google Scholar] [CrossRef]
- Kosfeld, A.; Jahreis, K. Characterization of the interaction between the small regulatory peptide SgrT and the EIICBGlc of the glucose-phosphotransferase system of E. coli K-12. Metabolites 2012, 2, 756–774. [Google Scholar] [CrossRef] [Green Version]
- Raina, M.; Storz, G. SgrT, a small protein that packs a sweet punch. J. Bacteriol. 2017, 199, e00130-17. [Google Scholar] [CrossRef] [Green Version]
- Lloyd, C.R.; Park, S.; Fei, J.; Vanderpool, C.K. The small protein SgrT controls transport activity of the glucose-specific phosphotransferase system. J. Bacteriol. 2017, 199, e00869-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Negrete, A.; Majdalani, N.; Phue, J.N.; Shiloach, J. Reducing acetate excretion from E. coli K-12 by over-expressing the small RNA SgrS. New Biotechnol. 2013, 30, 269–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wassarman, K.M.; Zhang, A.; Storz, G. Small RNAs in Escherichia coli. Trends Microbiol. 1999, 7, 37–45. [Google Scholar] [CrossRef]
- Vanderpool, C.K.; Balasubramanian, D.; Lloyd, C.R. Dual-function RNA regulators in bacteria. Biochimie 2011, 93, 1943–1949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richards, G.R.; Patel, M.V.; Lloyd, C.R.; Vanderpool, C.K. Depletion of glycolytic intermediates plays a key role in glucose-phosphate stress in Escherichia coli. J. Bacteriol. 2013, 195, 4816–4825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bobrovskyy, M.; Vanderpool, C.K. Diverse mechanisms of post-transcriptional repression by the small RNA regulator of glucose-phosphate stress. Mol. Microbiol. 2016, 99, 254–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, A.; Wassarman, K.M.; Rosenow, C.; Tjaden, B.C.; Storz, G.; Gottesman, S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003, 50, 1111–1124. [Google Scholar] [CrossRef] [Green Version]
- Bobrovskyy, M.; Vanderpool, C.K. The small RNA SgrS: Roles in metabolism and pathogenesis of enteric bacteria. Front. Cell. Infect. Microbiol. 2014, 4, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanderpool, C.K.; Gottesman, S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 2004, 54, 1076–1089. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, D.; Vanderpool, C.K. New developments in post-transcriptional regulation of operons by small RNAs. RNA Biol. 2013, 10, 337–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Vanderpool, C.K. Physiological consequences of multiple-target regulation by the small RNA SgrS in Escherichia coli. J. Bacteriol. 2013, 195, 4804–4815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanderpool, C.K. Physiological consequences of small RNA-mediated regulation of glucose-phosphate stress. Curr. Opin. Microbiol. 2007, 10, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Podkaminski, D.; Hinton, J.C.D.; Vogel, J. The ancestral SgrS RNA discriminates horizontally acquired Salmonella mRNAs through a single GU wobble pair. Proc. Natl. Acad. Sci. USA 2012, 109, 757–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Negrete, A.; Ng, W.I.; Shiloach, J. Glucose uptake regulation in E. coli by the small RNA SgrS: Comparative analysis of E. coli K-12 (JM109 and MG1655) and E. coli B (BL21). Microb. Cell Factories 2010, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Wadler, C.S.; Vanderpool, C.K. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc. Natl. Acad. Sci. USA 2007, 104, 20454–20459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawamoto, H.; Morita, T.; Shimizu, A.; Inada, T.; Aiba, H. Implication of membrane localization of target mRNA in the action of a small RNA: Mechanism of post-transcriptional regulation of glucose transporter in Escherichia coli. Genes Dev. 2005, 19, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Morita, T.; Maki, K.; Yagi, M.; Aiba, H. Analyses of mRNA Destabilization and Translational Inhibition Mediated by Hfq-Binding Small RNAs. Methods Enzymol. 2008, 447, 359–378. [Google Scholar] [CrossRef] [PubMed]
- Wadler, C.S.; Vanderpool, C.K. Characterization of homologs of the small RNA SgrS reveals diversity in function. Nucleic Acids Res. 2009, 37, 5477–5485. [Google Scholar] [CrossRef] [Green Version]
- Yu, E.K.; Saddler, J.N. Fed-batch approach to production of 2,3-butanediol by Klebsiella pneumoniae grown on high substrate concentrations. Appl. Environ. Microbiol. 1983, 46, 630–635. [Google Scholar] [CrossRef] [Green Version]
- Ge, J.; Wang, J.; Ye, G.; Sun, S.S.; Guo, R.; Song, G.; Ping, W. Disruption of the lactate dehydrogenase and acetate kinase genes in Klebsiella pneumoniae HD79 to enhance 2,3-butanediol production, and related transcriptomics analysis. Biotechnol. Lett. 2020, 42, 537–549. [Google Scholar] [CrossRef]
- Guo, X.; Cao, C.; Wang, Y.; Li, C.; Wu, M.; Chen, Y.; Zhang, C.; Pei, H.; Xiao, D. Effect of the inactivation of lactate dehydrogenase, ethanol dehydrogenase, and phosphotransacetylase on 2,3-butanediol production in Klebsiella pneumoniae strain. Biotechnol. Biofuels 2014, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Jung, M.Y.; Ng, C.Y.; Song, H.; Lee, J.; Oh, M.K. Deletion of lactate dehydrogenase in Enterobacter aerogenes to enhance 2,3-butanediol production. Appl. Microbiol. Biotechnol. 2012, 95, 461–469. [Google Scholar] [CrossRef]
- Nguyen, A.D.; Hwang, I.Y.; Lee, O.K.; Kim, D.; Kalyuzhnaya, M.G.; Mariyana, R.; Hadiyati, S.; Kim, S.M.; Lee, E.Y. Systematic metabolic engineering of Methylomicrobium alcaliphilum 20Z for 2,3-butanediol production from methane. Metab. Eng. 2018, 47, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Kimata, K.; Tanaka, Y.; Inada, T.; Aiba, H. Expression of the glucose transporter gene, ptsG, is regulated at the mRNA degradation step in response to glycolytic flux in Escherichia coli. EMBO J. 2001, 20, 3587–3595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richards, G.R.; Vanderpool, C.K. Induction of the Pho regulon suppresses the growth defect of an Escherichia coli sgrS mutant, connecting phosphate metabolism to the glucose-phosphate stress response. J. Bacteriol. 2012, 194, 2520–2530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice, J.B.; Vanderpool, C.K. The small RNA SgrS controls sugar–phosphate accumulation by regulating multiple PTS genes. Nucleic Acids Res. 2011, 39, 3806–3819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moons, P.; Van Houdt, R.; Vivijs, B.; Michiels, C.M.; Aertsen, A. Integrated regulation of acetoin fermentation by quorum sensing and pH in Serratia plymuthica RVH1. Appl. Environ. Microbiol. 2011, 77, 3422–3427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, D.; Xu, J.; Sun, J.; Shi, J.; Hao, J. 2-Ketogluconic acid production by Klebsiella pneumoniae CGMCC 1.6366. J. Ind. Microbiol. Biotechnol. 2013, 40, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.H.; Bennett, G.N.; San, K.Y. Effect of modified glucose uptake using genetic engineering techniques on high-level recombinant protein production in Escherichia coli dense cultures. Biotechnol. Bioeng. 1994, 44, 952–960. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Lee, S.; Jeong, D.; Yang, J.; Oh, M.K.; Lee, J. Redistribution of carbon flux toward 2,3-butanediol production in Klebsiella pneumoniae by metabolic engineering. PLoS ONE 2014, 9, e105322. [Google Scholar] [CrossRef] [PubMed]






| Strain | Characteristics | Source/Reference |
|---|---|---|
| E. coli DH5α | Ampr | Laboratory preservation |
| E. coli JM110 | Strr, dam dcm supE44Δ(lac-proAB) | Jiangsu Yugong Life |
| K. pneumoniae HD79 | Ampr, Original strain | Laboratory preservation |
| K. pneumoniae HD79-O | Ampr, Kanr, PT7lacO- sgrS:: RFP | Mutant from gene disruption; this study |
| K. pneumoniae HD79-K | Ampr, ΔsgrS:: EGFP | Mutant from gene disruption; this study |
| K. pneumoniae HD79-02 | Ampr, Cmr, Kanr, Δldh and Δack | Laboratory preservation |
| K. pneumoniae HD79-02-O | Ampr, Cmr, Kanr, Δldh and Δack and PT7lacO-sgrS:: RFP | Mutant from gene disruption; this study |
| K. pneumoniae HD79-02-K | Ampr, Cmr, Kanr, Δldh and Δack and ΔsgrS:: EGFP | Mutant from gene disruption; this study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, R.; Kang, J.; Wang, X.; Xiu, B.; Ping, W.; Ge, J. Enhancement of 2,3-Butanediol Production by Klebsiella pneumoniae: Emphasis on the Mediation of sRNA-SgrS on the Carbohydrate Utilization. Fermentation 2022, 8, 359. https://doi.org/10.3390/fermentation8080359
Sun R, Kang J, Wang X, Xiu B, Ping W, Ge J. Enhancement of 2,3-Butanediol Production by Klebsiella pneumoniae: Emphasis on the Mediation of sRNA-SgrS on the Carbohydrate Utilization. Fermentation. 2022; 8(8):359. https://doi.org/10.3390/fermentation8080359
Chicago/Turabian StyleSun, Rui, Jie Kang, Xuemeng Wang, Baolin Xiu, Wenxiang Ping, and Jingping Ge. 2022. "Enhancement of 2,3-Butanediol Production by Klebsiella pneumoniae: Emphasis on the Mediation of sRNA-SgrS on the Carbohydrate Utilization" Fermentation 8, no. 8: 359. https://doi.org/10.3390/fermentation8080359
APA StyleSun, R., Kang, J., Wang, X., Xiu, B., Ping, W., & Ge, J. (2022). Enhancement of 2,3-Butanediol Production by Klebsiella pneumoniae: Emphasis on the Mediation of sRNA-SgrS on the Carbohydrate Utilization. Fermentation, 8(8), 359. https://doi.org/10.3390/fermentation8080359





