Overexpression of LAS21 in Cellulase-Displaying Saccharomyces cerevisiae for High-Yield Ethanol Production from Pretreated Sugarcane Bagasse
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Plasmid and Recombinant Strain Construction
2.3. Enzyme Activity Assay on the Yeast Cell Surface
2.4. Determination of Gene Copy Number by Quantitative Real-Time PCR
2.5. Pretreatment of Lignocellulosic Materials
2.6. Ethanol Production from Pretreated Sugarcane Bagasse
2.7. Scale-Up of Ethanol Production in a 1 L Bioreactor
2.8. Statistical Method
3. Results and Discussion
3.1. Effect of GPI Biosynthesis and Remodeling Proteins on the Displayed BGL Activity
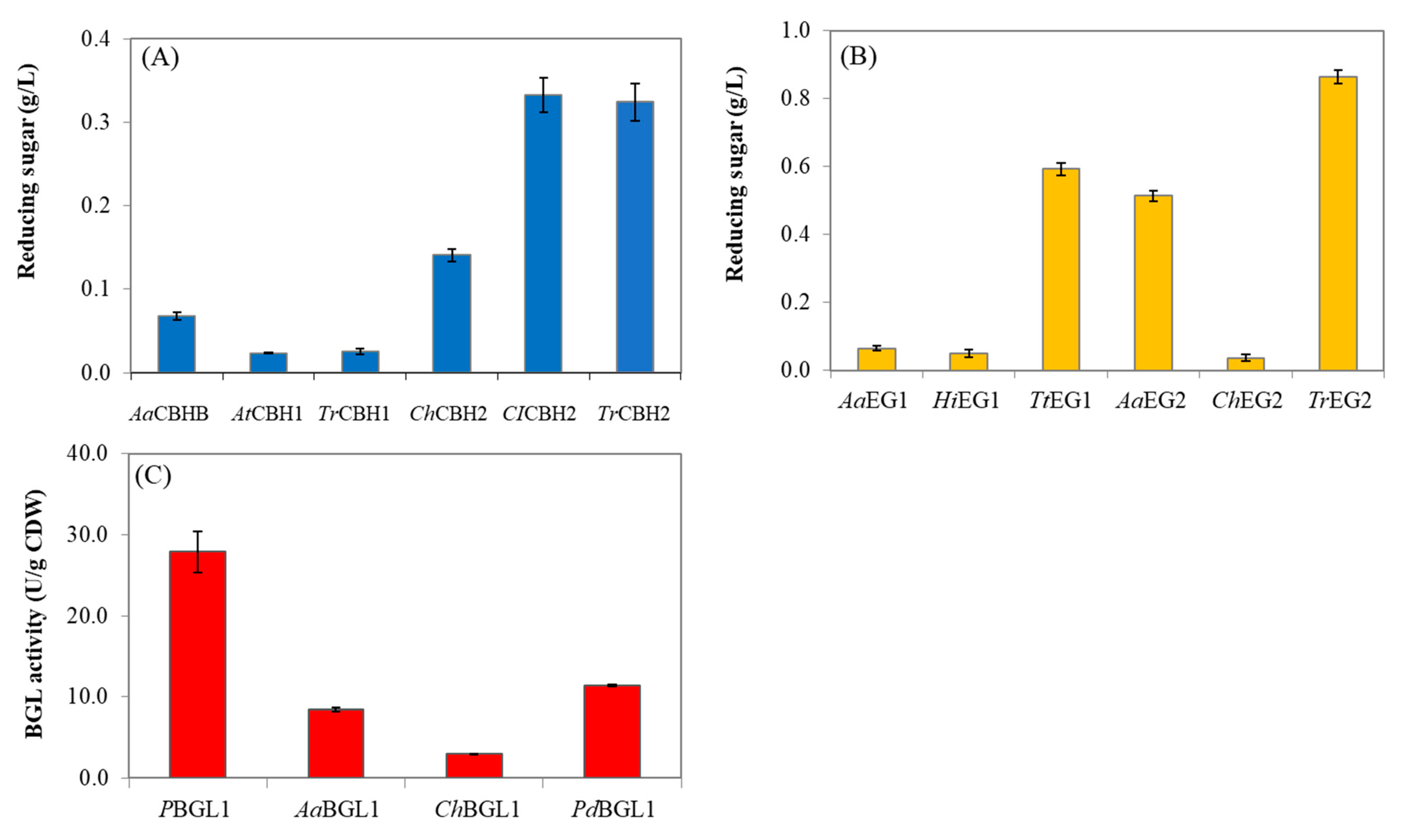
3.2. Selection of Cellulolytic Enzymes for Yeast Cell Surface Display
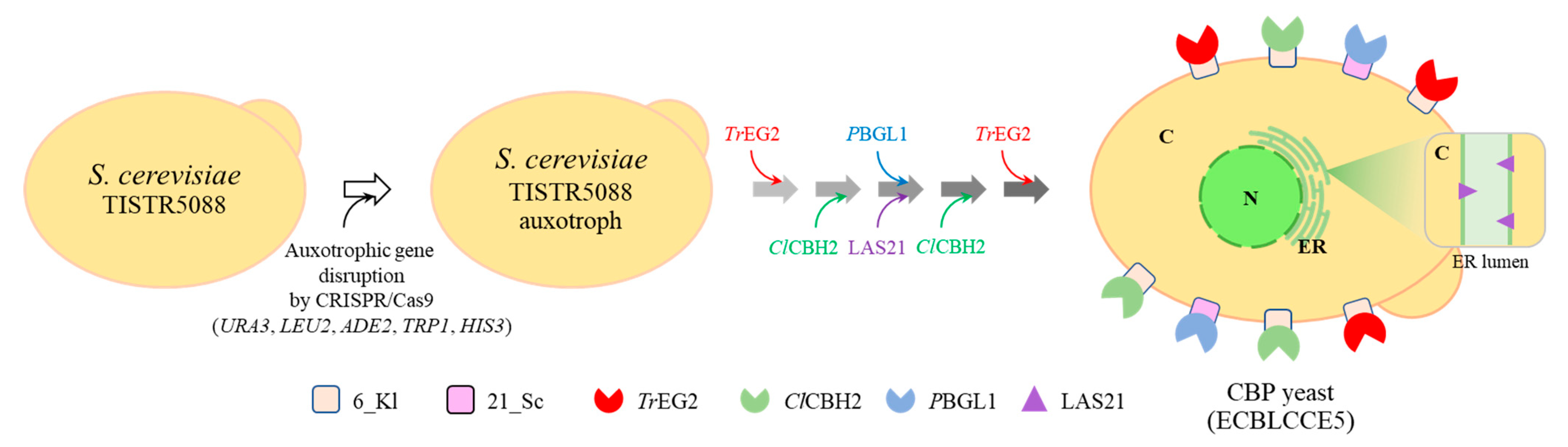
3.3. Construction of Thermotolerant Yeast Strains Displaying Cellulolytic Enzymes
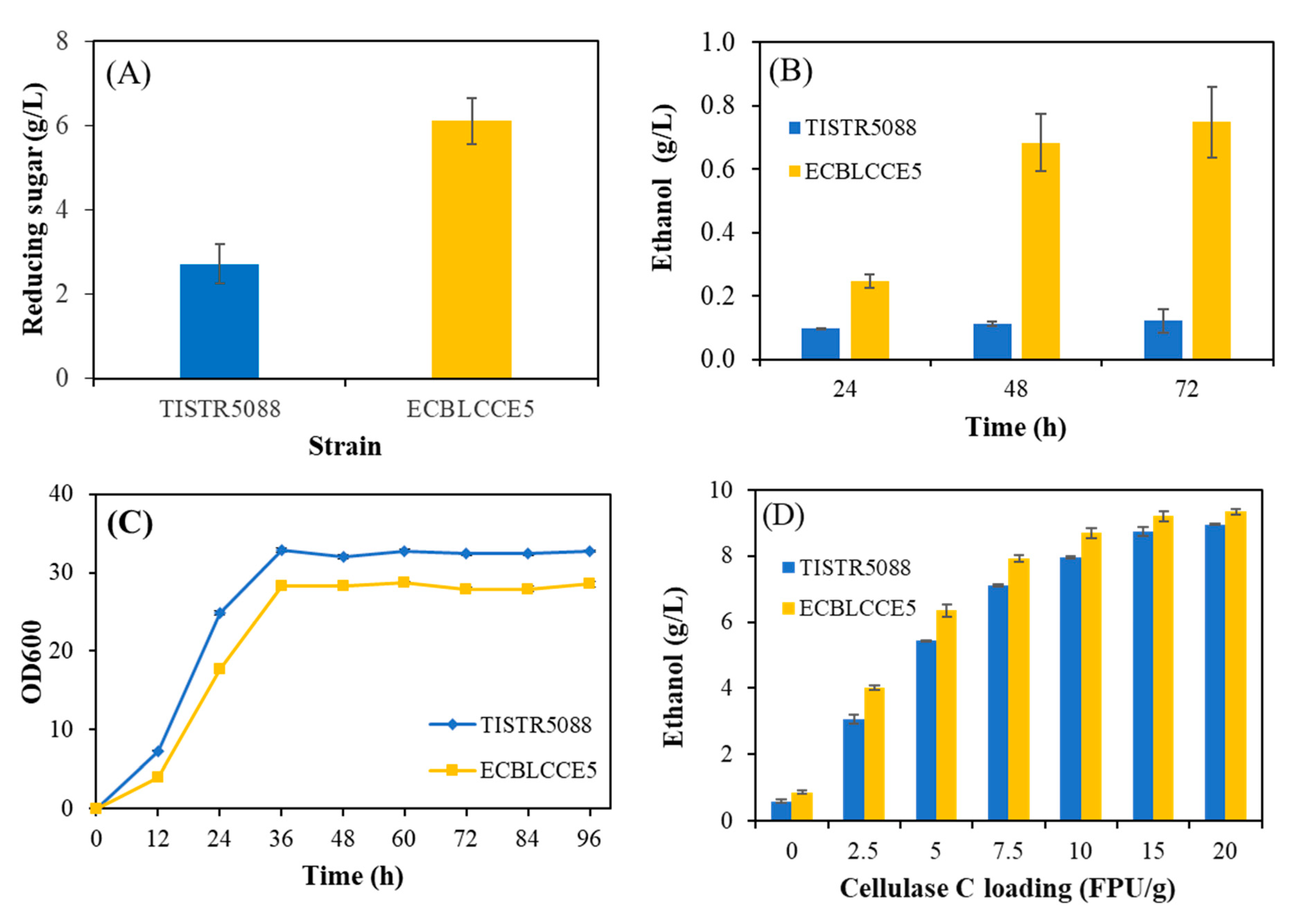
3.4. Effect of Exogenous Cellulase on Ethanol Production from Sugarcane Bagasse
3.5. Optimization of the Pretreated Sugarcane Bagasse Valorization Process with the ECBLCCE5 Strain
3.6. Scale-Up of Cellulosic Ethanol Production in a 1 L Bioreactor under Optimized Conditions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vasić, K.; Knez, Ž.; Leitgeb, M. Bioethanol production by enzymatic hydrolysis from different lignocellulosic sources. Molecules 2021, 26, 753. [Google Scholar] [CrossRef]
- Hoang, T.-D.; Nghiem, N. Recent developments and current status of commercial production of fuel ethanol. Fermentation 2021, 7, 314. [Google Scholar] [CrossRef]
- Rosales-Calderon, O.; Arantes, V. A review on commercial-scale high-value products that can be produced alongside cellulosic ethanol. Biotechnol. Biofuels 2019, 12, 240. [Google Scholar] [CrossRef]
- Cunha, J.T.; Gomes, D.G.; Romaní, A.; Inokuma, K.; Hasunuma, T.; Kondo, A.; Domingues, L. Cell surface engineering of Saccharomyces cerevisiae for simultaneous valorization of corn cob and cheese whey via ethanol production. Energy Convers. Manag. 2021, 243, 114359. [Google Scholar] [CrossRef]
- Wang, N.; Yan, Z.; Liu, N.; Zhang, X.; Xu, C. Synergy of cellulase systems between Acetivibrio thermocellus and Thermoclostridium stercorarium in consolidated-bioprocessing for cellulosic ethanol. Microorganisms 2022, 10, 502. [Google Scholar] [CrossRef]
- Chang, J.J.; Lin, Y.J.; Lay, C.H.; Thia, C.; Wu, Y.C.; Hou, Y.H.; Huang, C.C.; Li, W.H. Constructing a cellulosic yeast host with an efficient cellulase cocktail. Biotechnol. Bioeng. 2018, 115, 751–761. [Google Scholar] [CrossRef]
- Cunha, J.T.; Romaní, A.; Inokuma, K.; Johansson, B.; Hasunuma, T.; Kondo, A.; Domingues, L. Consolidated bioprocessing of corn cob-derived hemicellulose: Engineered industrial Saccharomyces cerevisiae as efficient whole cell biocatalysts. Biotechnol. Biofuels 2020, 13, 138. [Google Scholar] [CrossRef]
- Liu, Z.; Inokuma, K.; Ho, S.H.; den Haan, R.; van Zyl, W.H.; Hasunuma, T.; Kondo, A. Improvement of ethanol production from crystalline cellulose via optimizing cellulase ratios in cellulolytic Saccharomyces cerevisiae. Biotechnol. Bioeng. 2017, 114, 1201–1207. [Google Scholar] [CrossRef]
- Amoah, J.; Ishizue, N.; Ishizaki, M.; Yasuda, M.; Takahashi, K.; Ninomiya, K.; Yamada, R.; Kondo, A.; Ogino, C. Development and evaluation of consolidated bioprocessing yeast for ethanol production from ionic liquid-pretreated bagasse. Bioresour. Technol. 2017, 245, 1413–1420. [Google Scholar] [CrossRef]
- Lee, C.-R.; Sung, B.H.; Lim, K.-M.; Kim, M.-J.; Sohn, M.J.; Bae, J.-H.; Sohn, J.-H. Co-fermentation using recombinant Saccharomyces cerevisiae yeast strains hyper-secreting different cellulases for the production of cellulosic bioethanol. Sci. Rep. 2017, 7, 4428. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, Y.; Wei, N. Biocatalytic properties of cell surface display laccase for degradation of emerging contaminant acetaminophen in water reclamation. Biotechnol. Bioeng. 2020, 117, 342–353. [Google Scholar] [CrossRef]
- Tabañag, I.D.F.; Chu, I.M.; Wei, Y.-H.; Tsai, S.-L. The role of yeast-surface-display techniques in creating biocatalysts for consolidated bioprocessing. Catalysts 2018, 8, 94. [Google Scholar] [CrossRef]
- Lozančić, M.; Sk. Hossain, A.; Mrša, V.; Teparić, R. Surface display—An alternative to classic enzyme immobilization. Catalysts 2019, 9, 728. [Google Scholar] [CrossRef]
- Phienluphon, A.; Mhuantong, W.; Boonyapakron, K.; Deenarn, P.; Champreda, V.; Wichadakul, D.; Suwannarangsee, S. Identification and evaluation of novel anchoring proteins for cell surface display on Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2019, 103, 3085–3097. [Google Scholar] [CrossRef]
- Muñiz, M.; Zurzolo, C. Sorting of GPI-anchored proteins from yeast to mammals-common pathways at different sites? J. Cell Sci. 2014, 127, 2793–2801. [Google Scholar] [CrossRef]
- Pittet, M.; Conzelmann, A. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 2007, 1771, 405–420. [Google Scholar] [CrossRef]
- Laughery, M.F.; Hunter, T.; Brown, A.; Hoopes, J.; Ostbye, T.; Shumaker, T.; Wyrick, J.J. New vectors for simple and streamlined CRISPR-Cas9 genome editing in Saccharomyces cerevisiae. Yeast 2015, 32, 711–720. [Google Scholar] [CrossRef]
- Helmuth, M.; Altrock, W.; Böckers, T.M.; Gundelfinger, E.D.; Kreutz, M.R. An electrotransfection protocol for yeast two-hybrid library screening. Anal. Biochem. 2001, 293, 149–152. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Cui, J.; Lynd, L.R.; Kuang, L.R. A transition from cellulose swelling to cellulose dissolution by o-phosphoric acid: Evidence from enzymatic hydrolysis and supramolecular structure. Biomacromolecules 2006, 7, 644–648. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Leelatanawit, R.; Klanchui, A.; Uawisetwathana, U.; Karoonuthaisiri, N. Validation of reference genes for real-time PCR of reproductive system in the black tiger shrimp. PLoS ONE 2012, 7, e52677. [Google Scholar] [CrossRef]
- Suwannarangsee, S.; Arnthong, J.; Eurwilaichitr, L.; Champreda, V. Production and characterization of multi-polysaccharide degrading enzymes from Aspergillus aculeatus BCC199 for saccharification of agricultural residues. J. Microbiol. Biotechnol. 2014, 24, 1427–1437. [Google Scholar] [CrossRef]
- Technical Association of the Pulp and Paper Industry. TAPPI Test Method T264 cm-97. Preparation of Wood for Chemical Analysis. Available online: https://www.scribd.com/document/340758480/Tappi-T264-Cm-97# (accessed on 10 October 2022).
- Komath, S.S.; Singh, S.L.; Pratyusha, V.A.; Sah, S.K. Generating anchors only to lose them: The unusual story of glycosylphosphatidylinositol anchor biosynthesis and remodeling in yeast and fungi. IUBMB Life 2018, 70, 355–383. [Google Scholar] [CrossRef]
- Benachour, A.; Sipos, G.; Flury, I.; Reggiori, F.; Canivenc-Gansel, E.; Vionnet, C.; Conzelmann, A.; Benghezal, M. Deletion of GPI7, a yeast gene required for addition of a side chain to the glycosylphosphatidylinositol (GPI) core structure, affects GPI protein transport, remodeling, and cell wall integrity. J. Biol. Chem. 1999, 274, 15251–15261. [Google Scholar] [CrossRef]
- Dadwal, A.; Sharma, S.; Satyanarayana, T. Progress in ameliorating beneficial characteristics of microbial cellulases by genetic engineering approaches for cellulose saccharification. Front. Microbiol. 2020, 11, 1387. [Google Scholar] [CrossRef]
- Oh, E.J.; Jin, Y.-S. Engineering of Saccharomyces cerevisiae for efficient fermentation of cellulose. FEMS Yeast Res. 2020, 20, foz089. [Google Scholar] [CrossRef]
- Ilmén, M.; den Haan, R.; Brevnova, E.; McBride, J.; Wiswall, E.; Froehlich, A.; Koivula, A.; Voutilainen, S.P.; Siika-Aho, M.; la Grange, D.C.; et al. High level secretion of cellobiohydrolases by Saccharomyces cerevisiae. Biotechnol. Biofuels 2011, 4, 30. [Google Scholar] [CrossRef]
- Tang, H.; Song, M.; He, Y.; Wang, J.; Wang, S.; Shen, Y.; Hou, J.; Bao, X. Engineering vesicle trafficking improves the extracellular activity and surface display efficiency of cellulases in Saccharomyces cerevisiae. Biotechnol. Biofuels 2017, 10, 53. [Google Scholar] [CrossRef]
- Lopez, S.; Rodriguez-Gallardo, S.; Sabido-Bozo, S.; Muñiz, M. Endoplasmic reticulum export of GPI-anchored proteins. Int. J. Mol. Sci. 2019, 20, 3506. [Google Scholar] [CrossRef]
- Baruah, J.; Nath, B.K.; Sharma, R.; Kumar, S.; Deka, R.C.; Baruah, D.C.; Kalita, E. Recent trends in the pretreatment of lignocellulosic biomass for value-added products. Front. Energy Res. Bioenergy Biofuels 2018, 6, 141. [Google Scholar] [CrossRef]
- Ding, J.; Liang, G.; Zhang, K.; Hong, J.; Zou, S.; Lu, H.; Ma, Y.; Zhang, M. Extra metabolic burden by displaying over secreting: Growth, fermentation and enzymatic activity in cellobiose of recombinant yeast expressing β-glucosidase. Bioresour. Technol. 2018, 254, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Lopes, T.S.; De Wijs, I.J.; Steenhauer, S.I.; Verbakel, J.; Planta, R.J. Factors affecting the mitotic stability of high-copy-number integration into the ribosomal DNA of Saccharomyces cerevisiae. Yeast 1996, 12, 467–477. [Google Scholar] [CrossRef]
- Fang, C.; Wang, Q.; Selvaraj, J.N.; Zhou, Y.; Ma, L.; Zhang, G.; Ma, Y. High copy and stable expression of the xylanase XynHB in Saccharomyces cerevisiae by rDNA-mediated integration. Sci. Rep. 2017, 7, 8747. [Google Scholar] [CrossRef]
- Zhang, Q.; Weng, C.; Huang, H.; Achal, V.; Wang, D. Optimization of bioethanol production using whole plant of water hyacinth as substrate in simultaneous saccharification and fermentation process. Front. Microbiol. 2016, 6, 1411. [Google Scholar] [CrossRef]
- Mohapatra, S.; Jena, S.; Jena, P.K.; Badhai, J.; Acharya, A.N.; Thatoi, H. Partial consolidated bioprocessing of pretreated Pennisetum sp. by anaerobic thermophiles for enhanced bioethanol production. Chemosphere 2020, 256, 127126. [Google Scholar] [CrossRef]
- Gao, Y.; Xu, J.; Yuan, Z.; Jiang, J.; Zhang, Z.; Li, C. Ethanol production from sugarcane bagasse by fed-batch simultaneous saccharification and fermentation at high solids loading. Energy Sci. Eng. 2018, 6, 810–818. [Google Scholar] [CrossRef]
- Lou, H.; Zeng, M.; Hu, Q.; Cai, C.; Lin, X.; Qiu, X.; Yang, D.; Pang, Y. Nonionic surfactants enhanced enzymatic hydrolysis of cellulose by reducing cellulase deactivation caused by shear force and air-liquid interface. Bioresour. Technol. 2018, 249, 1–8. [Google Scholar] [CrossRef]
- Lee, W.G.; Lee, J.S.; Lee, J.P.; Shin, C.S.; Kim, M.S.; Park, S.C. Effect of surfactants on ethanol fermentation using glucose and cellulosic hydrolyzates. Biotechnol. Lett. 1996, 18, 299–304. [Google Scholar] [CrossRef]
- Pereira, J.D.C.; Giese, E.C.; SouzaMoretti, M.M.D.; Gomes, A.C.D.S.; Perrone, O.M.; Boscolo, M.; Silva, R.D.; Gomes, E.; Martins, D.A.B. Effect of metal ions, chemical agents and organic compounds on lignocellulolytic enzymes activities. Enzym. Inhib. Act. 2017, 29, 139–164. [Google Scholar] [CrossRef]
- Yang, J.; Huang, K.; Xu, X.; Miao, Y.; Lin, Y.; Han, S. Cell surface display of Thermomyces lanuginosus lipase in Pichia pastoris. Front. Bioeng. Biotechnol. 2020, 8, 544058. [Google Scholar] [CrossRef]
- Gruno, M.; Väljamäe, P.; Pettersson, G.; Johansson, G. Inhibition of the Trichoderma reesei cellulases by cellobiose is strongly dependent on the nature of the substrate. Biotechnol. Bioeng. 2004, 86, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Müller, G.; Chylenski, P.; Bissaro, B.; Eijsink, V.G.H.; Horn, S.J. The impact of hydrogen peroxide supply on LPMO activity and overall saccharification efficiency of a commercial cellulase cocktail. Biotechnol. Biofuels 2018, 11, 209. [Google Scholar] [CrossRef] [PubMed]
- Jugwanth, Y.; Sewsynker-Sukai, Y.; Gueguim Kana, E.B. Valorization of sugarcane bagasse for bioethanol production through simultaneous saccharification and fermentation: Optimization and kinetic studies. Fuel 2020, 262, 116552. [Google Scholar] [CrossRef]
- Inokuma, K.; Kurono, H.; den Haan, R.; van Zyl, W.H.; Hasunuma, T.; Kondo, A. Novel strategy for anchorage position control of GPI-attached proteins in the yeast cell wall using different GPI-anchoring domains. Metab. Eng. 2020, 57, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.V.; Nguyen, K.H.; Nguyen, N.L.; Ho, X.T.; Truong, P.H.; Thi Nguyen, K.C. Lychee-Derived, Thermotolerant yeasts for second-generation bioethanol production. Fermentation 2022, 8, 515. [Google Scholar] [CrossRef]
- Matano, Y.; Hasunuma, T.; Kondo, A. Cell recycle batch fermentation of high-solid lignocellulose using a recombinant cellulase-displaying yeast strain for high yield ethanol production in consolidated bioprocessing. Bioresour. Technol. 2013, 135, 403–409. [Google Scholar] [CrossRef]
- Louie, T.M.; Louie, K.; DenHartog, S.; Gopishetty, S.; Subramanian, M.; Arnold, M.; Das, S. Production of bio-xylitol from D-xylose by an engineered Pichia pastoris expressing a recombinant xylose reductase did not require any auxiliary substrate as electron donor. Microb. Cell Factories 2021, 20, 50. [Google Scholar] [CrossRef]



| Process | Properties of S. cerevisiae | Substrate | Substrate Loading (%w/v) | Commercial Cellulase Loading | Ethanol (g/L) | Ethanol Yield (g/g Biomass) | Ethanol Theoretical Yield (%) | References |
|---|---|---|---|---|---|---|---|---|
| SSF | Noncellulolytic | Sugarcane bagasse | 10.0 | 100 U/g Celluclast 1.5 L | 4.9 | 0.05 | - | [44] |
| CBP | Cellulolytic (YSD) | Rice straw | 2.5 | No addition | 0.80 | 0.03 | - | [8] |
| CBP | Cellulolytic (YSD) | Sugarcane bagasse | 0.5 | No addition | 0.93 | 0.19 | 91.2 | [9] |
| SSF | Cellulolytic (Secretion) | Rice straw | 5.0 | 10.0 FPU/g of Tec-mix | 14.0 | 0.28 | 79.0 | [10] |
| SSF | Cellulolytic (YSD) | Rice straw | 10.0 | 0.4 FPU/g of CTec2 | 8.0 | 0.08 | 33.0 | [45] |
| SSF | Cellulolytic (YSD) | Sugarcane bagasse | 10.0 | 7.5 FPU/g of CTec2 | 28.0 | 0.28 | 86.5 | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arnthong, J.; Bussadee, P.; Phienluphon, A.; Deenarn, P.; Tulsook, K.; Plupjeen, S.-n.; Siamphan, C.; Tachaapaikoon, C.; Champreda, V.; Suwannarangsee, S. Overexpression of LAS21 in Cellulase-Displaying Saccharomyces cerevisiae for High-Yield Ethanol Production from Pretreated Sugarcane Bagasse. Fermentation 2022, 8, 652. https://doi.org/10.3390/fermentation8110652
Arnthong J, Bussadee P, Phienluphon A, Deenarn P, Tulsook K, Plupjeen S-n, Siamphan C, Tachaapaikoon C, Champreda V, Suwannarangsee S. Overexpression of LAS21 in Cellulase-Displaying Saccharomyces cerevisiae for High-Yield Ethanol Production from Pretreated Sugarcane Bagasse. Fermentation. 2022; 8(11):652. https://doi.org/10.3390/fermentation8110652
Chicago/Turabian StyleArnthong, Jantima, Piyada Bussadee, Apisan Phienluphon, Pacharawan Deenarn, Kan Tulsook, Sa-ngapong Plupjeen, Chatuphon Siamphan, Chakrit Tachaapaikoon, Verawat Champreda, and Surisa Suwannarangsee. 2022. "Overexpression of LAS21 in Cellulase-Displaying Saccharomyces cerevisiae for High-Yield Ethanol Production from Pretreated Sugarcane Bagasse" Fermentation 8, no. 11: 652. https://doi.org/10.3390/fermentation8110652
APA StyleArnthong, J., Bussadee, P., Phienluphon, A., Deenarn, P., Tulsook, K., Plupjeen, S.-n., Siamphan, C., Tachaapaikoon, C., Champreda, V., & Suwannarangsee, S. (2022). Overexpression of LAS21 in Cellulase-Displaying Saccharomyces cerevisiae for High-Yield Ethanol Production from Pretreated Sugarcane Bagasse. Fermentation, 8(11), 652. https://doi.org/10.3390/fermentation8110652






