Screening of Hanseniaspora Strains for the Production of Enzymes with Potential Interest for Winemaking
Abstract
:1. Introduction
2. Experimental Section
2.1. Yeast Strains
2.2. Molecular Characterization
2.2.1. DNA Extraction
2.2.2. PCR Amplification and Restriction Profiles
2.2.3. Sequencing
2.3. Physiological Characterization
2.4. Qualitative Tests of Enzymatic Activities
2.4.1. β-Glucosidase Activity
2.4.2. Protease Activity
2.4.3. Lipase and Esterase Activities
2.4.4. Pectinase and Polygalacturonase Activities
2.5. Quantitative Tests of Enzymatic Activities
2.5.1. Glycosidase Activities
2.5.2. Lipase and Esterase Activities
2.5.3. Protease Activity
2.6. Effect of Glucose and Ethanol on Glycosidase Activities
2.7. Winemaking and Determination of Volatile Compounds
3. Results
3.1. Molecular Analysis
3.2. Physiological Characterization
| Group | Species and Laboratory Code | N° CECT | Amplified rDNA Size (bp) | Size of Restriction Fragments (bp) | ||
|---|---|---|---|---|---|---|
| HaeIII | HinfI | CfoI | ||||
| H. uvarum | ||||||
| I | HU1 | 1444 | 748 | 748, 500, 413, 156 | 320 | 295 |
| II | HU2-HU4-HU7-HU8 | 10387, 10509, 11106, 11107 | 786 | 335, 234, 136 | 372 | 335, 303 |
| III | HU3-HU9 | 10389, 11156 | 748 | 748 | 345, 201, 177 | 321, 132 |
| IV | HU5 | 10603 | 855 | 307, 233, 176 | 345 | 337, 307 |
| V | HU6 | 11105 | 372 | 372, 273, 120 | 206, 190 | 136, 129 |
| H. vineae | ||||||
| I | HV1-HV3 | 11326, 11338 | 577 | 577 | 293 | 520 |
| II | HV2 | 11330 | 673 | 288, 194, 104 | 319 | 300, 270, 96 |
| H. guilliermondii | ||||||
| I | HG1-HG3 | 11102, 11104 | 489 | 489 | 265 | 480 |
| II | HG2 | 11103 | 363 | 363, 269 | 194, 183 | 200 |
| III | HG4-HG5 | 11027, 11029 | 755 | 755 | 356, 212, 190 | 336, 147 |
| H. valbyensis | ||||||
| HVA1-HVA2-HVA3 | 1445, 10122, 11339 | 763 | 763 | 239, 211, 165, 123 | 646, 123 | |
| H. osmophila | ||||||
| HO1-HO2-HO3 | 1119,11206, 11207 | 769 | 434, 170, 153 | 361 | 279, 184, 157 | |
| H. occidentalis | ||||||
| HOC1-HOC3-HOC4 | 11341, 11472, 11329 | 795 | 673, 128 | 276, 239, 134 | 340, 125 | |
| Strain | Substrate | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glucose | Glycerol | 2-Ketoglutarate | Arabinose | Xylose | Adonitol | Xylitol | Galactose | Inositol | Sorbitol | Methyl-α-d-glucopyranoside | N-acethyl-glucosamine | Cellobiose | Lactose | Maltose | Sucrose | Trehalose | Melezitose | Raffinose | |
| HU1 | + | + | + | - | - | - | - | + | - | - | - | + | + | - | - | + | - | - | - |
| HU2 | + | + | - | - | - | - | + | - | - | + | + | - | - | - | - | + | - | + | + |
| HU3 | + | - | + | - | - | - | - | - | - | - | - | - | + | - | - | + | + | - | + |
| HU4 | + | + | - | - | + | + | + | + | - | + | + | + | + | - | - | + | + | + | - |
| HU5 | + | + | - | - | + | - | - | + | - | - | - | - | + | - | - | + | - | + | + |
| HU6 | + | + | + | - | - | + | + | + | - | - | - | - | + | - | - | + | + | + | + |
| HU7 | + | + | + | - | - | - | - | - | - | - | - | - | + | - | - | + | - | - | + |
| HU8 | + | + | + | - | - | + | - | - | - | - | - | - | + | - | - | + | - | - | + |
| HU9 | + | - | + | - | - | - | - | - | - | - | - | - | + | - | - | - | - | - | - |
| HV1 | + | - | - | - | - | - | - | - | - | - | - | - | + | - | - | - | - | - | + |
| HV2 | + | - | - | - | - | - | - | - | - | - | - | - | + | - | - | - | - | - | - |
| HV3 | + | + | - | - | - | - | - | - | - | + | + | - | + | - | - | + | - | + | + |
| HG1 | + | - | + | - | - | - | - | - | - | - | - | - | + | - | - | + | + | + | + |
| HG2 | + | + | + | - | - | + | + | + | - | + | + | + | + | - | - | + | - | + | - |
| HG3 | + | + | + | - | - | + | + | - | - | + | + | + | + | - | - | + | - | + | + |
| HG4 | + | - | + | - | - | - | - | - | - | - | - | - | + | - | - | + | - | - | - |
| HG5 | + | - | + | - | - | - | - | - | - | - | - | - | + | - | - | + | + | - | + |
| HVA1 | + | - | - | - | - | - | - | - | - | - | - | - | + | - | - | - | - | + | + |
| HVA2 | + | - | - | - | - | - | - | - | - | - | - | - | + | - | - | - | - | - | - |
| HVA3 | + | - | - | - | - | - | - | - | - | - | - | - | + | - | - | - | - | - | - |
| HO1 | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| HO2 | + | - | - | - | - | - | - | - | - | - | - | - | + | - | - | - | - | - | - |
| HO3 | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| HOC1 | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| HOC3 | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| HOC4 | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Strain | Substrate | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glucose | Maltose | Sucrose | Trehalose | Raffinose | Fatty acid ester | N-Acetyl-glucosamine | α-Glucoside | β-Glucoside | β-Galactoside | α-Galactoside | β-Fucoside | Phosphate | Phosphatidylcholine | Urea | Proline-β-naphthylamide | Histidine β-naphthylamide | Leucyl-glycine β-naphthylamide | |
| HU1 | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + | − |
| HU2 | + | − | + | − | + | − | − | + | − | − | − | − | − | − | − | + | + | + |
| HU3 | + | + | + | + | + | − | − | − | + | − | − | − | − | − | − | − | + | − |
| HU4 | + | − | − | − | − | − | − | + | + | − | − | − | − | − | − | − | + | − |
| HU5 | + | − | + | − | + | − | − | − | − | − | − | − | − | − | − | − | + | − |
| HU6 | + | + | − | − | + | − | − | + | − | − | − | − | − | − | − | + | + | + |
| HU7 | + | − | − | − | + | − | − | − | + | − | − | − | − | − | − | − | − | − |
| HU8 | + | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − |
| HU9 | + | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | + | − |
| HV1 | + | + | + | − | + | − | − | + | + | − | − | + | − | − | − | + | + | + |
| HV2 | + | − | − | − | − | − | − | + | + | − | − | + | − | − | − | − | + | + |
| HV3 | + | + | + | − | + | − | − | + | + | − | − | + | − | − | − | − | + | + |
| HG1 | + | + | + | + | + | − | − | + | + | − | − | + | − | − | − | − | + | + |
| HG2 | + | − | − | − | − | − | − | + | + | − | − | − | − | − | − | + | + | + |
| HG3 | + | + | + | + | + | − | − | + | + | − | − | + | − | − | − | − | + | + |
| HG4 | + | + | + | + | − | − | − | + | + | − | − | + | − | − | − | − | + | − |
| HG5 | + | + | + | + | + | − | − | − | + | − | − | + | − | − | − | − | − | − |
| HVA1 | + | − | − | − | − | − | − | + | + | − | − | + | − | − | − | − | − | − |
| HVA2 | + | − | − | − | − | − | − | + | + | − | − | + | − | − | − | − | − | − |
| HVA3 | + | + | + | + | − | − | − | − | + | − | − | + | − | − | − | − | − | − |
| HO1 | + | − | − | − | − | − | − | + | + | − | − | − | − | − | − | − | + | + |
| HO2 | + | + | − | − | − | − | − | + | + | − | − | − | − | − | − | − | + | + |
| HO3 | + | + | + | + | − | − | − | + | + | − | − | − | − | − | − | − | + | + |
| HOC1 | + | − | + | − | − | − | − | + | + | − | − | + | − | − | − | − | + | + |
| HOC3 | + | − | + | − | − | − | − | + | + | − | − | + | − | − | − | − | + | + |
| HOC4 | + | + | + | + | + | − | − | − | + | − | − | + | − | − | − | − | + | + |
| Protease | Esterase | Lipase | β-Glucosidase | Polygalacturonase | Pectinase | |
|---|---|---|---|---|---|---|
| HU1 | − | + | − | + | − | + |
| HU2 | − | + | + | ++ | − | − |
| HU3 | + | − | − | + | − | − |
| HU4 | + | + | + | ++ | − | + |
| HU5 | − | + | + | + | − | − |
| HU6 | − | − | − | ++ | + | − |
| HU7 | + | + | + | ++ | + | + |
| HU8 | + | + | + | ++ | − | − |
| HU9 | + | − | − | ++ | − | − |
| HV1 | + | + | + | ++ | + | − |
| HV2 | − | − | − | ++ | − | − |
| HV3 | + | + | + | ++ | + | + |
| HG1 | − | + | − | + | − | − |
| HG2 | ++ | + | − | ++ | − | − |
| HG3 | + | + | − | ++ | + | + |
| HG4 | + | − | − | ++ | − | − |
| HG5 | + | − | − | ++ | + | − |
| HVA1 | + | − | − | + | − | − |
| HVA2 | − | − | − | ++ | − | − |
| HVA3 | − | − | − | +++ | − | − |
| HO1 | + | − | − | + | − | − |
| HO2 | − | − | − | +++ | − | − |
| HO3 | + | − | − | + | − | − |
| HOC1 | + | − | − | + | − | − |
| HOC3 | + | + | − | +++ | − | + |
| HOC4 | − | − | − | +++ | − | − |
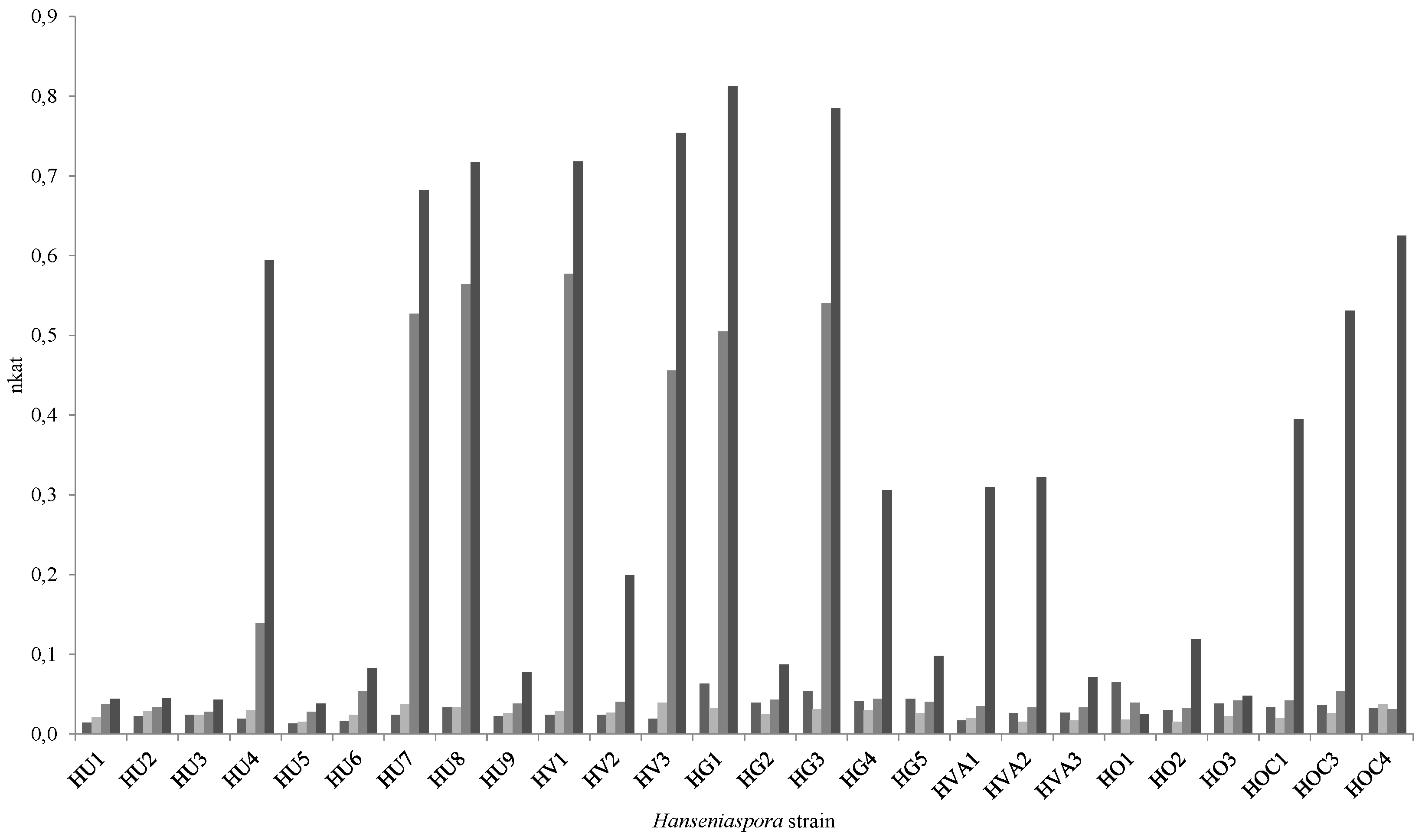
| Hanseniaspora Isolate | Activity (nkat) |
|---|---|
| HG1 | 20.7 ± 2.2 |
| HG3 | 37.9 ± 1.4 |
| HVA1 | 35.8 ± 1.1 |
| HOC3 | 40.5 ± 1.8 |
| HOC4 | 31.7 ± 2.1 |
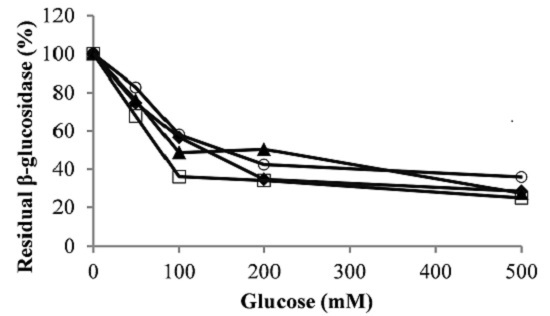
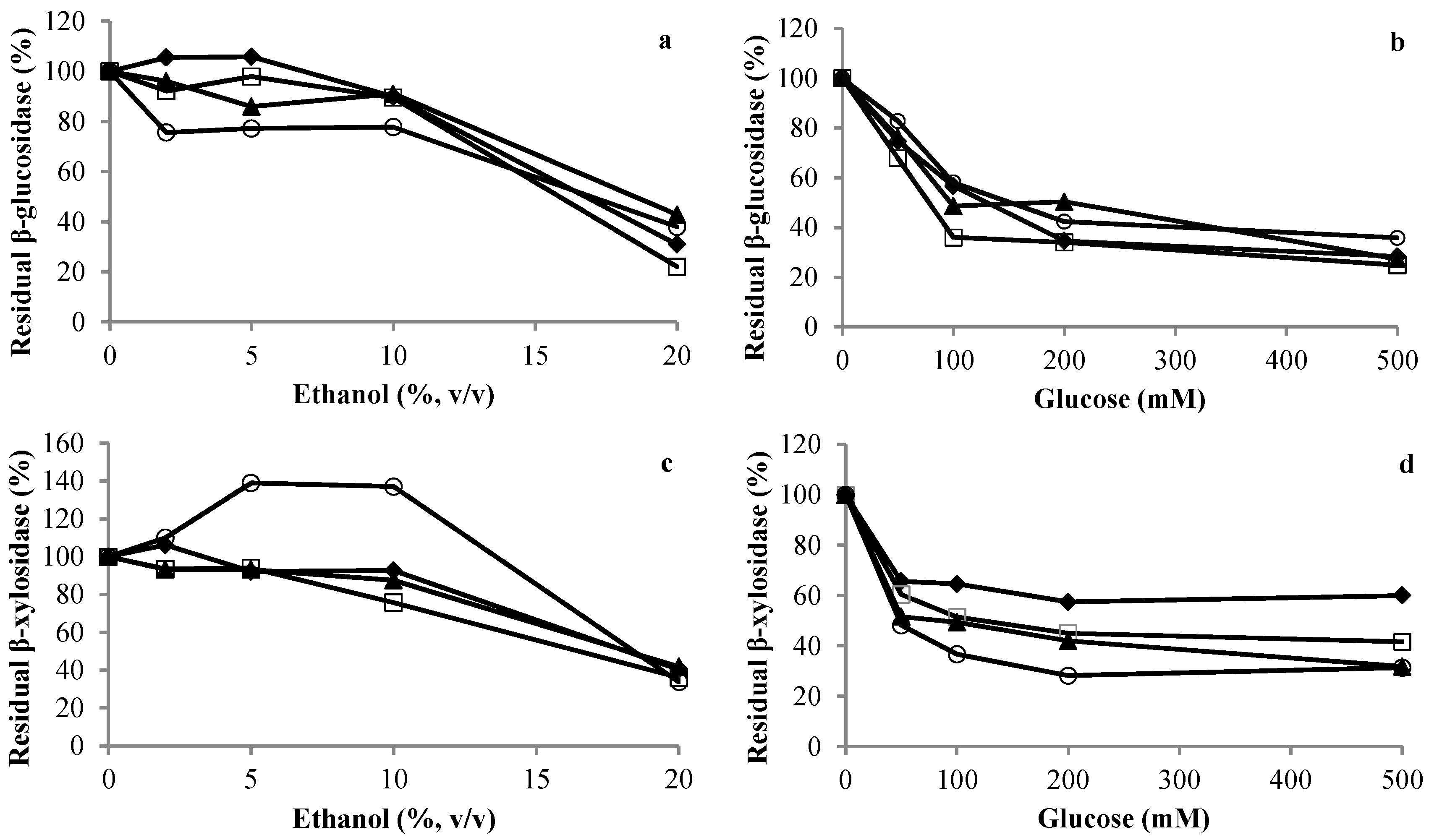
3.3. Effect of Sugars and Ethanol on Glycosidase Activities
3.4. Determination of Volatile Compounds Liberated from Wine
| Compound | Control b | Non-Saccharomyces Yeast Inoculated a | |||
|---|---|---|---|---|---|
| H. uvarum HU8 | H. uvarum HU7 | H. vineae HV1 | H. vineae HV3 | ||
| TERPENES | |||||
| cis-5-Vinyltetrahydro-1,1,5-trimethyl-2-furanmethanol | 29.7 (1.2) | 30.3 (3.1) | 33.6 (3.3) | 33.9 (3.3) | 35.9 (3.3) |
| trans-5-Vinyltetrahydro-1,1,5-trimethyl-2-furanmethanol | nd | nd | nd | 3.8 * (1.3) | 6.3 * (3.3) |
| Linalool | 20.0 (0.9) | 30.3 * (3.9) | 36.3 * (3.3) | 38.8 * (3.6) | 33.6 * (3.9) |
| Ho-trienol | 24.0 (3.2) | 51.3 * (5.3) | 35.1 * (3.3) | 38.0 (3.3) | 30.0 (3.6) |
| cis-6-Vinyltetrahydro-2,2,6-trimethyl-2H-pyran-3-ol | nd | 36.1 * (3.6) | nd | 3.9 (3.6) | nd |
| trans-6-Vinyltetrahydro-2,2,6-trimethyl-2H-pyran-3-ol | nd | 36.3 * (1.3) | nd | 3.9 (0.9) | nd |
| Terpineol | 53.3 (3.4) | 66.3 * (3.6) | 65.1 * (1.3) | 51.1 (5.6) | 36.3 (3.3) |
| Nerol | 24.6 (2.8) | 35.8 (1.1) | 33.3 (3.1) | 35.1 (3.5) | 36.1 (1.3) |
| Geraniol | 59.8 (5.0) | 61.3 (3.6) | 56.9 (1.6) | 59.1 (3.3) | 58.3 (3.3) |
| 2,6-Dimethyl-3,7-octadien-2,6-diol | 43.2 (4.7) | 86.9 * (3.1) | 80.3 * (3.1) | 68.3 * (3.3) | 69.0 * (3.8) |
| 2,6-Dimethyl-7-octene-2,6-diol | nd | 58.8 * (3.1) | 53.0 * (3.3) | 35.6 * (3.6) | 39.6 * (3.3) |
| 2,6-Dimethyl-2,7-octadien-1,6-diol | 12.0 (0.6) | 13.3 (0.9) | 6.8 (3.6) | 10.3 (3.6) | 5.8 (1.5) |
| OTHER VOLATILE COMPOUNDS | |||||
| 4-Vinylphenol | 63.2 (1.2) | 89.6 * (3.3) | 65.6 * (5.8) | 56.9 (3.5) | 59.3 (3.1) |
| 2-Methoxy-4-vinylphenol | 89.0 (6.1) | 103.0 * (5.3) | 105.3 * (6.5) | 88.6 (3.9) | 83.6 (3.3) |
| 2-Phenylethanol | 1890.2 (43.4) | 3056.5 * (39.8) | 3636.8 * (36.8) | 3308.8 * (36.6) | 3333.3 * (33.6) |
| 2-Phenylethyl acetate | 28.0 (4.1) | 56.3 * (6.3) | 33.3 (1.3) | 33.6 (3.3) | 16.3 (3.5) |
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cadez, N.P.; Raspor, A.W.A.M.; de Cock, T.; Boekhout, T.; Smith, M.T. Molecular identificacion and genetic diversity whitin species of genera Hanseniaspora and Kloeckera. FEMS Yeast Res. 2002, 1, 279–289. [Google Scholar] [PubMed]
- Strauss, M.C.A.; Jolly, N.P.; Lambrechts, M.G.; van Rensburg, P. Screening for the production of extracellular hydrolytic enzymes by non-Saccharomyces wine yeasts. J. Appl. Microbiol. 2001, 91, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Esteve-Zarzoso, B.; Belloch, C.; Uruburu, F.; Querol, A. Identification of yeasts by RFLP analysis of the 5.85 rRNA gene and the two ribosomal internal transcribed spacers. Int. J. Syst. Bacteriol. 1999, 49, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny of ascomycetous yeast from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef] [PubMed]
- Mingorance-Cazorla, L.; Clemente-Jiménez, J.M.; Martínez-Rodríguez, S.; las Heras-Vázquez, F.J.; Rodríguez-Vico, F. Contribution of different natural yeasts to the aroma of two alcoholic beverages. World J. Microb. Biotechnol. 2003, 19, 297–304. [Google Scholar] [CrossRef]
- Mendoza, L.; Farías, M.E. Improvement of wine organoleptic characteristics by non-Saccharomyces yeasts. In Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology (II); Méndez-Vilas, M., Ed.; Formatex: Badajoz, Spain, 2010; pp. 908–919. [Google Scholar]
- Mateo, J.J.; Peris, L.; Ibáñez, C.; Maicas, S. Characterization of glycolytic activities from non-Saccharomyces yeasts isolated from Bobal musts. J. Ind. Microbiol. Biotechnol. 2011, 38, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Pando, R.; Lastra, A.; Suarez, B. Screening of enzymatic activities in non-Saccharomyces cider yeast. J. Food Biochem. 2012, 36, 683–689. [Google Scholar] [CrossRef]
- Ugliano, M. Enzymes in winemaking. In Wine Chemistry and Biochemistry; Moreno-Arribas, M.V., Polo, C., Eds.; Springer Science-Business Media: Adelaide, Australia, 2009; pp. 103–126. [Google Scholar]
- Romano, P.; Fiore, C.; Paraggio, M.; Caruso, M.; Capece, A. Function of yeast species and strains in wine flavor. Int. J. Food Microbiol. 2003, 86, 169–180. [Google Scholar] [CrossRef]
- Paraggio, M. Biodiversity of a natural population of Saccharomyces cerevisiae and Hanseniaspora uvarum from Aglianico del Vulture. Food Technol. Biotechnol. 2004, 42, 165–168. [Google Scholar]
- Arévalo, M.; Úbeda, J.F.; Briones, A.I. Glucosidase activity in wine yeasts: Application in enology. Enzyme Microb. Technol. 2007, 40, 420–425. [Google Scholar]
- Swangkeaw, J.; Vichitphan, S.; Butzke, C.E.; Vichitphan, K. Characterization of β-glucosidases from Hanseniaspora sp. and Pichia anomala with potentially aroma-enhancing capabilities in juice and wine. World J. Microbiol. Biotechnol. 2011, 27, 423–430. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; O’Donell, K.; Smith, M.T. Phylogeny of the yeast genera Hanseniaspora (anamorph Kloeckera), Dekkera (anamorph Brettanomyces), and Eeniella as inferred from partial 26s ribosomal DNA nucleotide sequences. Int. J. Syst. Bacteriol. 1994, 44, 781–786. [Google Scholar]
- Basic local alignment search tool. Available online: http://www.ncbi.nlm.nih.gov/BLAST/ (accessed on 21 December 2015).
- Manzanares, P.; Ramon, D.; Querol, A. Screening of non-Saccharomyces wine yeasts for the production of β-d-xylosidase activity. Int. J. Food Microbiol. 1999, 46, 105–112. [Google Scholar] [CrossRef]
- Rodríguez, F.; Arroyo, F.N.; Lopez, A.; Bautista, J.; Garrido, A. Lipolytic activity of the yeast species associated with the fermentation/storage phase of ripe olive processing. Food Microbiol. 2010, 27, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Madrigal, T.; Maicas, S.; Mateo, J.J. Glucose and ethanol tolerant enzymes produced by Pichia (Wickerhamomyces) isolates from enological ecosystems. Am. J. Enol. Vitic. 2013, 64, 126–133. [Google Scholar] [CrossRef]
- Oliveira, R.Q.; Rosa, C.A.; Uetanabaro, A.P.; Azeredo, A.; Neto, A.G.; Assis, S.A. Polygalacturonase secreted by yeasts from Brazilian semi-arid environments. Int. J. Food Sci. Nutr. 2009, 60 (Suppl. S7), 72–80. [Google Scholar] [CrossRef] [PubMed]
- Tremacoldi, C.R.; Carmona, E.C. Production of extracellular alkaline proteases by Aspergillus clavatus. World J. Microbiol. Biotechnol. 2005, 21, 169–172. [Google Scholar] [CrossRef]
- López, S.; Mateo, J.J.; Maicas, S. Characterization of Hanseniaspora isolates with aroma-enhancing properties in Muscat wine. S. Afr. J. Enol. Vitic. 2014, 35, 292–303. [Google Scholar]
- Maicas, S.; Mateo, J.J. Hydrolysis of terpenyl glycosides in grape juice and other fruit juices: A review. Appl. Microbiol. Biotechnol. 2005, 67, 322–355. [Google Scholar] [CrossRef] [PubMed]
- Gunata, Z.; Bitteur, S.; Brillouet, J.M.; Bayonove, C.; Cordonnier, R. Sequential enzymic hydrolysis of potentially aromatic glycosides from grape. Carbohydr. Res. 1988, 184, 139–149. [Google Scholar] [CrossRef]
- Dubourdieu, D.P.; Darriet, P.; Chatonnet, F.; Boidron, J.M. Identification of volatile compounds in Bordeaux wine varieties. In Proceedings of the Fourth International Enology Symposium, Bourdeaux, France; 1989; pp. 151–159. [Google Scholar]
- Moreira, N.; Mendes, F.; Hogg, T.; Vasconcelos, I. Alcohols, esters and heavy sulphur compounds production by pure and mixed cultures of apiculate wine yeasts. Int. J. Food Microbiol. 2005, 103, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Rojas, V.; Gil, J.V.; Pinaga, F.; Manzanares, P. Studies on acetate ester production by non-Saccharomyces wine yeasts. Int. J. Food Microbiol. 2001, 70, 283–289. [Google Scholar] [CrossRef]
- Viana, F.; Gil, J.V.; Vallés, S.; Manzanares, P. Increasing the levels of 2-phenylethyl acetate in wine through the use of a mixed culture of Hanseniaspora osmophila and Saccharomyces cerevisiae. Int. J. Food. Microbiol. 2009, 135, 6–75. [Google Scholar] [CrossRef] [PubMed]
- Charoenchai, C.; Fleet, G.H.; Henschke, P.A.; Todd, B.E.N. Screening of non-Saccharomyces wine yeasts for the presence of extracellular hydrolytic enzymes. Aust. J. Grape Wine Res. 1997, 3, 2–8. [Google Scholar] [CrossRef]
- Bujdosó, G.; Egli, C.M.; Henick-Kling, T. Inter- and intra-specific differentiation of natural wine strains of Hanseniaspora (Kloeckera) by fhysiological and molecular methods. Food Technol. Biotechnol. 2001, 39, 19–28. [Google Scholar]
- Suarez, B.; Pando, R.; Fernández, N.; Querol, A.; Rodríguez, R. Yeast species associated with the spontaneous fermentation of cider. Food Microbiol. 2007, 24, 25–31. [Google Scholar]
- Guillamón, J.M.; Sabaté, J.; Barrio, E.; Cano, J.; Querol, A. Rapid identification of wine yeasts species based on RFLP analysis of the ribosomal ITS regions. Arch. Microbiol. 1998, 169, 387–392. [Google Scholar]
- Lomolino, G.; Zocca, F.; Spettoli, P.; Lante, A. Detection of β-glucosidase and esterase activities in wild yeast in a distillery environment. J Int. Brew. 2006, 112, 97–100. [Google Scholar] [CrossRef]
- Medina, K.; Ferreri, L.; Fariña, L.; Boido, E.; Dellacassa, E.; Gaggero, C.; Carrau, F.M. Aplicación de la levadura Hanseniaspora vineae en cultivos mixtos con Saccharomyces cerevisiae en la vinificación. Rev. Enol. 2008, 6, 1–6. [Google Scholar]
- De Benedictis, M.; Bleve, G.; Grieco, F.; Tristezza, M.; Tufariello, M.; Grieco, F. An optimized procedure for the enological selection of non-Sacchraromyces starter cultures. Antonie Leeuwenhoek 2011, 99, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Alexandre, H.; Heintz, D.; Chassagne, D.; Guilloux-Benatier, M.; Charpentier, C.; Feuillat, M. Protease A activity and nitrogen fractions released during alcoholic fermentation and autolysis in enological conditions. J. Ind. Microb. Biotechnol. 2001, 26, 235–240. [Google Scholar] [CrossRef]
- Buzzini, P.; Martini, A. Extracellular enzymatic activity profiles in yeast and yeast-like strains isolated from tropical environments. J. Appl. Microbiol. 2002, 93, 1020–1025. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, A.; Bartowsky, E.; Jiranek, V. A survey of glycosidase activities of commercial wine strains of Oenococcus oeni. Int. J. Food Microbiol. 2005, 105, 233–234. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, J.M.; Oliveira, P.; Baumes, R.L.; Maia, M.O. Volatile and glycosidically bound composition of Loureiro and Alvarinho wines. Food Sci. Technol. Int. 2008, 14, 341–353. [Google Scholar] [CrossRef] [Green Version]
- Swiegers, J.H.; Bartowsky, E.J.; Henschke, P.A.; Pretorius, I.S. Yeast and bacterial modulation of wine aroma and flavour. Austr. J. Grape Wine Res. 2005, 11, 139–173. [Google Scholar] [CrossRef]
- Fernández-González, M.; Di Stefano, R.; Briones, A. Hydrolysis and transformation of terpene glycosides from muscat must by different yeast species. Food Microbiol. 2003, 20, 35–41. [Google Scholar] [CrossRef]
- Sefton, M.A.; Williams, P.J. Generation of oxidation artifacts during the hydrolysis of norisoprenoid glycosides by fungal enzyme preparations. J. Agric. Food Chem. 1991, 39, 1994–1997. [Google Scholar] [CrossRef]
- Gunata, Z. Etude et exploitation par voie enzymatize des precurseurs d’arômes du raisin de nature glycosidique. Rev. Oenol. 1993, 74, 22–27. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
López, S.; Mateo, J.J.; Maicas, S. Screening of Hanseniaspora Strains for the Production of Enzymes with Potential Interest for Winemaking. Fermentation 2016, 2, 1. https://doi.org/10.3390/fermentation2010001
López S, Mateo JJ, Maicas S. Screening of Hanseniaspora Strains for the Production of Enzymes with Potential Interest for Winemaking. Fermentation. 2016; 2(1):1. https://doi.org/10.3390/fermentation2010001
Chicago/Turabian StyleLópez, Sandra, José Juan Mateo, and Sergi Maicas. 2016. "Screening of Hanseniaspora Strains for the Production of Enzymes with Potential Interest for Winemaking" Fermentation 2, no. 1: 1. https://doi.org/10.3390/fermentation2010001
APA StyleLópez, S., Mateo, J. J., & Maicas, S. (2016). Screening of Hanseniaspora Strains for the Production of Enzymes with Potential Interest for Winemaking. Fermentation, 2(1), 1. https://doi.org/10.3390/fermentation2010001