Abstract
Antimicrobial resistance represents one of the most critical public health challenges of the 21st century. The emergence of multidrug resistance (MDR) in bacterial and fungal pathogens to diverse chemical agents severely impedes the effective treatment of diseases such as cancer and systemic infections. The rapid escalation of microbial resistance underscores the urgent need for the discovery of novel antimicrobial agents and innovative approaches to drug development. In both clinical and industrial contexts, the identification of new antibiotics and antifungals remains pivotal for pathogen control. Current research efforts focus on the development of alternative formulations that offer high efficacy, reduced resistance potential, minimal side effects, and synergistic interactions, particularly those derived from natural sources. Filamentous fungi originating from extreme environments have evolved to thrive under harsh conditions, making them promising reservoirs of bioactive metabolites with unique structural and functional properties. These fungi exhibit potent antimicrobial activity through diverse mechanisms that disrupt essential cellular processes in pathogens. Despite their remarkable potential, the bioprospecting of extremophilic filamentous fungi for drug development remains underexplored. This highlights the necessity for expanded research into the efficacy and safety of their derived compounds. This review aims to emphasize the capacity of extremophilic fungi to produce antimicrobial agents, elucidate resistance mechanisms, characterize fungal bioactive extracts, and analyze their molecular actions in the context of their extreme ecological niches.
1. Introduction
Throughout their lives, humans are continually exposed to a vast array of microorganisms, ranging from beneficial commensal bacteria to potentially lethal pathogens. The complex and dynamic interactions among the host immune system, resident microbiota, and invading pathogens are critical determinants of health and disease progression. Infections arise when pathogenic organisms proliferate within the host, causing cellular and tissue damage and eliciting an immune response [1].
The advent of antibiotics marked a paradigm shift in modern medicine, significantly enhancing life expectancy, improving quality of life, and enabling the safe implementation of complex medical procedures [2,3]. Beyond clinical use, antibiotics have also played a pivotal role in food safety and public health [2]. However, the widespread and often inappropriate use of antimicrobial agents has accelerated the emergence of antimicrobial resistance (AMR), defined as the ability of microorganisms—including bacteria, viruses, fungi, protozoa, and helminths—to withstand the effects of therapeutic agents specifically designed to inhibit or kill them. As a result, treatments become ineffective, infection control is compromised, and the potential for transmission increases [4]. Methicillin-resistant Staphylococcus aureus (MRSA) is among the leading causes of death from antimicrobial-resistant infections in the world. Globally, 3.5% of new and 18% of treated cases of tuberculosis are caused by multidrug-resistant bacteria. Over the years, the misuse of antibiotics, especially at the wrong dose and duration, has led to the selection of resistant bacteria. In addition to medicine, the use of antibiotics in animal feed in developing countries has also contributed significantly to antimicrobial resistance. This requires stricter control to limit the spread of drug-resistant bacteria. Antibiotic resistance affects human health in both the treatment and prevention of infections [4]. Despite the historic efficacy of antibiotics, genetically encoded resistance mechanisms have become increasingly prevalent, particularly in the early 21st century, coinciding with a marked decline in the discovery of novel antibiotic classes. Notably, certain infectious diseases have never been effectively treated with antibiotics and continue to represent significant causes of morbidity and mortality, often exceeding the clinical burden posed by resistant infections [5].
According to the World Health Organization (WHO), AMR is currently responsible for approximately 700,000 deaths annually, with projections suggesting this figure could rise to 10 million by 2050 in the absence of concerted global intervention [6].
According to Yam et al. [7], the fight against AMR cannot rely solely on traditional pharmaceutical approaches [7]. Instead, multisectoral collaboration is needed, along with the exploration of alternative antimicrobial compounds. The global AMR crisis requires urgent research into new antimicrobial agents, especially those derived from natural sources, which offer promising potential in the search for new and effective treatments.
Fungi inhabiting extreme environments are exposed to harsh environmental conditions to which mesophilic fungi are not adapted. As a result, it is hypothesized that many of the compounds they synthesize are specific to their unique adaptive mechanisms.
Extremophilic fungi are therefore considered promising candidates for the isolation of novel bioactive compounds. Several natural products with unusual structural characteristics have already been isolated from such fungi. Approximately 40% of these compounds are new natural products, including, for example, spiromastixones A–O—a group of depsidone-based derivatives isolated from Spiromastix sp., which feature n-propyl substituents, a structural element rarely found in natural products. Another notable discovery includes four derivatives of asterinic acid containing an uncommon nitro group, identified in the Antarctic fungus Pseudogymnoascus sp. [8].
In comparison, the proportion of new natural products with similarly unusual features in typical mesophilic fungal groups is around 13%. These findings provide compelling evidence that fungi isolated from extreme environments possess significant potential as producers of unique bioactive compounds with novel structures [9].
The present review aims to address the problem of antibiotic resistance and the potential of filamentous fungi from extremophilic habitats to overcome it. It summarizes the recent scientific data focused on this problem. Particular attention is paid to the relationship between harsh living conditions and the production of new and effective antimicrobial agents by filamentous fungi inhabiting them.
2. Antimicrobial Resistance
Bacterial resistance mechanisms are diverse and rapidly evolving, posing substantial challenges to contemporary therapeutic strategies. These mechanisms include alterations to antibiotic target sites, enzymatic degradation of drugs, reduced membrane permeability, and the activation of efflux pumps [10]. Such adaptations compromise the efficacy of existing treatments, transforming otherwise manageable infections into major public health threats.
2.1. Contributing Factors to Antibiotic Failure and Underlying Mechanisms
2.1.1. Biofilm Formation
Beyond well-characterized mechanisms of antimicrobial resistance, emerging pathways, such as biofilm formation, present significant therapeutic challenges. Biofilms are structured microbial communities embedded within a self-produced extracellular polymeric matrix, typically adhered to surfaces [11]. These formations serve as a physical and biochemical barrier, protecting resident bacteria from both antimicrobial agents and host immune responses [12].
Biofilms are implicated in approximately 65% of all human infections and are particularly prevalent in device-associated and implant-related infections [13]. They are also central to the pathogenesis of chronic infections affecting the lungs, bladder, wounds, oral cavity, skin, and upper respiratory tract, including sinusitis and otitis media [14]. Within biofilms, bacterial cells engage in quorum sensing and horizontal gene transfer, which facilitate the dissemination of resistance genes [15]. Furthermore, the presence of persister cells—metabolically dormant subpopulations that exhibit transient antibiotic tolerance—compounds the difficulty of eradicating biofilm-associated infections [16]. These factors underscore the limitations of conventional antibiotics and highlight the need for alternative therapeutic strategies targeting biofilm-specific mechanisms.
2.1.2. Adaptive Resilience and Non-Genetic Resistance
In addition to biofilm-associated resistance, bacteria can exhibit adaptive resistance, a transient, non-heritable state of reduced antibiotic susceptibility. While related to the phenomena of tolerance and persistence—where bacteria survive but do not proliferate in the presence of antibiotics—adaptive resistance is distinct in that it is modulated by environmental and physiological conditions rather than stable genetic changes [17].
Adaptive resistance arises in response to various external stimuli, such as surface attachment, nutrient limitation, oxidative stress, and subinhibitory concentrations of antimicrobials. Unlike acquired genetic resistance, this phenotype is reversible upon removal of the inducing factors [18,19]. Altered gene expression plays a central role in adaptive resistance, with increasing evidence suggesting that epigenetic modifications, such as DNA methylation, may also contribute to its regulation [19]. Understanding these mechanisms is crucial for developing strategies that prevent transient resistance phenotypes from becoming fixed and heritable.
2.1.3. Modification of Antibiotic Target Sites
One of the most common and effective bacterial resistance strategies involves alterations to antibiotic binding sites. Many antibiotics exert their effects by targeting essential bacterial enzymes or structural components—such as ribosomal subunits, penicillin-binding proteins, or DNA gyrases—to disrupt key cellular processes. Resistance can develop through point mutations or enzymatic modifications that alter the structure of these targets, thereby reducing antibiotic affinity and efficacy [20].
These modifications not only diminish the effectiveness of current treatment regimens but can also confer cross-resistance to multiple drug classes, particularly when mutations occur in highly conserved regions. Consequently, target site alteration remains a critical focus in the development of next-generation antimicrobials and resistance diagnostics.
2.1.4. Enzymatic Degradation of Antibiotics
Bacteria can produce enzymes that chemically modify or degrade antibiotics, neutralizing their effects. Some of the best-documented mechanisms of such degradation include:
- β-lactamases, including extended-spectrum β-lactamases (ESBLs) and carbapenemases (e.g., KPC, NDM-1, OXA-48), which hydrolyze the β-lactam ring, rendering penicillins, cephalosporins, and carbapenems ineffective [12].
- Aminoglycoside-modifying enzymes (AMEs), which inactivate aminoglycosides by acetylation, phosphorylation, or nucleotidylation [20].
- Macrolide resistance enzymes modify the structure of macrolide antibiotics, preventing their inhibitory effects [21].
2.1.5. Reduced Membrane Permeability
Gram-negative bacteria possess an outer membrane that serves as a selective barrier, limiting the entry of antibiotics. Modifications of this membrane significantly contribute to the development of antibiotic resistance.
- Porin mutations reduce the permeability of the bacterial cell wall, limiting the uptake of antibiotics. This has been observed in fluoroquinolone-resistant Escherichia coli and Klebsiella pneumoniae [22].
- Loss of outer membrane proteins (OMPs), particularly in Acinetobacter baumannii, has been associated with resistance to carbapenems [10].
2.1.6. Efflux Pumps
Efflux pumps actively efflux antibiotics from bacterial cells before they reach their target sites. This reduces intracellular drug concentrations and reduces antibiotic efficacy [23].
- The Resistance-Nodulation-Division (RND) family in Gram-negative bacteria efficiently removes β-lactams, fluoroquinolones, tetracyclines, and macrolides [24].
- The major facilitator superfamily (MFS) and ATP-binding cassette (ABC) transporters contribute to multidrug resistance by effluxing a broad spectrum of antibiotics [25].
2.1.7. Factors Accelerating the Rate of AMR
- ✓
- Misuse and Overuse of Antibiotics
Studies around the world show that many patients strongly believe that antibacterial agents will help with viral illnesses such as the common cold or flu. Furthermore, in many developing countries, where diagnostics are not yet well established, the treatment of patients depends largely on prescribing antibiotics. In India and Vietnam, many antibiotics are also available over-the-counter and of poor quality [26]. The use of antibiotics when they are not actually needed for treatment is another common example of their misuse [26,27].
- ✓
- Agricultural Use of Antibiotics
The use of antibiotics in agriculture is a key factor in antimicrobial resistance in humans. In the United States, about 80% of antibiotics sold are used in animal feed [28]. In 2010, global antibiotic use in livestock reached 63,200 tons—more than in humans. In addition to treatment, antibiotics are administered prophylactically to healthy animals through feed and water to prevent disease, promote growth, and increase feed efficiency. One commonly used antibiotic for this purpose is colistin—a vital last-line drug for the treatment of severe human infections [29].
- ✓
- Increase in Income Levels
As a result of rising incomes, there has been an overconsumption of antibiotics, with global use increasing by 65% between 2000 and 2015. Rising living standards in low- and middle-income countries are associated with increased antibiotic use. Rising incomes in developing countries also lead to higher consumption of animal protein, which often necessitates greater use of antibiotics in livestock farming [30].
- ✓
- Easy Travel Routes
Global travel of people, animals, and goods significantly contributes to the spread of antimicrobial resistance. Travelers often become colonized with resistant pathogens and carry them back to their home countries. For example, European tourists who visited India without contact with the health system there have returned with carbapenemase-producing Enterobacteriaceae [31].
- ✓
- Biological Factors
Antibiotic resistance may happen spontaneously through mutation and bacterial evolution [27]. Additionally, plasmids—small DNA fragments—can acquire resistance genes and pass them between different bacterial species through horizontal gene transfer, accelerating the spread of resistance in a population [32].
Figure 1 presents the impact of antibiotic misuse.
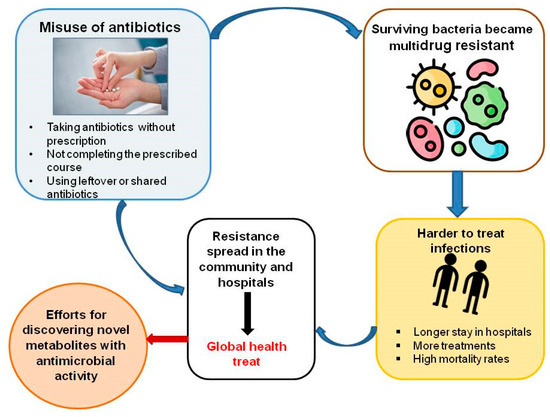
Figure 1.
Impact of antibiotic misuse.
2.2. Clinical and Economic Impact of Antimicrobial Resistance
The growing prevalence of AMR represents a critical threat to both public health and global economic stability. Infections caused by multidrug-resistant (MDR) organisms are associated with significantly longer hospitalizations, increased morbidity and mortality, and escalating healthcare costs [33]. The burden is especially pronounced in low- and middle-income countries, where surveillance, diagnostic capacity, and access to second-line therapies are often limited.
Of particular concern is the rising resistance to empirical antibiotic regimens used in the treatment of severe infections such as neonatal and pediatric sepsis and meningitis. A recent multicenter study by Williams et al. [34], encompassing 11 countries across Southeast Asia and the Pacific, revealed alarming rates of resistance to first-line antibiotics, including aminopenicillins, gentamicin, and third-generation cephalosporins. The study identified carbapenems as the most effective remaining option for many of these infections; however, the increased reliance on carbapenems raises the risk of selecting carbapenem-resistant organisms, further narrowing therapeutic options. These findings underscore the urgent need for the development of new empirical treatment strategies tailored to pediatric populations in high-burden regions.
AMR also threatens the safety and success of life-saving medical interventions, such as organ transplantation, cancer chemotherapy, and major surgical procedures, all of which rely heavily on effective prophylactic and therapeutic antibiotic regimens [35]. Patients undergoing these procedures are at heightened risk of acquiring infections that may no longer be treatable with available antimicrobials. Moreover, the efficacy of global disease control programs for tuberculosis (TB), human immunodeficiency virus (HIV), and malaria is increasingly compromised by the emergence of resistant strains, adding complexity to already challenging public health efforts [36].
2.3. Strategies to Combat Antimicrobial Resistance
Effectively addressing the global challenge of antimicrobial resistance (AMR) necessitates a comprehensive, multidisciplinary strategy that integrates antimicrobial stewardship, international surveillance, and the development of novel therapeutic agents. Central to this approach is the implementation of evidence-based prescribing practices and the reduction in inappropriate antibiotic use in both clinical and agricultural settings [12]. Overuse and misuse of antibiotics remain key drivers of resistance, and interventions such as clinician education, prescription audits, and public awareness campaigns have demonstrated measurable success in curbing unnecessary consumption [27].
Global coordination is equally essential. International collaborations facilitate the standardization of resistance surveillance, enable the rapid sharing of epidemiological data, and support the deployment of targeted interventions in regions facing high resistance burdens [22]. Initiatives such as the World Health Organization’s Global Antimicrobial Resistance Surveillance System (GLASS) exemplify the importance of cross-border data integration in tracking resistance trends and informing public health policy.
Beyond stewardship and surveillance, the discovery and development of new antimicrobial compounds remain critical. Natural sources—particularly those derived from extremophilic organisms—represent a promising frontier in antibiotic innovation. Extremophilic fungi, which thrive in extreme environments characterized by high temperature, salinity, acidity, or radiation, produce unique bioactive secondary metabolites that may serve as scaffolds for next-generation antimicrobials [37]. These organisms have evolved biochemical adaptations that confer resistance to environmental stressors, and in doing so, generate compounds with potent antimicrobial activity. Preliminary studies have highlighted the potential of extremophilic fungi to produce novel enzymes and metabolites with clinical relevance, offering a valuable reservoir for antibiotic discovery [38]. Figure 2 presents the role of extremophilic filamentous fungi in solving the problems with AMR.
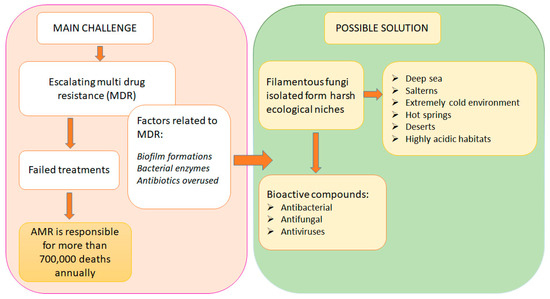
Figure 2.
Extremophilic fungi vs. antimicrobial resistance.
3. Extremophilic Fungi: Diversity, Adaptations, and Ecological Roles
The term extremophile was first introduced by MacElroy in 1974 to describe organisms capable of surviving and thriving under extreme environmental conditions [39]. Among these, thermophiles and hyperthermophiles—organisms that grow optimally at temperatures ranging from 50 °C to above 80 °C—are primarily isolated from deep-sea hydrothermal vents, hot springs, and decomposing plant matter [40,41]. Conversely, psychrophiles inhabit cold ecosystems such as glacial environments, the Arctic, and Antarctica [42]. Piezophiles—microorganisms that can withstand high pressure within an environment—should also be mentioned here, which can be both psychrophilic and thermophilic [43]. Halophiles thrive in environments with salt concentrations exceeding 8.8% NaCl and are often associated with salt-preserved foods [44]. Notably, microbial life has been documented in environments with temperatures as high as +121 °C and as low as −25 °C, as well as in saturated saline conditions, underscoring the extraordinary adaptive capacity of extremophiles [45,46]. Microorganisms inhabiting extreme habitats are subjected to multiple stresses as a result of the action of multiple adverse environmental factors, which is why they are also known as polyextremophiles [47]. Investigations on polyextremophiles and extremozymes attract scientific attention to microbial biochemical diversity and look for new or improved applications [48].
For many years, extreme environments were believed to be dominated almost exclusively by prokaryotic organisms. However, recent discoveries have revealed that fungi also occupy these niches. Fungi are among the most adaptable lineages in the Tree of Life, exhibiting remarkable morphological plasticity and ecological versatility [49]. They have been identified in nearly all known ecosystems, functioning as parasites, symbionts, or saprotrophs [41,50]. In extreme habitats, extremophilic fungi contribute to ecosystem resilience and nutrient cycling by acting as decomposers and mediating critical biogeochemical processes, particularly carbon and nitrogen fluxes [51].
Extreme environmental conditions, such as thermal fluctuations and osmotic stress, pose significant challenges to cellular integrity. These conditions can induce the formation of reactive oxygen species (ROS), cause protein misfolding, disrupt the osmotic balance, and lead to cellular damage [52,53]. In response, extremophilic fungi have evolved a suite of biochemical and structural adaptations. These include the production of protective pigments such as melanin in the cell wall, enhanced osmotic regulation through glycerol retention, meristematic growth patterns, secretion of extracellular polysaccharides (EPS), and modifications in membrane composition and fluidity to maintain homeostasis under saline stress [54].
Despite their resilience, the growth of extremophilic and extremotolerant fungi is often slow due to low nutrient availability, complex morphological demands, and high energy requirements for stress adaptation. However, this is not universally true; some fungi isolated from extreme environments exhibit rapid growth when cultured under more moderate conditions [55].
Although extreme environments are often thought of as distant and exotic, similar conditions can be found in homes. We encounter these fungi in extreme environments in our households—in kitchen sinks, dishwashers, refrigerators, between bathroom tiles, washing machines—with fluctuating pH, detergents, and conditions where they are exposed to occasional high temperatures, ranging from oligotrophic to eutrophic. Some of these fungi are known to be rare pathogens of susceptible individuals [41].
3.1. Thermophilic Fungi: Classification, Ecology, and Thermotolerance Mechanisms
The thermophilic fungus Rhizomucor pusillus was first isolated from bread by Lindt in 1886 [56], marking one of the earliest recorded identifications of a fungus capable of thriving at elevated temperatures. Thermophilic fungi are the only known eukaryotes capable of sustained growth at temperatures approaching 60 °C [57]. Their growth temperature ranges from 20 to 62 °C, with the optimum at 35–55 °C, while thermotolerants are the ones with a growth temperature range of 20–55 °C [58,59].
Thermophilic fungi inhabit a variety of microhabitats, especially those subject to microbial self-heating, such as decomposing herbivore litter, dung, and compost—environments where temperatures naturally exceed 45 °C due to microbial metabolism [60]. Notable examples include Myceliophthora thermophila, an endophyte isolated from the desert tree Parkinsonia microphylla [61], and Chaetomium thermophilum, commonly found in compost and known for its ability to grow at temperatures up to 60 °C [31]. While Rhizomucor pusillus is typically an environmental saprotroph, it is also recognized as an opportunistic pathogen in immunocompromised individuals [62,63].
Taxonomically, thermophilic fungi are distributed primarily within two fungal phyla: Mucoromycota and Ascomycota. Within Mucoromycota, thermophilic representatives are found in the orders Calcarisporiellales [64] and Mucorales [65]. Ascomycota thermophiles are primarily classified within the orders Onygenales, Sordariales, and Eurotiales.
Adaptations to high-temperature environments are comparatively rare among fungi. Out of an estimated 100,000 known fungal species [66], only about 75 are currently identified as thermophilic [65]. Nevertheless, those that do inhabit thermophilic niches display specialized physiological adaptations that allow them to survive conditions that are lethal to most other organisms.
Thermophilic and thermotolerant fungi exhibit a range of molecular and cellular mechanisms to withstand elevated temperatures. These include the production of heat shock proteins (HSPs), unique lipid storage systems, slower overall metabolic rates, rapid turnover of key enzymes, and an increased proportion of saturated fatty acids in membrane phospholipids, which contributes to membrane stability at high temperatures [67,68]. Despite these adaptations, fungi generally exhibit a preference for lower temperatures. The maximum known growth temperature for fungi slightly exceeds 60 °C, whereas some archaeal species can grow at temperatures as high as 123 °C. This thermal preference may reflect the evolutionary history of fungi, which likely diversified and adapted to terrestrial ecosystems during the globally cold “Snowball Earth” periods.
3.2. Psychrophilic Fungi: Adaptations to Cold Environments
The terms psychrophiles and psychrotrophs were first introduced by Richard Morita [69] to describe organisms capable of thriving in cold ecosystems. Psychrophilic and psychrotolerant fungi are characterized by their ability to grow at temperatures as low as 0 °C. Psychrophiles typically have an optimal growth temperature below 15 °C and fail to grow at temperatures exceeding 20 °C [70]. For instance, Rhodotorula frigidalcoholis, isolated from Antarctica, can grow at temperatures as low as −10 °C, while Pseudogymnoascus pannorum has been found to thrive at −20 °C [41]. Similarly, Mrakia frigida is described as a psychrophile, with optimal growth at 4 °C and 15 °C, and poor growth at 20 °C [71]. Mesophilic-psychrotolerant fungi, which combine mesophilic behavior with psychrotolerant adaptation, are classified as strains that do not grow at 5 °C but show optimal growth at 35 °C and can grow even at 45 °C [72].
Earth hosts numerous extreme cold environments, such as the polar regions and high mountain ranges, where conditions are harsh and generally inhospitable for most life forms. Nevertheless, cold-adapted fungi have been isolated from a variety of cryotic environments, including glacial ice [73], permafrost [74], ice sheets, ice shelves, glaciers, sea ice, freshwater ice, and cold-water bodies in both polar regions [75]. Antarctica, one of the driest and most inhospitable places on Earth, is home to fungi that thrive in the cold water of the deep sea, permafrost, and glacial ice, despite the lack of liquid water and extremely low temperatures [41].
Fungi from a diverse range of taxonomic groups have been isolated from these cold environments. Species from the phyla Zygomycota, Ascomycota, Deuteromycota, and Basidiomycota—including genera such as Penicillium, Mucor, Aspergillus, Cladosporium, Botrytis, Alternaria, Rhizopus, Lecanicillium, Geomyces, and Monodictys—have been identified in cold habitats [76,77,78].
The environments that support psychrophilic fungi are often characterized by extreme challenges, such as limited water and nutrient availability, exposure to ultraviolet radiation, and freeze–thaw cycles that can damage cellular structures through ice crystal formation. These stressors can lead to reduced enzyme activity, altered membrane fluidity, and disruption of cellular processes. To overcome these harsh conditions, psychrophilic and psychrotolerant fungi have evolved various survival strategies. One key adaptation is the production of antifreeze proteins (AFPs), which are synthesized by fungi to prevent the formation of ice within their cells at subzero temperatures [79]. AFPs are found across all cold-adapted taxa, and their primary function is to inhibit ice formation, allowing the organism to remain hypothermic and maintain cellular integrity even as external temperatures drop below freezing [80].
3.3. Halophilic Fungi
Low temperatures are typically associated with reduced water availability due to high solute concentrations, desiccation, or the formation of ice crystals that expel solutes. Halophilic fungi, however, can thrive in environments with high salinity, including temperatures below 50 °C in sea ice and brines with salinities up to 20% NaCl. The presence of fungi in hypersaline environments was first documented in 1999 by Zalar et al. [81,82] and Gunde-Cimerman et al. [83], who identified fungi as active inhabitants of man-made solar salt pans located at the border between Slovenia and Croatia. Halophilic fungi are distinguished from halotolerant fungi by their ability to grow in environments containing more than 17% NaCl (w/v), while halotolerant species can endure but not necessarily grow in such conditions.
Notable halophilic fungi include Aspergillus atacamensis, Aspergillus baarnensis, Wallemia ichthyophaga, Phialosimplex salinarum, W. muriae, and A. salisburgensis, which require NaCl concentrations ranging from 5 to 10% (w/v). One particularly rare species, W. ichthyophaga, has been exclusively isolated from salt-preserved foods such as ham, fish, and brine in salt pans [84]. The halotolerant fungi Aspergillus sydowii and the black yeast Hortaea werneckii are capable of growing across almost the entire salinity spectrum. H. werneckii has been isolated from a variety of hypersaline habitats, including salty foods, sea sponges, human skin, brine in salt pans, and deep-sea environments [85].
Halophilic and halotolerant fungi from the phylum Ascomycota are typically classified within the orders Eurotiales, Dothideales, and Capnodiales. Dominant halophilic species include Aureobasidium pullulan, Trimmatostroma salinum, H. werneckii, and Phaetotheca triangularis. Halotolerant species include A. sydowii, A. niger, Penicillium chrysogenum, Eurotium amstelodami, A. tubingensis, A. flavus, Eurotium herbariorum, A. versicolor, Penicillium steckii, and Penicillium citrinum [71]. Several species, including A. versicolor, A. sydowii, and various Penicillium species such as P. brevicompactum and P. chrysogenum, have been isolated from hypersaline environments. Additionally, Candida parapsilosis, Candida glabrata, Candida krusei, and Candida tropicalis have been found in the hypersaline waters of the Dead Sea [86].
In hypersaline environments, fungi employ a strategy of accumulating “compatible solutes,” such as free amino acids and polyols, to maintain intracellular NaCl concentrations below toxic levels [87]. Melanin, a key component in many halophilic fungi, provides protection against UV radiation, desiccation, high salt concentrations, heavy metals, and radionuclides. This pigment helps fungi survive in extreme environments such as high altitudes, deserts, and on plant surfaces [88,89]. In the presence of high salt concentrations, halophilic and halotolerant fungi respond through the high osmolarity glycerol (HOG) signaling pathway, which leads to changes in protein ubiquitination, transcription, and translation. Transporters regulate the intracellular concentrations of inorganic ions, thus balancing osmotic pressure. The identification of the MAP kinase homolog, HwHog1, in H. werneckii confirms the presence of a signaling pathway similar to that of Saccharomyces cerevisiae’s HOG pathway, which is involved in sensing and responding to elevated NaCl levels [90].
Fungi from hypersaline and acidic environments, such as those found in Lake Magic in Western Australia, are being studied as models for survival in space. Conditions on planets such as Venus, with its hot and acidic surface, and Mars, characterized by cold deserts, present extreme challenges. Studying extremophilic fungi from Earth could provide valuable insights into potential forms of extraterrestrial life.
3.4. Acidophiles and Alkaliphiles: Adaptations to Extreme pH Environments
Acidophiles are microorganisms that thrive in environments with a pH of 3 or lower while maintaining a near-neutral internal pH. In contrast, alkaliphiles are adapted to live in environments with pH levels ranging from 9 to 11, while their internal pH remains around 9 [91,92]. Survival in such extreme pH conditions necessitates complex cellular adaptations to counteract the various ecophysiological challenges associated with these environments.
Both acidophiles and alkaliphiles exhibit unique structural and functional modifications to regulate their internal pH. Despite the extreme acidity of their external environment, acidophiles cannot tolerate such low pH levels within their cells without risking macromolecular instability. As a result, these organisms have evolved several mechanisms to expel excess protons and maintain internal conditions that are neutral to slightly acidic [93,94]. To preserve cellular pH homeostasis, acidophiles implement multiple adaptive strategies, including: enhancing proton efflux, upregulating secondary transporter expression, reducing proton permeability in the cell membrane, and improving cytoplasmic buffering [93,95,96]. In the case of the obligate alkaliphilic fungus Sodiomyces alkaline (Ascomycota), it has been discovered that the organism maintains its internal pH by accumulating polyamines (PA) within its cytoplasm, allowing it to survive in the extremely high pH conditions of soda lakes [97].
3.5. Other Extremes
It is important to note that many extremophilic organisms do not fall into the categories discussed previously. For example, radiotolerant species are known for their remarkable ability to withstand radiation, thanks to their efficient DNA repair mechanisms, which make them potential candidates for bioremediation in radioactive environments. Additionally, organisms that can thrive in environments with high concentrations of heavy metals or exposure to toxic substances like hexachlorobenzenes have been considered for their potential role in bioremediation. Another group of extremophiles includes piezophiles, or occasionally barophiles, which are adapted to survive under extreme barometric pressure. Lastly, “xerophiles” are specialized organisms capable of proliferating in arid, moisture-deprived conditions [98].
Most of the extreme microorganisms are polyextremophiles or poly-extreme tolerant and are capable of surviving under a wide range of stresses. In marine environments, thermopiezophilic microorganisms, psychrophile-piezophilic, etc., can be found [99].
4. Extremophilic Fungi as Producers of Antimicrobial Substances
With the increasing prevalence of multidrug-resistant pathogens, the search for novel antibiotics and bioactive microbial metabolites has become an urgent priority. In response, researchers are turning their attention to extremophilic microorganisms as promising sources of new antimicrobial compounds [100]. Under environmental stress, extremophiles synthesize a variety of specialized organic molecules—collectively known as extremolytes—including carbohydrates, amino acids, and exopolysaccharides. These compounds have found valuable applications in the pharmaceutical and cosmetic industries due to their stability and bioactivity under extreme conditions.
Extremophilic organisms, particularly extremophilic fungi, are increasingly being explored for their potential to produce novel natural products with potent antimicrobial activity, especially against MDR pathogens [101]. These fungi have been shown to secrete a diverse array of secondary metabolites, such as steroids, peptides, terpenes, alkaloids, and polyketides, which exhibit a broad spectrum of biological activities, including antibacterial, antifungal, antiviral, anticancer, and cytotoxic effects [102]. Table 1 summarizes the antimicrobial properties of selected extremophilic filamentous fungi.

Table 1.
Antimicrobial activity of some natural products from extremophilic fungi.
These findings indicate that extreme environments hold promise for the discovery of new and valuable microorganisms with significant pharmaceutical potential [101].
5. Characterization of the Active Substances Produced by Extremophilic Fungi
Filamentous fungi are recognized for producing a wide array of molecules used in antibacterial therapies, especially in cases where traditional antibiotics fail to be effective. As noted by compounds with significant pharmaceutical promise [128]. Table 2 presents some antimicrobial substances produced by extremophilic fungi.

Table 2.
Some antimicrobial substances produced by extremophilic fungi.
Filamentous extremophilic fungi are recognized for generating a wide variety of antibacterial compounds, characterized by their distinct structural traits and modes of action. These substances hold promise for the development of new antimicrobial treatments, especially in cases where traditional antibiotics are less effective [128]. Some of them are presented below.
β-Lactam antibiotics, such as penicillins and cephalosporins, are traditionally produced by filamentous fungi like Penicillium and Acremonium. Recent research has focused on enhancing their production through genetic engineering and optimizing fermentation conditions. While no entirely new β-lactam structures have been reported from filamentous fungi in the past year, advancements have been made in increasing yields and expanding the spectrum of activity of existing compounds. Penicillins, such as benzylpenicillin (penicillin G), contain a β-lactam ring fused to a thiazolidine ring and are renowned for their efficacy against Gram-positive bacteria [143].
Ergosteroids are a group of sterol derivatives primarily produced by fungi, known for their diverse biological properties, including significant antimicrobial activity. A halotolerant fungus, Aspergillus flocculosus PT05-1, isolated from hypersaline sediment in China and cultured in a 10% saline medium, yielded newly identified ergosterol compounds. Some of them demonstrated notable antimicrobial activity against Enterobacter aerogenes, P. aeruginosa, and C. albicans [144]. Such findings highlight the pharmaceutical promise of ergosteroids derived from extremophilic fungi.
Terremides are antimicrobial secondary metabolites produced by the salt-tolerant fungus A. terreus. According to the article by Pócsi et al. [130], these compounds were noted for their antimicrobial properties against E. aerogenes, P. aeruginosa, S. aureus, and C. albicans [130].
Anthraquinones are a class of aromatic polyketides characterized by a three-ring core structure. Representing the largest group of natural pigments of quinoid nature, these compounds are frequently found in marine-derived and halophilic fungi. Notably, halogenated derivatives, such as chlorinated and brominated anthraquinones, are commonly produced by these fungi, and such modifications are often associated with enhanced biological activity. For instance, 2-(dimethoxymethyl)-1-hydroxyanthracene-9,10-dione, isolated from A. versicolor, demonstrated potent antibacterial effects against multidrug-resistant S. aureus strains, and moderate activity against Vibrio species [131]. Furthermore, the obligate halophilic marine fungus Asteromyces cruciatus KMM 4696 exhibits antimicrobial activity against S. aureus, attributed to anthraquinone-derived metabolites [133].
Carotenoids. The antimicrobial activity of carotenoids derived from cold-adapted fungi has been demonstrated, representing a significant additional benefit and potential avenue for use in food preservation. In terms of agricultural applications, carotenoids can be incorporated into feeds as precursors to vitamins and coloring substances, including, for example, aquarium feeds and those used on a large scale in salmon farming [75,145]. Torulene is a carotenoid pigment produced by the cold-adapted fungus Rhodotorula mucilaginosa, known for its antioxidant and antimicrobial properties. This pigment has potential applications in food preservation and natural colorant production due to its stability and bioactivity under low-temperature conditions.
Melanins. Melanin is a distinctive class of insoluble pigments that exhibit amorphous characteristics. They are secondary metabolites, produced by black fungi in extreme environments, that possess photoprotective, osmoprotective, antioxidant, and antimicrobial properties with a significant role in a variety of cosmetic and medicinal uses. The halophilic marine fungi Trimmatostroma salinum and Phaeotheca triangularis, located on the eastern Adriatic Sea coast, generate melanin in highly saline (saturated NaCl) environments, exhibiting antimicrobial properties [136]
A total of 52 fungal strains were isolated from soil samples collected near the southeastern edge of the Collins Glacier. In addition, 106 previously collected filamentous fungi from the glacier’s western edge were examined for their ability to produce extracellular pigments. Five strains demonstrated pigment production and were identified through ITS sequencing as Talaromyces cnidii, Pseudogymnoascus shaanxiensis, and an unidentified Pseudogymnoascus species. Extracts from Pseudogymnoascus strains SC04.P3, SC3.P3, SC122. P3, and ACF093 showed inhibitory activity against S. aureus ATCC6538, while two strains (SC12.P3 and SC32.P3) also exhibited activity against Leishmania (L.) infantum, Leishmania amazonensis, and Trypanosoma cruzi. Chemical analysis of the extracts using UPLC-ESI QToF identified biologically active compounds including asterric acid, violaceol, mollicellin, psegynamide A, diorcinol, and thailandolide A [135].
Eutypellazines P–S Eutypellazines P–S are thiodiketopiperazine alkaloids produced by the deep-sea fungus Eutypella sp. MCCC 3A00281, isolated from sediment in the South Atlantic Ocean. These compounds exhibit antibacterial activity, particularly against S. aureus and vancomycin-resistant Enterococci. Structurally, they belong to a broader family of eutypellazines known for their complex and sulfur-containing ring systems. As marine-derived metabolites, they exemplify the untapped drug discovery potential of extremophilic fungi [139].
Polyketides. Polyketides are a diverse class of secondary metabolites with significant antimicrobial properties. Fungi like the Fusarium species produce polyketides such as fusarithioamide B, which has shown potent activity against S. aureus, B. cereus, and E. coli [146]. Other polyketides include cytosporones Y and Z from Trichoderma species, exhibiting antifungal properties [147]. Recent discoveries include the following: Cladrioides A–N: Fourteen new australifungin analogs from Cladosporium cladosporioides LD-8, exhibiting strong antifungal activities against phytopathogenic fungi. Compounds 7, 10, and 16 showed IC50 values ranging from 1.71 to 16.63 μg/mL, outperforming the commercial fungicide hymexazol in some cases [148]. Penialdin F and Citromycetin: Polyketides from Penicillium bissettii and P. glabrum demonstrated inhibitory activity against methicillin-resistant S. aureus and Mycobacterium species [149]. Two highly oxygenated polyketides, penilactones A and B, isolated from the Antarctic deep-sea-derived fungus P. crustosum, were found to inhibit nuclear factor-κB (NF-κB) activity [150]. Additionally, a psychrophilic fungal strain from an Antarctic marine funfus, identified as Pseudogymnoascus sp., yielded four novel nitroasteric acid derivatives—pseudogymnoascins—and 3-nitroasterric acid. These compounds represent the first nitro-substituted derivatives of the known fungal metabolite asterric acid. Although most previously reported asterric acid derivatives exhibit antibacterial and antifungal properties, the present nitro-derivatives were inactive, possibly due to the presence of the nitro group [8]. Bio-guided fractionation of the culture broth of the Antarctic soil-derived Aspergillus ochraceopetaliformis led to the isolation of five new highly oxygenated polyketides, ochraceopones, along with a new double bond isomer of asteltoxin, isoasteltoxin, as well as asteltoxin B. Among these metabolites, ochraceopones and isoasteltoxin showed promising antiviral activity against the influenza viruses H1N1 and H3N2 [105]. Penicilones are polyketide-derived secondary metabolites isolated from different fungi. Some scientific data present their antibacterial or antifungal efficacy [137,138].
Terpenes. A psychrophilic fungal Eutypella strain isolated from Arctic soil on London Island was the source of four new diterpenes: scoparasin B, libertellenone H and eutypenoids A and C [151]. From the culture broth of the Antarctic moss-derived fungus Penicillium funiculosum, the meroterpenoids chrodrimanins I and J and five known structurally related chrodrimanins were purified. The novel compounds showed weak inhibitory activity against the influenza A virus H1N1 [152]. However, it is now becoming apparent that bacteria and fungi adapted to extreme conditions in these habitats, notably low temperatures (−1–4 °C), dearth of nutrients, and high hydrostatic pressure, provide interesting targets for bioprospecting campaigns [153,154,155,156]. A novel chloro-trinoreremophilane sesquiterpene, along with three new chlorinated eremophilane sesquiterpenes, was isolated from the Antarctic deep-sea-derived fungus Penicillium sp. PR19N-1. These compounds exhibited moderate cytotoxic activity against HL-60 and A549 cancer cell lines [157]. Subsequent chemical analysis of the same strain resulted in the discovery of five additional cytotoxic eremophilane-type sesquiterpenes [158].
Terpenoids and Meroterpenoids. Terpenoids and meroterpenoids are hybrid natural products combining terpenoid and polyketide moieties, exhibiting diverse biological activities. Helvolic acid, a fusidane-type triterpenoid, is synthesized by A. fumigatus and exhibits antibacterial, antifungal, and antiprotozoal activities. Similarly, pyripyropene A, a meroterpenoid from A. fumigatus, has demonstrated insecticidal properties and the potential to inhibit cholesterol acyltransferase [159]. Recent studies have identified several novel compounds from marine-derived and endophytic fungi: Aspergillactone: A meroterpenoid from Aspergillus sp. CSYZ-1, showing potent antimicrobial activity against Helicobacter pylori and S. aureus with MIC values of 1–4 μg/mL and 2–16 μg/mL, respectively [160]. Miniolutelides D and E: New meroterpenoids from Talaromyces purpureogenus, isolated from pomegranate tissues, demonstrated antibacterial activity against S. aureus and methicillin-resistant S. aureus strains [161]. Chrodrimanins A and B: Isolated from Talaromyces funiculosus, these compounds exhibited broad-spectrum antibacterial activity, notably against S. aureus, Mycobacterium phlei, and E. coli [162]. Recently, Mo et al. [163] isolated four undescribed meroterpenoids from the fungus Penicillium sp. GDGJ-285 and one of them suppressed LPS-induced expression of TNF-α, IL-1β and iNOS by inhibiting the NF-κB signaling pathway.
Bisvertinolone is a secondary metabolite produced by certain Aspergillus species, notably A. versicolor (previously referred to as A. protuberus), which are often Isolated from cold, saline environments. This compound belongs to the class of polyketides and has attracted attention due to its notable antimicrobial activity. Bisvertinolone has been reported to inhibit the growth of various pathogenic microorganisms. The production of bisvertinolone by extremophilic fungi underscores the pharmaceutical potential of organisms adapted to harsh ecological niches [125].
Bisdechlorogeodin and secondary metabolites were extracted from the filamentous fungus Pseudogymnoascus sp. LAMAI 2784, isolated from soil sediments in Antarctica. It showed antibacterial activity against Xanthomonas citri subsp. citri, the cause of citrus canker. These fungal-derived metabolites are nucleoside transport inhibitors and have the potential to be used in the control of bacteria that cause diseases in agriculturally important crops [140].
Exopolysaccharides (EPS) are among the key bioactive compounds produced by psychrophilic and psychrotolerant filamentous fungi, particularly those inhabiting the extreme environments of continental Antarctica. These biopolymers can be used alone or alongside other bioactive agents in a wide variety of pharmaceutical applications, thanks to their diverse biological activities, including antioxidant, antimicrobial, anti-inflammatory, antiviral, antidiabetic, and anticoagulant properties. As highlighted by Zucconi et al. [164], the fungal component of these oligotrophic soils is predominantly composed of mesophilic-psychrotolerant and psychrophilic species that have evolved various adaptive strategies.
Alkaloids and Quinones. Alkaloids and quinones produced by filamentous fungi have shown promising antimicrobial activities. Compounds like fusarubin and bostrycoidin, produced by Fusarium solani, possess antimicrobial and antioxidant activities, contributing to the chemical defense arsenal of these fungi [146].
Marine-Derived Alkaloids: Over the past five years, 35 new antimicrobial alkaloids have been identified from marine fungi, with 22 exhibiting activity against Gram-negative bacteria. These compounds offer potential as narrow-spectrum antibiotics [165]. Liao et al. [166] isolated two new quinazoline alkaloids, versicomides G–H (1 and 2), along with seven known compounds, from A. versicolor HYQZ-215, obtained from the sediment of Qarhan Salt Lake. The antimicrobial activity of these compounds was evaluated against seven agricultural pathogenic fungi and eight clinically drug-resistant bacteria.
Fungal Quinones: Quinones such as emodins, fumigatins, and sorbicillinoids, produced by genera like Aspergillus, Penicillium, and Fusarium, have demonstrated antibacterial and antifungal properties. Their mechanisms include membrane disruption and inhibition of biofilm formation [167,168].
Peptides and Cyclodepsipeptides. Cyclodepsipeptides are cyclic compounds containing both peptide and ester bonds, known for their antimicrobial and cytotoxic activities. While specific new compounds were not detailed in the provided sources, ongoing research continues to explore marine-derived fungi for novel peptides and cyclodepsipeptides with potential therapeutic applications. Beauvericin, a cyclodepsipeptide from F. oxysporum, displays antimicrobial activity against methicillin-resistant S. aureus and B. subtilis [146]. The main characteristics of some of the mentioned biologically active substances are presented in Table 3.

Table 3.
Comparison of antibiotics and other antimicrobial compounds produced by filamentous fungi.
The potential of extremophilic fungi in antimicrobial drug discovery is vast, with numerous studies highlighting their ability to produce bioactive compounds with significant medicinal value. These compounds, often produced under harsh ecological conditions, represent a largely untapped resource for the development of novel therapeutic agents.
6. Mechanisms of Action of Antimicrobial Compounds from Filamentous Fungi
The antimicrobial efficacy of these fungal metabolites arises from various mechanisms targeting essential cellular processes in pathogens (Figure 3):
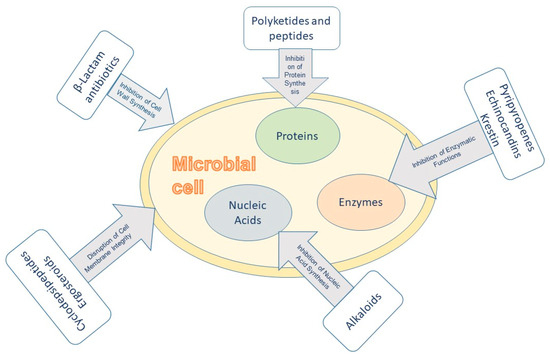
Figure 3.
Mechanisms of action of fungal antimicrobial metabolites.
6.1. Inhibition of Cell Wall Synthesis
β-Lactam antibiotics, such as penicillins, inhibit bacterial cell wall synthesis by binding to penicillin-binding proteins (PBPs), thereby disrupting peptidoglycan cross-linking and leading to cell lysis [143]. Cephalosporins, inhibit bacterial cell wall formation by preventing peptidoglycan bond connections. They bind to enzymes like carboxypeptidase and transpeptidase, blocking their function. Produced using strains of Paecilomyces persicinius, cephalosporins effectively treat bacterial infections from both Gram-positive and Gram-negative bacteria, including Staphylococcus aureus and Escherichia coli [143].
6.2. Disruption of Cell Membrane Integrity
An antibiotic peptide extracted from the culture medium of alkalophilic Emericellopsis alkalina, named emericellipsin A, exhibits strong antifungal activity against C. albicans, A. niger, and various clinical isolates of human pathogens. Additionally, it has demonstrated antimicrobial effects against both Gram-positive and Gram-negative bacteria. Its mechanism of action is linked to its impact on cell membranes, positioning it as a promising candidate for antimicrobial applications [169] as well as the mechanism of action of new sterol, 3,7-diketo-cephalosporin P1, in the culture liquid of the deep-sea A. fumigatus SCSIO 41012 [170].
Several antifungal and antibacterial agents produced by filamentous fungi exert their activity by disrupting the integrity of the microbial cell membrane. One notable example is Beauvericin, a cyclodepsipeptide isolated from various Fusarium species such as F. oxysporum and F. proliferatum. Beauvericin exhibits broad-spectrum antimicrobial activity, including efficacy against Gram-positive bacteria and fungi. Its mechanism involves the formation of ion-selective channels within the lipid bilayer of microbial membranes, thereby disrupting the membrane potential. These channels facilitate uncontrolled transmembrane fluxes of essential cations, particularly Ca2+, Na+, and K+, leading to osmotic imbalance, oxidative stress, and eventual cell lysis. In eukaryotic cells, this disruption can also trigger apoptotic pathways, making beauvericin a candidate for both antimicrobial and anticancer applications [171].
6.3. Inhibition of Protein Synthesis
Some fungal metabolites interfere with protein synthesis. For instance, certain polyketides and peptides can bind to ribosomal subunits or translation factors, hindering the translation process. One such compound is Aurovertin B, a flavonoid polyketide isolated from the marine-derived fungus Calcarisporium arbuscula. Although its primary target is not the ribosome itself, Aurovertin B potently inhibits mitochondrial ATP synthase by binding to the F1 subunit. This inhibition impairs cellular ATP production, which is essential for all stages of protein synthesis, including initiation, elongation, and termination. Consequently, Aurovertin B acts as an indirect inhibitor of protein synthesis, leading to energy starvation and suppressed growth in sensitive microorganisms [172]. A more direct example is Gliotoxin, an epipolythiodioxopiperazine-type metabolite produced by A. fumigatus. Gliotoxin interferes with protein biosynthesis by binding to thiol-containing enzymes and disrupting ribosomal function. In addition, it induces oxidative stress via the production of reactive oxygen species (ROS), which further impairs translation processes and leads to cellular dysfunction. The dual action of gliotoxin, through enzymatic inhibition and ROS-mediated damage, contributes to its potent antimicrobial and immunosuppressive activities [173].
6.4. Inhibition of Nucleic Acid Synthesis
Alkaloids like bostrycoidin may intercalate into DNA or inhibit topoisomerases, thereby obstructing DNA replication and transcription [146]. Griseofulvin is an antifungal agent that alters microtubules and DNA synthesis, respectively [174].
6.5. Inhibition of Enzymatic Functions
Pyripyropene A inhibits acyl-coenzyme A: cholesterol acyltransferase (ACAT2), an enzyme involved in lipid metabolism, which may contribute to its antimicrobial and insecticidal effects [143]. Echinocandins are the precursor leads of semisynthetic antifungal drugs against systemic infections such as aspergillosis and candidiasis. These drugs act by noncompetitively inhibiting the enzyme 1,3-β-D-glucan synthase required for the synthesis of fungal cell wall component β-glucan polymers. Echinocandin B isolated from Aspergillus nidulans was the first drug lead of class echinocandin [175]. Krestin is a protein-bound polysaccharide extracted from Basidiomycetes with anti-HIV properties. It was reported to inhibit the activity of the reverse transcriptase enzyme of the avian myeloblastosis virus [176].
7. The Relationship Between Extreme Environmental Conditions and the Production of Bioactive Compounds by Extremophilic Fungi
Extremophilic fungi inhabit Extreme habitats, such as polar regions, caves, glaciers, deep seas, deserts, hot springs, and volcanoes, that provide unique ecosystems for microorganisms. The microorganisms’ biochemical and genetic adaptations that enable survival under these conditions often correlate with the production of structurally unique and biologically active secondary metabolites. These compounds have significant applications in pharmaceuticals, cosmeceuticals, nutraceuticals, and biotechnology [177,178,179]. The environmental stressors present in such habitats impose selective pressures that drive the evolution of novel biosynthetic capabilities. Extremophilic fungi have demonstrated a remarkable capacity to generate secondary metabolites with diverse pharmacological properties, including antimicrobial, anticancer, and antioxidant activities. Notably, recent surveys indicate that fungi isolated from extreme environments yield a higher proportion of previously undescribed bioactive molecules compared to those from temperate ecosystems. They often produce unique metabolites to protect DNA, proteins, and lipids from damage [180]. The production of bioactive compounds in extremophiles is frequently linked to protective mechanisms against environmental stress. For example, certain strains synthesize melanin-like pigments that serve not only as a defense against radiation and oxidative damage but also exhibit inhibitory effects on microbial growth. Additionally, these fungi often produce osmoregulatory compounds, stress-responsive proteins, and enzymes with broad functional stability, which may indirectly or directly contribute to their bioactivity. Natural extreme conditions include fluctuating environmental factors such as salinity, dehydration, UV radiation, air pressure, pH, temperature, inorganic oxidants, and low organic carbon [181,182,183,184]. Technological advances in genome mining and metabolomics have facilitated the identification of biosynthetic gene clusters responsible for the production of these secondary metabolites. High-throughput sequencing and analytical chemistry have enabled the discovery of numerous new compound classes and scaffolds from extremophilic fungal strains, supporting the hypothesis that harsh habitats promote metabolic diversification.
In essence, the challenging conditions present in extreme environments act as evolutionary drivers, fostering metabolic innovations in filamentous fungi. These innovations not only ensure survival but also result in the synthesis of compounds with significant biotechnological and therapeutic potential. As our understanding of these organisms deepens, extremophilic fungi are increasingly recognized as promising sources of novel agents to combat antimicrobial resistance and other global health concerns. The search for new bioactive compounds from these environments is based on the idea that harsh abiotic conditions are selected for microorganisms that produce novel chemistry [185].
7.1. Low-Temperature Environments—Psychrophilic and Psychrotrophic Fungi
Some of the most frigid and least explored regions on Earth include the polar zones, permafrost layers, and deep-sea sediments and waters. These environments are difficult to access but harbor diverse microbial life, especially microorganisms adapted to extreme cold. Antarctica, the coldest and most remote continent, presents extreme conditions—low temperatures, intense seasonal light variation, and persistent ice cover—that have driven the evolution of unique biological communities producing a wide array of bioactive compounds [186].
Arctic and Antarctic marine ecosystems are increasingly recognized as valuable reservoirs of structurally diverse natural products with potential biomedical applications. Microorganisms in these cold environments have evolved various survival strategies, including entering dormant states and producing specialized cold-adapted metabolites and proteins [187]. Cryophilic organisms can minimize their metabolic activity and nutrient uptake by accumulating energy reserves such as polyphosphates, triglycerides, wax esters, and glycogen [188]. To maintain membrane fluidity in freezing conditions, they adjust the composition of fatty acids, increasing the proportion of branched and saturated forms [189].
Numerous studies have outlined the physiological, biochemical, and ecological adaptations that enable cold-tolerant fungi to persist in these harsh climates [190,191,192]. One such adaptation is the synthesis of cold-active enzymes by extremophilic fungi, which are thought to play a crucial role in their ability to function under extreme conditions [193,194,195].
Research efforts increasingly focus on isolating and characterizing natural products from these fungi, especially those with well-defined structures and proven biological activity against human pathogens. Among the most commonly reported compounds from cold-adapted fungi are terpenes, terpenoids, and their derivatives. Notably, antimicrobial activity is the most prevalent biological property observed among these metabolites [196].
Fungal communities in Arctic permafrost are mainly composed of Ascomycota, Basidiomycota, and Chytridiomycota, with common genera including Aspergillus, Cladosporium, Geomyces, and Penicillium [115,197]. The survival of shrubs in these areas is often supported by ectomycorrhizal fungi [198,199]. Antarctic soil systems also harbor diverse fungal species, with Cladosporium and Geomyces being potentially endemic [200]. The fungal diversity in Antarctic lichens on King George Island includes Arthonimycetes, Eurotiomycetes, Leoanoromycetes, Leotiomycetes, and Sordariomycetes (Ascomycota), as well as Cystobasidiomycetes and Tremellomycetes (Basidiomycota) [201].
Marine fungi are also found in cold deep-sea sediments, with diverse fungal assemblages observed in sediments from the northern Antarctic Peninsula and the East Indian Ocean [153,156]. Cold-adapted yeasts, including Candida, Cryptococcus, Pichia, and Rhodotorula, dominate the deep Polar Sea [202]. Additionally, deep-sea sediments in the Central Indian Basin have yielded filamentous fungi and yeasts from the Ascomycota [203,204]. Graphium species, which are barotolerant at high pressures (100 bar), have been recovered from the Northern Antarctic Peninsula [156].
Marine fungi are a rich source of bioactive compounds such as polyketide griseofulvin, terpenoid fusidic acid, and cephalosporins [205]. The isolation of new bioactive molecules from marine environments has been increasing, with 448 new compounds reported in 2017 alone [206].
7.2. Permafrost Soils and Cold-Adapted Fungi
Permafrost refers to ground that remains at or below 0 °C for at least two consecutive years and constitutes approximately 25% of Earth’s terrestrial surface [207]. These frozen layers, along with the seasonally thawed active layer above them, serve as unique ecological niches for psychrophilic and psychrotolerant microbial communities [208,209,210]. Unlike other cold ecosystems such as sea ice or deep-sea environments, permafrost is characterized by pronounced structural heterogeneity due to variations in soil composition, ice content, and organic material along horizontal and vertical gradients [207]. Microorganisms that thrive within the permafrost and tolerate temperature ranges from –17 °C to +10 °C are classified as cryophiles [193].
The genus Penicillium has emerged as a prolific producer of bioactive secondary metabolites, particularly from strains isolated in extreme environments. P. griseofulvum MCCC 3A00225, derived from Indian Ocean deep-sea sediment, yielded 10 novel and 26 known compounds as reported by Xing et al. [211]. These compounds were screened for anti-allergic activity using an immunoglobulin E (IgE)-mediated rat basophilic leukemia (RBL-2H3) cell model. Among them, (-)-cyclopenol exhibited significant activity by inhibiting degranulation with an IC50 value of 60.3 μM, outperforming the positive control, loratadine (IC50 = 91.6 μM).
7.3. High-Temperature Environments: Thermophilic and Hyperthermophilic Fungi
Thermophilic and hyperthermophilic microorganisms inhabit a range of extreme environments, including deserts, hot springs, deep-sea hydrothermal vents, and anthropogenic niches such as compost heaps [212,213]. Thermophiles typically grow optimally at approximately 55 °C, while hyperthermophiles thrive at temperatures exceeding 80 °C. Some hypertermophiles like a hypertermophilic archea Pyrodyctium occultum known as strain 121 are able to grow at 121 °C [214]. These organisms may also encounter and adapt to additional stressors such as low pH or high salinity, which influence their environmental distribution. One adaptive trait of hyperthermophiles is the production of polyamines—long-chain functional polymers that contribute to thermal stability [215]. Their ability to withstand such conditions makes them promising sources for biotechnological applications, including thermostable enzymes and biofuel production [216,217].
Fungi exhibit a broad spectrum of adaptations that enable survival in high-temperature and arid environments, such as those found in deserts. These include the synthesis of melanin, which provides protection against high ultraviolet (UV) radiation [218]. A preliminary survey of the Atacama Desert identified 13 fungal genera, with Alternaria and Ulocladium being among the most prominent [219]. Further investigations have revealed that desert fungal communities harbor a rich and largely unexplored reservoir of biodiversity [220]. For instance, Grishkan and Nevo [221] identified 135 novel ascomycetous species in Makhtesh Ramon desert soils, while Castillo and Beck [222] documented 77 lichenoid fungi, including four new species, along altitudinal transects in Alto Patache. High-altitude rocks in the Atacama Desert were found to host fungal genera such as Cladosporium, Neocatenulostroma, and Penicillium [223]. Two halophilic fungi, Aspergillus atacamensis and Aspergillus salisburgensis, were isolated from a cave in the Coastal Range of the Atacama Desert [224]. Additional novel desert fungi, including Diverispora omaniana, Septoglomus nakheelum, and Rhizophagus arabicus, have been discovered in Oman [225].
Fungi of the genus Wallemia, previously categorized as halophiles [226], have since been recognized for their metabolic versatility. Wallemia sebi, isolated from the Atacama Desert, produces diverse secondary metabolites such as wallimidione, 15-azasterol, and 24,28-dihydro-15-azasterol [227]. These compounds exhibit antitumor activity in vivo and antiproliferative effects against HeLa S3 cells, as well as antimicrobial activity against Saccharomyces cerevisiae and Gram-positive bacteria. Moreover, the cyclopentanopyridine alkaloid from W. sebi inhibits Enterobacter aerogenes [228], and the fungus also synthesizes unique terpenes like walleminone and walleminol [227].
Although thermophilic fungi have historically garnered limited attention, recent studies have reported thermotolerant species such as Aspergillus clavatus [229] and Talaromyces thermophilus [230,231], underscoring their potential importance in both ecological and applied microbiology.
7.4. Desert Ecosystems
Despite covering approximately 20% of Earth’s land surface, deserts have historically received limited attention in microbiological research. These ecosystems pose significant challenges to microbial life, primarily due to extreme aridity and the persistent scarcity of water. Among the most well-studied desert systems is the Atacama Desert, a temperate, non-polar desert whose unique abiotic characteristics have been detailed in several comprehensive reviews [232,233,234]. The polar desert areas, such as the Dry Valleys in Antarctica, should also be mentioned [235].
Microbiological surveys in the Atacama Desert have concentrated on isolating and characterizing extremophilic microorganisms, particularly actinobacteria, from its hyperarid and extreme hyperarid regions. These regions exhibit exceptionally low moisture levels, with the ratio of mean annual precipitation to evaporation as low as 0.05% and 0.02%, respectively. For many years, it was assumed that such extreme conditions rendered Atacama’s core region virtually lifeless [236]. However, recent studies have revealed that a diverse array of microorganisms has adapted to thrive under these inhospitable conditions [237].
Interestingly, a rare and intense rainfall event in the hyperarid core of the Atacama Desert paradoxically resulted in the destruction of surface microbial communities. This counterintuitive outcome was attributed to the osmotic shock and metabolic disruption experienced by microorganisms unaccustomed to sudden water availability [238].
7.5. Highly Acidic Habitats: Acidophilic Fungi
Extreme acidophiles are generally defined as microorganisms that grow optimally at pH levels below 3 [239]. These organisms are commonly isolated from highly acidic environments such as acid lakes, acid sulfate soils, and acid mine drainage sites [240,241]. Acidophilic microorganisms have evolved specialized adaptations that enable them to survive and function in such harsh conditions, with membrane integrity and ion transport systems playing key roles in pH homeostasis [93,242]. In prokaryotic acidophiles, such as certain archaea and bacteria, the composition and properties of the cell membrane are critical for maintaining intracellular pH [243,244].
Despite their ecological significance, relatively little is known about fungal species inhabiting acidic environments, although they represent an integral component of microbial communities in these biomes [244]. Notably, several acidophilic fungi have demonstrated potential for biotechnological applications. For example, Penicillium spp. isolated from an abandoned open-pit mine containing acid-metal waste in Montana, USA, were found to produce novel bioactive compounds [119]. Among these, the P. rubrum strain produced unique meroterpenoids—including berkeleyones, berkeleydione, and berkeleytrione—as well as preaustinoids A and A1, all of which inhibited in vitro production of interleukin-1β [245]. In addition, Penicillium solitum, another fungal strain isolated from the same acid mine environment, was found to produce two new drimane sesquiterpene lactones, berkedrimanes A and B, along with a novel tricarboxylic acid. These compounds exhibited anti-inflammatory activity by inhibiting caspase-1 and caspase-3 enzymes in vitro [246].
7.6. Saline and Hypersaline Habitats: Halotolerant and Halophilic Fungi
Saline environments are typically defined as habitats with salt concentrations comparable to seawater, approximately 3.5% (w/v) of total dissolved salts [247]. In contrast, hypersaline environments—such as Antarctic biomes and hypersaline lakes—contain significantly higher salt concentrations, exceeding 100 g/L [248]. These extreme habitats present unique physicochemical challenges, including elevated osmotic pressure and reduced water activity. Halophilic microorganisms are well-adapted to such conditions through the evolution of specialized molecular and cellular mechanisms [177,249,250]. In comparison, halotolerant fungi are capable of growing in non-saline environments but can withstand high salinity when necessary.
Extremely halotolerant and halophilic fungi have been isolated from solar salterns across diverse geographic regions. Notable examples include melanized species within the genera Aspergillus, Cladosporium, and Penicillium, as well as Emericella and Eurotium. Additionally, non-melanized yeasts and species of Wallemia have also been identified in these salt-rich environments [250].
8. Conclusions
Antimicrobial resistance remains one of the most serious challenges facing modern medicine and global public health. It results from a complex interplay of factors, including the uncontrolled use of antibiotics and broad socio-economic changes, that accelerate the spread of resistant pathogens. As a result, the need to discover new antimicrobial agents is more urgent than ever. In this context, extremophilic microorganisms, which possess exceptional adaptive mechanisms for surviving in harsh environments, offer unique biomedical potential.
Particular attention should be given to extremophilic and extremotolerant filamentous fungi, which are capable of synthesizing bioactive compounds with significant antimicrobial activity. These compounds not only hold promise for the development of novel therapeutic agents but may also prove crucial in overcoming resistance to existing drugs. Moreover, research into these organisms contributes both to the expansion of fundamental knowledge in microbiology and ecology and to the advancement of innovative biotechnological approaches.
Based on current scientific evidence, it can be concluded that extremophilic filamentous fungi represent a valuable and still underexplored resource in the fight against antimicrobial resistance. Strategic investment in targeted research and the development of biotechnological platforms focused on their exploration and utilization could significantly contribute to the creation of effective and sustainable therapies in the future.
Author Contributions
Conceptualization, E.K.; methodology, E.K., R.A., J.M.-S., V.D., M.A. and B.S.; software, J.M.-S. and G.S.; validation, R.A. and Y.G.; formal analysis, M.A.; investigation, E.K. and R.A.; resources, E.K., R.A., J.M.-S., Y.G., G.S., V.D., L.Y. and B.S.; data curation, R.A.; writing—original draft preparation, E.K.; writing—review and editing, E.K.; visualization, R.A.; supervision, E.K. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Bulgarian Ministry of Education and Science through the National Centre for Polar Studies, and Sofia University “St. Kliment Ohridski” in the framework of the National Program for Polar Studies 2022–2025.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| MDR | Multidrug Resistance |
| AMR | Antimicrobial resistance |
References
- Libertucci, J.; Young, V.B. The role of the microbiota in infectious diseases. Nat. Microbiol. 2018, 4, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Muteeb, G.; Rehman, M.T.; Shahwan, M.; Aatif, M. Origin of Antibiotics and Antibiotic Resistance, and Their Impacts on Drug Development: A Narrative Review. Pharmaceuticals 2023, 16, 1615. [Google Scholar] [CrossRef] [PubMed]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A. Antimicrobial resistance: A growing serious threat for global public health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef]
- de la Fuente-Nunez, C.; Cesaro, A.; Hancock, R.E. Antibiotic failure: Beyond antimicrobial resistance. Drug Resist. Updates 2023, 71, 101012. [Google Scholar] [CrossRef]
- Adebisi, Y.A.; Ogunkola, I.O. The global antimicrobial resistance response effort must not exclude marginalised populations. Trop. Med. Health 2023, 51, 33. [Google Scholar] [CrossRef] [PubMed]
- Yam, E.L.Y.; Hsu, L.Y.; Yap, E.P.-H.; Yeo, T.W.; Lee, V.; Schlundt, J.; Lwin, M.O.; Limmathurotsakul, D.; Jit, M.; Dedon, P.; et al. Antimicrobial resistance in the Asia Pacific region: A meeting report. Antimicrob. Resist. Infect. Control 2019, 8, 202. [Google Scholar] [CrossRef]
- Figueroa, L.; Jiménez, C.; Rodríguez, J.; Areche, C.; Chávez, R.; Henríquez, M.; de la Cruz, M.; Díaz, C.; Segade, Y.; Vaca, I. 3-Nitroasterric acid derivatives from an Antarctic sponge-derived Pseudogymnoascus sp. fungus. J. Nat. Prod. 2015, 78, 919–923. [Google Scholar] [CrossRef]
- Chávez, R.; Fierro, F.; García-Rico, R.O.; Vaca, I. Filamentous fungi from extreme environments as a promising source of novel bioactive secondary metabolites. Front. Microbiol. 2015, 6, 903. [Google Scholar] [CrossRef]
- Ramírez-Castillo, F.Y.; Moreno-Flores, A.C.; Avelar-González, F.J.; Márquez-Díaz, F.; Harel, J.; Guerrero-Barrera, A.L. An evaluation of multidrug-resistant Escherichia coli isolates in urinary tract infections from Aguascalientes, Mexico: Cross-sectional study. Ann. Clin. Microbiol. Antimicrob. 2018, 17, 34. [Google Scholar] [CrossRef]
- Davies, D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2003, 2, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Founou, R.C.; Founou, L.L.; Essack, S.Y. Clinical and economic impact of antibiotic resistance in developing countries: A systematic review and meta-analysis. PLoS ONE 2017, 12, e0189621. [Google Scholar] [CrossRef] [PubMed]
- Khatoon, Z.; McTiernan, C.D.; Suuronen, E.J.; Mah, T.F.; Alarcon, E.I. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon 2018, 4, e01067. [Google Scholar] [CrossRef]
- Hancock, R.E.W.; Alford, M.A.; Haney, E.F. Antibiofilm activity of host defence peptides: Complexity provides opportunities. Nat. Rev. Microbiol. 2021, 9, 786–797. [Google Scholar] [CrossRef]
- Chatterjee, S.; Poonawala, H.; Jain, Y. Drug-resistant tuberculosis: Is India ready for the challenge? BMJ Glob. Health 2018, 3, e000971. [Google Scholar] [CrossRef]
- Bassetti, M.; Peghin, M.; Vena, A.; Giacobbe, D.R. Treatment of infections due to MDR gram-negative bacteria. Front. Med. 2019, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Kaldalu, N.; Hauryliuk, V.; Turnbull, K.J.; La Mensa, A.; Putrinš, M.; Tenson, T. In vitro studies of persister cells. Microbiol. Mol. Biol. Rev. 2020, 84, e00070-20. [Google Scholar] [CrossRef]
- Sandoval-Motta, S.; Aldana, M. Adaptive resistance to antibiotics in bacteria: A systems biology perspective. WIREs Syst. Biol. Med. 2016, 8, 253–267. [Google Scholar] [CrossRef]
- Rizi, K.S.; Ghazvini, K.; Noghondar, M. Adaptive antibiotic resistance: Overview and perspectives. J. Infect. Dis. Ther. 2018, 6, 363. [Google Scholar] [CrossRef]
- Annavajhala, M.K.; Gomez-Simmonds, A.; Uhlemann, A.-C. Multidrug-resistant Enterobacter cloacae complex emerging as a global, diversifying threat. Front. Microbiol. 2019, 10, 44. [Google Scholar] [CrossRef]
- Llor, C.; Bjerrum, L. Antimicrobial resistance: Risk associated with antibiotic overuse and initiatives to reduce the problem. Ther. Adv. Drug Saf. 2014, 5, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Chokshi, A.; Sifri, Z.; Cennimo, D.; Horng, H. Global contributors to antibiotic resistance. J. Glob. Infect. Dis. 2019, 11, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef]
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.; Wertheim, H.F.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance—The need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef]
- Chaw, P.S.; Höpner, J.; Mikolajczyk, R. The knowledge, attitude and practice of health practitioners towards antibiotic prescribing and resistance in developing countries—A systematic review. J. Clin. Pharm. Ther. 2018, 43, 606–613. [Google Scholar] [CrossRef]
- Dadgostar, P. Antimicrobial resistance: Implications and costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, J.G.; Gilbert, D.N.; Spellberg, B. Seven ways to preserve the miracle of antibiotics. Clin. Infect. Dis. 2013, 56, 1445–1450. [Google Scholar] [CrossRef]
- Rhouma, M.; Beaudry, F.; Thériault, W.; Letellier, A. Colistin in pig production: Chemistry, mechanism of antibacterial action, microbial resistance emergence, and one health perspectives. Front. Microbiol. 2016, 7, 1789. [Google Scholar] [CrossRef]
- Klein, E.Y.; Van Boeckel, T.P.; Martinez, E.M.; Laxminarayan, R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 2018, 115, E3463–E3470. [Google Scholar] [CrossRef]
- Ruppé, E.; Armand-Lefèvre, L.; Estellat, C.; El-Mniai, A.; Boussadia, Y.; Consigny, P.H.; Girard, P.M.; Vittecoq, D.; Bouchaud, O.; Pialoux, G.; et al. Acquisition of carbapenemase-producing Enterobacteriaceae by healthy travellers to India, France, February 2012 to March 2013. Eurosurveillance 2014, 19, 20768. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Jeannot, K.; Xiao, Y.; Knapp, C.W. Editorial: Horizontal gene transfer mediated bacterial antibiotic resistance. Front. Microbiol. 2019, 10, 1933. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.C.M.; Jones, M.; Snelling, T.L.; Duguid, R.; Moore, N.; Dickson, B.; Wu, Y.; Saunders, J.; Wijeratne, P.; Douangnouvong, A.; et al. Coverage gaps in empiric antibiotic regimens used to treat serious bacterial infections in neonates and children in Southeast Asia and the Pacific. Lancet Reg. Health Southeast Asia 2023, 22, 100291. [Google Scholar] [CrossRef]
- Bassetti, S.; Tschudin-Sutter, S.; Egli, A.; Osthoff, M. Optimizing antibiotic therapies to reduce the risk of bacterial resistance. Eur. J. Intern. Med. 2022, 99, 7–12. [Google Scholar] [CrossRef]
- Friedrich, M.J. Drug-resistant tuberculosis predicted to increase in high-burden countries. JAMA 2017, 318, 231. [Google Scholar] [CrossRef]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef]
- Hao, H.; Cheng, G.; Iqbal, Z.; Ai, X.; Hussain, H.I.; Huang, L.; Dai, M.; Wang, Y.; Liu, Z.; Yuan, Z. Benefits and risks of antimicrobial use in food-producing animals. Front. Microbiol. 2014, 5, 288. [Google Scholar] [CrossRef]
- MacElroy, R.D. Some comments on the evolution of extremophiles. Biosystems 1974, 6, 74–75. [Google Scholar] [CrossRef]
- Arbab, S.; Ullah, H.; Khan, M.I.U.; Khattak, M.N.K.; Zhang, J.; Li, K. Diversity and distribution of thermophilic microorganisms and their applications in biotechnology. J. Basic Microbiol. 2022, 62, 95–108. [Google Scholar] [CrossRef]
- Gostinčar, C.; Stajich, J.E.; Gunde-Cimerman, N. Extremophilic and extremotolerant fungi. Curr. Biol. 2023, 33, 752–756. [Google Scholar] [CrossRef] [PubMed]
- Kanekar, P.P.; Kanekar, S.P. Psychrophilic, Psychrotrophic, and Psychrotolerant Microorganisms. In Diversity and Biotechnology of Extremophilic Microorganisms. In Diversity and Biotechnology of Extremophilic Microorganisms from India; Kanekar, P.P., Kanekar, S.P., Eds.; Springer Nature: Singapore, 2022; pp. 215–249. ISBN 978-981-19157-3-4. [Google Scholar]
- Das, T.; Ray, P.; Nandy, S.; Al-Tawaha, A.R.; Pandey, D.K.; Kumar, V.; Dey, A. Piezophilic fungi: Sources of novel natural products with preclinical and clinical significance. In Extremophilic Fungi: Ecology, Physiology and Applications; Springer: Singapore, 2022; pp. 523–545. [Google Scholar]
- Coleine, C.; Stajich, J.E.; Selbmann, L. Fungi are key players in extreme ecosystems. Trends Ecol. Evol. 2022, 37, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Merino, N.; Aronson, H.S.; Bojanova, D.P.; Feyhl-Buska, J.; Wong, M.L.; Zhang, S.; Giovannelli, D. Living at the extremes: Extremophiles and the limits of life in a planetary context. Front. Microbiol. 2019, 10, 780. [Google Scholar] [CrossRef]
- Hallsworth, J.E.; Mancinelli, R.L.; Conley, C.A.; Dallas, T.D.; Rinaldi, T.; Davila, A.F.; Benison, K.C.; Rapoport, A.; Cavalazzi, B.; Selbmann, L.; et al. Astrobiology of life on Earth. Environ. Microbiol. 2021, 23, 3335–3344. [Google Scholar] [CrossRef]
- Basu, B.; Roy, C.; Saha, S.; Tirkey, A.S.; Biswas, R.; Dey, A.; Mitra, A.K. Extremophiles to polyextremophiles: Survival of the fittest. In Extremophiles: A Paradox of Nature with Biotechnological Implications; De Gruyter: Berlin, Germany; Boston, MA, USA, 2022; Volume 1, p. 221. [Google Scholar]
- Fenice, M.; Khare, S.K.; Gorrasi, S. Mining, designing, mechanisms and applications of extremophilic enzymes. Front. Microbiol. 2021, 12, 709377. [Google Scholar] [CrossRef] [PubMed]
- Naranjo-Ortiz, M.A.; Gabaldón, T. Fungal evolution: Major ecological adaptations and evolutionary transitions. Biol. Rev. Camb. Philos. Soc. 2019, 94, 1443–1476. [Google Scholar] [CrossRef]
- Selbmann, L.; Egidi, E.; Isola, D.; Onofri, S.; Zucconi, L.; de Hoog, G.S.; Varese, G.C. Biodiversity, evolution and adaptation of fungi in extreme environments. Plant Biosyst. 2013, 147, 237–246. [Google Scholar] [CrossRef]
- Drake, H.; Ivarsson, M. The role of anaerobic fungi in fundamental biogeochemical cycles in the deep biosphere. Fungal Biol. Rev. 2018, 32, 20–25. [Google Scholar] [CrossRef]
- Miteva-Staleva, J.; Krumova, E.; Stefanova, T.; Angelova, M. Age-related changes in reactive oxygen species production in the filamentous fungus Penicillium rugulosum T35 under cold stress conditions. Comptes Rendus L’académie Bulg. Sci. 2015, 68, 1123–1129. [Google Scholar]
- Gao, Y.; Fang, H.; Fang, L.; Liu, D.; Liu, J.; Su, M.; Jiao, H. The modification and design of antimicrobial peptide. Curr. Pharm. Des. 2018, 24, 904–910. [Google Scholar] [CrossRef]
- Gunde-Cimerman, N.; Plemenitaš, A.; Oren, A. Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol. Rev. 2018, 1, 353–375. [Google Scholar] [CrossRef]
- Gostinčar, C.; Gunde-Cimerman, N.; Grube, M. Polyextremotolerance as the fungal answer to changing environments. In Microbial Evolution Under Extreme Conditions; Bakermans, C., Ed.; De Gruyter: Berlin, Germany; Boston, MA, USA, 2015; pp. 185–208. [Google Scholar] [CrossRef]
- Lindt, W. Mitteilungen über einige neue pathogene Schimmelpilze. Arch. Exp. Pathol. Pharmakol. 1886, 21, 269–298. [Google Scholar] [CrossRef]
- Tansey, M.R.; Brock, T.D. The upper temperature limit for eukaryotic organisms. Proc. Natl. Acad. Sci. USA 1972, 69, 2426–2428. [Google Scholar] [CrossRef]
- Maheshwari, R.; Bharadwaj, G.; Bhat, M.K. Thermophilic fungi: Their physiology and enzymes. Microbiol. Mol. Biol. Rev. 2000, 64, 461–488. [Google Scholar] [CrossRef] [PubMed]
- Patel, H.; Rawat, S. Thermophilic fungi: Diversity, physiology, genetics, and applications. In New and Future Developments in Microbial Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2021; pp. 69–93. [Google Scholar]
- Cooney, D.G.; Emerson, R. Thermophilic Fungi; W.H. Freeman and Company: San Francisco, CA, USA, 1964; Volume 27. [Google Scholar]
- Massimo, N.C.; Devan, M.N.; Arendt, K.R.; Wilch, M.H.; Riddle, J.M.; Furr, S.H.; Steen, C.; U’Ren, J.M.; Sandberg, D.C.; Arnold, A.E. Fungal endophytes in aboveground tissues of desert plants: Infrequent in culture, but highly diverse and distinctive symbionts. Mycologia 2015, 70, 61–76. [Google Scholar] [CrossRef] [PubMed]
- St-Germain, G.; Robert, A.; Ishak, M.; Tremblay, C.; Claveau, S. Infection due to: Report of four cases in patients with leukemia and review. Clin. Infect. Dis. 1993, 16, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Qiu, M.; Xu, H.; Guo, P.; Peng, Y. Application of Metagenomics Next-Generation Sequencing on Diagnosis of Disseminated Infection Caused by Rhizomucor pusillus in an Acute Lymphoblastic Leukemia Patient. Infect. Drug Resist. 2024, 17, 5707–5713. [Google Scholar] [CrossRef]
- Tedersoo, L.; Sánchez-Ramírez, S.; Kõljalg, U.; Bahram, M.; Döring, M.; Schigel, D.; May, T.; Ryberg, M.; Abarenkov, K. High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Divers. 2018, 90, 135–159. [Google Scholar] [CrossRef]
- Salar, R.K. Thermophilic fungi: Basic Concepts and Biotechnological Applications, 1st ed.; CRC Press: Boca Raton, FA, USA, 2018. [Google Scholar] [CrossRef]
- Kirk, P.M.; Cannon, P.F.; Minter, D.W.; Stalpers, J.A. Dictionary of the Fungi, 10th ed.; CABI: Wallingford, UK, 2008. [Google Scholar]
- Ianutsevich, E.A.; Danilova, O.A.; Groza, N.V.; Kotlova, E.R.; Tereshina, V.M. Heat shock response of thermophilic fungi: Membrane lipids and soluble carbohydrates under elevated temperatures. Microbiology 2016, 162, 989–999. [Google Scholar] [CrossRef]
- Almaguer Chávez, M. Thermotolerance and Adaptation to Climate Change. In The Impact of Climate Change. In The Impact of Climate Change on Fungal Diseases; Springer International Publishing: Cham, Switzerland, 2022; pp. 37–71. [Google Scholar] [CrossRef]
- Hassan, N.; Rafiq, M.; Hayat, M.; Shah, A.A.; Hasan, F. Psychrophilic and psychrotrophic fungi: A comprehensive review. Rev. Environ. Sci. Biotechnol. 2016, 15, 147–172. [Google Scholar] [CrossRef]
- Morita, R.Y. Psychrophilic bacteria. Bacteriol. Rev. 1975, 39, 144–167. [Google Scholar] [CrossRef] [PubMed]
- Di Menna, M.E. Three new yeasts from Antarctic soils: Candida nivalis, Candida gelida and Candida frigida spp. Antonie Van Leeuwenhoek 1966, 32, 25–28. [Google Scholar] [CrossRef] [PubMed]
- Abrashev, R.; Miteva-Staleva, J.; Gocheva, Y.; Stoyancheva, G.; Dishliyska, V.; Spasova, B.; Krumova, E.; Angelova, M. Cell Response to Oxidative Stress in Antarctic Filamentous Fungi. Appl. Sci. 2025, 15, 5149. [Google Scholar] [CrossRef]
- Ma, F.; Hanna, M.A. Biodiesel production: A review. Bioresour. Technol. 1999, 70, 1–15. [Google Scholar] [CrossRef]
- Golubev, V.I. Rhodotorula creatinovora and R. yakutica—New species of basidiomycetous yeasts, extracted from permafrost soils of eastern Siberian Arctic. Mikol. Fitopatol. 1998, 32, 8–13. [Google Scholar]
- Jodłowska, I.; Białkowska, A.M. Cold-Adapted Fungi: Goldmine of Biomolecules Applicable in Industry. Appl. Sci. 2024, 14, 11950. [Google Scholar] [CrossRef]
- Yadav, A.N.; Yadav, N.; Sachan, S.G.; Saxena, A.K. Biodiversity of psychrotrophic microbes and their biotechnological applications. J. Appl. Biol. Biotechnol. 2019, 7, 99–108. [Google Scholar] [CrossRef]
- Miteva-Staleva, J.; Krumova, E.; Stoyancheva, G.; Kostadinova, N.; Grozdanov, P.; Spassova, B.; Angelova, M. Isolation, Identification and Proteolytic Activity of Filamentous Fungi from Alaska. Acta Microbiol. Bulg. 2022, 38, 26–30. [Google Scholar]
- Tsuji, M. Survey on Fungi in Antarctica and High Arctic Regions, and Their Impact on Climate Change. Climate 2023, 11, 195. [Google Scholar] [CrossRef]
- Yusof, N.A.; Hashim, N.H.F.; Bharudin, I. Cold Adaptation Strategies and the Potential of Psychrophilic Enzymes from the Antarctic Yeast, Glaciozyma antarctica PI12. J. Fungi 2021, 7, 528. [Google Scholar] [CrossRef]
- Bowman, J.P. Genomics of Psychrophilic Bacteria and Archaea. In Psychrophiles: From Biodiversity to Biotechnology; Margesin, R., Ed.; Springer: Cham, Switzerland, 2017; pp. 201–220. [Google Scholar]
- Zalar, P.; de Hoog, G.S.; Gunde-Cimerman, N. Trimmatostroma salinum, a new species from hypersaline water. Stud. Mycol. 1999, 43, 61–67. [Google Scholar]
- Zalar, P.; de Hoog, G.S.; Gunde-Cimerman, N. Taxonomy of the endoconidial black yeast genera Phaeotheca and Hyphospora. Stud. Mycol. 1999, 43, 49–56. [Google Scholar]
- Gunde-Cimerman, N.; Zalar, P.; de Hoog, G.S.; Plemenitaš, A. Hypersaline waters in salterns—Natural ecological niches for halophilic black yeasts. FEMS Microbiol. Ecol. 2000, 32, 235–240. [Google Scholar] [CrossRef]
- Zajc, J.; Gunde-Cimerman, N. The genus Wallemia—From contamination of food to health threat. Microorganisms 2018, 6, 46. [Google Scholar] [CrossRef]
- Gunde-Cimerman, N.; Plemenitaš, A. Ecology and molecular adaptations of the halophilic black yeast Hortaea werneckii. Rev. Environ. Sci. Biotechnol. 2007, 5, 323–331. [Google Scholar] [CrossRef]
- Gunde-Cimerman, N.; Ramos, J.; Plemenitaš, A. Halotolerant and halophilic fungi. Mycol. Res. 2009, 113, 1231–1241. [Google Scholar] [CrossRef]
- Hohmann, S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002, 66, 300–372. [Google Scholar] [CrossRef] [PubMed]
- Kejžar, A.; Gobec, S.; Plemenitaš, A.; Lenassi, M. Melanin is crucial for growth of the black yeast Hortaea werneckii in its natural hypersaline environment. Fungal Biol. 2013, 117, 368–379. [Google Scholar] [CrossRef]
- Rafiq, M.; Hassan, N.; Rehman, M.; Hasan, F. Adaptation mechanisms and applications of psychrophilic fungi. In Fungi in Extreme Environments: Ecological Role and Biotechnological Significance; Tiquia-Arashiro, S., Grube, M., Eds.; Springer: Cham, Switzerland, 2019; pp. 145–160. [Google Scholar] [CrossRef]
- Kejžar, A.; Grötli, M.; Tamás, M.J.; Plemenitaš, A.; Lenassi, M. HwHog1 kinase activity is crucial for survival of Hortaea werneckii in extremely hyperosmolar environments. Fungal Genet. Biol. 2015, 74, 45–58. [Google Scholar] [CrossRef]
- Jin, Q.; Kirk, M.F. pH as a primary control in environmental microbiology: 1. Thermodynamic perspective. Front. Environ. Sci. 2018, 6, 21. [Google Scholar] [CrossRef]
- Krulwich, T.A.; Sachs, G.; Padan, E. Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 2011, 9, 330–343. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Dopson, M. Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 2007, 15, 165–171. [Google Scholar] [CrossRef]
- Matin, A. pH homeostasis in acidophiles. Novartis Found. Symp. 2007, 221, 152–166. [Google Scholar]
- Buetti-Dinh, A.; Dethlefsen, O.; Friedman, R.; Dopson, M. Transcriptomic analysis reveals how a lack of potassium ions increases Sulfolobus acidocaldarius sensitivity to pH changes. Microbiology 2016, 162, 1422–1434. [Google Scholar] [CrossRef]
- Christel, S.; Herold, M.; Bellenberg, S.; El Hajjami, M.; Buetti-Dinh, A.; Pivkin, I.V.; Sand, W.; Wilmes, P.; Poetsch, A.; Dopson, M. Multi-omics reveals the lifestyle of the acidophilic, mineral-oxidizing model species Leptospirillum ferriphilum. Appl. Environ. Microbiol. 2018, 84, e02091-17. [Google Scholar] [CrossRef]
- Sahay, S. Extremophilic fungi: Ecology, physiology, and applications. In Extremophilic Fungi; Springer: Singapore, 2022. [Google Scholar] [CrossRef]
- Goswami, S.; Das, M. Extremophiles: A clue to the origin of life and biology of other planets. Everyman’s Sci. 2016, 51, 17–25. [Google Scholar]
- Poli, A.; Finore, I.; Romano, I.; Gioiello, A.; Lama, L.; Nicolaus, B. Microbial diversity in extreme marine habitats and their biomolecules. Microorganisms 2017, 5, 25. [Google Scholar] [CrossRef] [PubMed]
- Belyagoubi, L.; Belyagoubi-Benhammou, N.; Jurado, V.; Dupont, J.; Lacoste, S.; Djebbah, F.; Saiz-Jimenez, C. Antimicrobial activities of culturable microorganisms (actinomycetes and fungi) isolated from Chaabe Cave, Algeria. Int. J. Speleol. 2018, 47, 189–199. [Google Scholar] [CrossRef]
- Meglali, A.; Ghellai, L. An overview of extremophiles: Microbial diversity, adaptive strategies, and potential applications. Microbiol. Biotechnol. Lett. 2024, 52, 233–254. [Google Scholar] [CrossRef]
- Gonçalves, N.S.; Mello, T.M.S.P.; Mizuno, C.S.; Haider, S.; Santos, R.A. Cis-trimethoxystilbene exhibits higher genotoxic and antiproliferative effects than its isomer trans-trimethoxystilbene in MCF-7 and MCF-10A cell lines. Genet. Mol. Biol. 2021, 44, e20200477. [Google Scholar] [CrossRef]
- Fredimoses, M.; Zhou, X.; Lin, X.; Tian, X.; Ai, W.; Wang, J.; Liao, S.; Liu, J.; Yang, B.; Yang, X.; et al. New prenylxanthones from the deep-sea derived fungus Emericella sp. SCSIO 05240. Mar. Drugs 2014, 12, 4218–4234. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Wang, J.; Zhang, X.; Nong, X.; Xu, X.; Qi, S. Cytotoxic polyketides from the deep-sea-derived fungus Engyodontium album DFFSCS021. Mar. Drugs 2014, 12, 2774–2790. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; He, W.; Huang, X.; Tian, X.; Liao, S.; Yang, B.; Wang, F.; Zhou, X.; Liu, Y. Antifungal new oxepine-containing alkaloids and xanthones from the deep-sea-derived fungus Aspergillus versicolor SCSIO 05879. J. Agric. Food Chem. 2016, 64, 2910–2916. [Google Scholar] [CrossRef]
- Wang, W.; Chen, R.; Luo, Z.; Wang, W.; Chen, J. Antimicrobial activity and molecular docking studies of a novel anthraquinone from a marine-derived fungus Aspergillus versicolor. Nat. Prod. Res. 2017, 32, 558–563. [Google Scholar] [CrossRef]
- Wang, W.; Li, S.; Chen, Z.; Li, Z.; Liao, Y.; Chen, J. Secondary metabolites produced by the deep-sea-derived fungus Engyodontium album. Chem. Nat. Compd. 2017, 53, 224–226. [Google Scholar] [CrossRef]
- Luo, X.; Lin, X.; Salendra, L.; Pang, X.; Dai, Y.; Yang, B.; Liu, J.; Wang, J.; Zhou, X.; Liu, Y. Isobenzofuranones and isochromenones from the deep-sea-derived fungus Leptosphaeria sp. SCSIO 41005. Mar. Drugs 2017, 15, 204. [Google Scholar] [CrossRef]
- Niu, S.; Liu, D.; Shao, Z.; Proksch, P.; Lin, W. Eutypellazines a-m, thiodiketopiperazine-type alkaloids from deep-sea-derived fungus Eutypella sp. MCCC 3A00281. RSC Adv. 2017, 7, 33580–33590. [Google Scholar] [CrossRef]
- Niu, S.; Liu, D.; Shao, Z.; Proksch, P.; Lin, W. Eutypellazines n–s, new thiodiketopiperazines from a deep-sea sediment-derived fungus Eutypella sp. with anti-VRE activities. Tetrahedron Lett. 2017, 58, 3695–3699. [Google Scholar] [CrossRef]
- Takahashi, K.; Sakai, K.; Nagano, Y.; Orui-Sakaguchi, S.; Lima, A.O.; Pellizari, V.H.; Iwatsuki, M.; Takishita, K.; Nonaka, K.; Fujikura, K. Cladomarine, a new anti-saprolegniasis compound isolated from the deep-sea fungus Penicillium coralligerum YK-247. J. Antibiot. 2017, 70, 911–914. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Y.-X.; Zhang, J.-P.; Yu, H.-B.; Liu, X.-Y.; Lu, X.-L.; Jiao, B.-H. A new sesquiterpene lactone from fungus Eutypella sp. D-1. Nat. Prod. Res. 2017, 31, 1676–1681. [Google Scholar] [CrossRef]
- Li, Y.; Sun, B.; Liu, S.; Jiang, L.; Liu, X.; Zhang, H.; Che, Y. Bioactive asterric acid derivatives from the Antarctic ascomycete fungus Geomyces sp. J. Nat. Prod. 2008, 71, 1643–1646. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Wiese, J.; Labes, A.; Kramer, A.; Schmaljohann, R.; Imhoff, J.F. Lindgomycin, an unusual antibiotic polyketide from a marine fungus of the Lindgomycetaceae. Mar. Drugs 2015, 13, 4617–4632. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhu, M.-L.; Sun, G.-Y.; Li, N.; Gu, Q.-Q.; Li, D.-H.; Che, Q.; Zhu, T.-J. Exopisiod B and Farylhydrazone C, two new alkaloids from the Antarctic-derived fungus Penicillium sp. HDN14-431. J. Asian Nat. Prod. Res. 2016, 18, 959–965. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wei, X.; Qin, X.; Tian, X.; Liao, L.; Li, K.; Zhou, X.; Yang, X.; Wang, F.; Zhang, T.; et al. Antiviral merosesquiterpenoids produced by the Antarctic fungus Aspergillus ochraceopetaliformis SCSIO 05702. J. Nat. Prod. 2016, 79, 59–65. [Google Scholar] [CrossRef]
- Pan, C.; Shi, Y.; Auckloo, N.B.; Chen, X.; Chen, A.C.-T.; Tao, X.; Wu, B. An unusual conformational isomer of verrucosidin backbone from a hydrothermal vent fungus, Penicillium sp. Y-50-10. Mar. Drugs 2016, 14, 156. [Google Scholar] [CrossRef]
- Shaaban, M.; El-Metwally, M.M.; Abdel-Razek, A.A.; Laatsch, H. Terretonin M: A new meroterpenoid from the thermophilic Aspergillus terreus TM8 and revision of the absolute configuration of penisimplicins. Nat. Prod. Res. 2018, 32, 2437–2446. [Google Scholar] [CrossRef]
- Zhang, X.; Li, S.J.; Li, J.J.; Liang, Z.Z.; Zhao, C.Q. Novel natural products from extremophilic fungi. Mar. Drugs 2018, 16, 194. [Google Scholar] [CrossRef]
- Zheng, J.; Zhu, H.; Hong, K.; Wang, Y.; Liu, P.; Wang, X.; Peng, X.; Zhu, W. Novel cyclic hexapeptides from marine-derived fungus Aspergillus sclerotiorum PT06-1. Org. Lett. 2009, 11, 5262–5265. [Google Scholar] [CrossRef]
- Bao, J.; Zhai, H.; Zhu, K.; Yu, J.-H.; Zhang, Y.; Wang, Y.; Jiang, C.-S.; Zhang, X.; Zhang, Y.; Zhang, H. Bioactive pyridone alkaloids from a deep-sea-derived fungus Arthrinium sp. UJNMF0008. Mar. Drugs 2018, 16, 174. [Google Scholar] [CrossRef]
- Elhosainy, A.M. Evaluation of Some Biological Activities of Phialosimplex asmahalo. Egypt. Acad. J. Biol. Sci. G Microbiol. 2020, 12, 9–19. [Google Scholar] [CrossRef]
- Oh, H.; Lee, Y.; Kim, Y.; Seo, Y.; Kang, J.; Park, E.; Yoon, Y. Development of antimicrobial hydrogel with edible formulations to control foodborne pathogens on food surfaces consumed raw. Innov. Food Sci. Emerg. Technol. 2021, 74, 102845. [Google Scholar] [CrossRef]
- Ballav, S.; Kerkar, S.; Thomas, S.; Augustine, N. Halophilic and halotolerant actinomycetes from a marine saltern of Goa, India producing anti-bacterial metabolites. J. Biosci. Bioeng. 2015, 119, 323–330. [Google Scholar] [CrossRef]
- Corral, P.; Esposito, F.P.; Tedesco, P.; Falco, A.; Tortorella, E.; Tartaglione, L.; de Pascale, D. Identification of a sorbicillinoid-producing Aspergillus strain with antimicrobial activity against Staphylococcus aureus: A new polyextremophilic marine fungus from Barents Sea. Mar. Biotechnol. 2018, 20, 502–511. [Google Scholar] [CrossRef]
- Wingfield, M.J.; Pham, N.Q.; Marincowitz, S.; Duong, T.A.; Wingfield, B.D.; Wilson, A.M. Blast from the past: A study of decades-old fungal cultures resolves a long-standing tree disease mystery. J. Plant Pathol. 2024, 106, 377–384. [Google Scholar] [CrossRef]
- Wang, H.; Li, Y.; Wang, Y.; Shi, T.; Wang, B. Penicillium janthinellum: A Potential Producer of Natural Products. Fermentation 2024, 10, 157. [Google Scholar] [CrossRef]
- Agrawal, S.; Chavan, P.; Dufossé, L. Hidden treasure: Halophilic fungi as a repository of bioactive lead compounds. J. Fungi 2024, 10, 290. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Bae, Y.M.; Jung, K.S.; Heu, S.; Lee, S.Y. Antimicrobial activity of natural antimicrobial substances against spoilage bacteria isolated from fresh produce. Food Control 2013, 32, 665–672. [Google Scholar] [CrossRef]
- Pócsi, I.; Dijksterhuis, J.; Houbraken, J.; de Vries, R.P. Biotechnological potential of salt-tolerant and xerophilic species of Aspergillus. Appl. Microbiol. Biotechnol. 2024, 108, 521. [Google Scholar] [CrossRef]
- Hafez Goran, S.; Taktaz, F.; Ayatollahi, S.A.; Kijjoa, A. Anthraquinones and their analogues from marine-derived fungi: Chemistry and biological activities. Mar. Drugs 2022, 20, 474. [Google Scholar] [CrossRef]
- Deng, J.; Li, Y.; Yuan, Y.; Yin, F.; Chao, J.; Huang, J.; Liu, Z.; Wang, K.; Zhu, M. Secondary metabolites from the genus Eurotium and their biological activities. Foods 2023, 12, 4452. [Google Scholar] [CrossRef]
- Zhuravleva, O.I.; Chingizova, E.A.; Oleinikova, G.K.; Starnovskaya, S.S.; Antonov, A.S.; Kirichuk, N.N.; Menshov, A.S.; Popov, R.S.; Kim, N.Y.; Berdyshev, D.V.; et al. Anthraquinone derivatives and other aromatic compounds from marine fungus Asteromyces cruciatus KMM 4696 and their effects against Staphylococcus aureus. Mar. Drugs 2023, 21, 431. [Google Scholar] [CrossRef] [PubMed]
- Pacelli, C.; Cassaro, A.; Maturilli, A.; Timperio, A.M.; Gevi, F.; Cavalazzi, B.; Onofri, S. Multidisciplinary characterization of melanin pigments from the black fungus Cryomyces antarcticus. Appl. Microbiol. Biotechnol. 2020, 104, 6385–6395. [Google Scholar] [CrossRef]
- Cavalcante, S.B.; da Silva, A.F.; Pradi, L.; Lacerda, J.W.F.; Tizziani, T.; Sandjo, L.P.; Robl, D. Antarctic fungi produce pigment with antimicrobial and antiparasitic activities. Braz. J. Microbiol. 2024, 55, 1251–1263. [Google Scholar] [CrossRef]
- Afroz, T.M.; Rahman, M.H.; Rahman, M.S.; Arif, M.; Nazir, K.H.M.N.H.; Dufossé, L. Fungal pigments: Carotenoids, riboflavin, and polyketides with diverse applications. J. Fungi 2023, 9, 454. [Google Scholar] [CrossRef]
- Chen, M.; Zheng, Y.Y.; Chen, Z.Q.; Shen, N.X.; Shen, L.; Zhang, F.M.; Wang, C.Y. NaBr-induced production of brominated azaphilones and related tricyclic polyketides by the marine-derived fungus Penicillium janthinellum HK1-6. J. Nat. Prod. 2019, 82, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Xu, J. Fungal pigments and their roles associated with human health. J. Fungi 2020, 6, 280. [Google Scholar] [CrossRef] [PubMed]
- Giddings, L.A.; Newman, D.J. Extremophilic fungi from marine environments: Underexplored sources of antitumor, anti-infective, and other biologically active agents. Mar. Drugs 2022, 20, 62. [Google Scholar] [CrossRef] [PubMed]
- Ferrarezi, J.H.; Marin, V.R.; Vieira, G.; Ferreira, H.; Sette, L.D.; Sass, D.C. Bisdechlorogeodin from Antarctic Pseudogymnoascus sp. LAMAI 2784 for citrus canker control. J. Appl. Microbiol. 2024, 135, lxae093. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Ikuta, M.; Nishimura, S.; Sugiyama, R.; Yoshimura, A.; Kakeya, H. Amphiol, an antifungal fungal pigment from Pseudogymnoascus sp. PF1464. J. Nat. Prod. 2021, 84, 986–992. [Google Scholar] [CrossRef]
- Zucconi, L.; Canini, F.; Isola, D.; Caneva, G. Fungi affecting wall paintings of historical value: A worldwide meta-analysis of their detected diversity. Appl. Sci. 2022, 12, 2988. [Google Scholar] [CrossRef]
- Sułkowska-Ziaja, K.; Trepa, M.; Olechowska-Jarząb, A.; Nowak, P.; Ziaja, M.; Kała, K.; Muszyńska, B. Natural compounds of fungal origin with antimicrobial activity—Potential cosmetics applications. Pharmaceuticals 2023, 16, 1200. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wang, Y.; Wang, J.; Liu, P.; Li, J.; Zhu, W. Antimicrobial ergosteroids and pyrrole derivatives from halotolerant Aspergillus flocculosus PT05-1 cultured in a hypersaline medium. Extremophiles 2013, 17, 963–971. [Google Scholar] [CrossRef]
- Konuray, G.; Erginkaya, Z. Antimicrobial and antioxidant properties of pigments synthesized from microorganisms. In The Battle Against Microbial Pathogens: Basic Science, Technological Advances and Educational Programs; Formatex Research Center: Norristown, PA, USA, 2015. [Google Scholar]
- Conrado, R.; Gomes, T.C.; Roque, G.S.C.; De Souza, A.O. Overview of bioactive fungal secondary metabolites: Cytotoxic and antimicrobial compounds. Antibiotics 2022, 11, 1604. [Google Scholar] [CrossRef]
- Azami, H.; Watanabe, Y.; Sakai, K.; Nakahara, H.; Kojima, H.; Tokiwa, T.; Nonaka, K.; Noguchi, Y.; Nagano, Y.; Hirose, T.; et al. Antifungal profile against Candida auris clinical isolates of tyroscherin and its new analog produced by the deep-sea-derived fungal strain Scedosporium apiospermum FKJ-0499. J. Antibiot. 2024, 77, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Wu, P.; Yang, J.; Weng, Y.; Lin, Y.; Chen, Z.; Yu, F.; Lv, X.; Ni, L. Development of a novel hybrid antimicrobial peptide for enhancing antimicrobial spectrum and potency against food-borne pathogens. J. Appl. Microbiol. 2024, 135, lxae023. [Google Scholar] [CrossRef] [PubMed]
- Cadelis, M.M.; Nipper, N.S.; Grey, A.; Geese, S.; van de Pas, S.J.; Weir, B.S.; Copp, B.R.; Wiles, S. Antimicrobial Polyketide Metabolites from Penicillium bissettii and P. glabrum. Mol. 2021, 27, 240. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, F.; Huang, Z.; Liu, H.; Xie, C.; Zhang, J.; Thacker, P.A.; Qiao, S. Effects of the antimicrobial peptide cecropin AD on performance and intestinal health in weaned piglets challenged with Escherichia coli. Peptides 2012, 35, 225–230. [Google Scholar] [CrossRef]
- Liu, J.T.; Hu, B.; Gao, Y.; Zhang, J.P.; Jiao, B.H.; Lu, X.L.; Liu, X.Y. Bioactive tyrosine-derived cytochalasins from fungus Eutypella sp. D-1. Chem. Biodivers. 2014, 11, 800–806. [Google Scholar] [CrossRef]
- Zhou, H.; Li, L.; Wang, W.; Che, Q.; Li, D.; Gu, Q.; Zhu, T. Chrodrimanins I and J from the Antarctic moss-derived fungus Penicillium funiculosum GWT2-24. J. Nat. Prod. 2015, 78, 1442–1445. [Google Scholar] [CrossRef]
- Zhang, S.D.; Barbe, V.; Garel, M.; Zhang, W.-J.; Chen, H.; Santini, C.-L.; Murat, D.; Jing, H.; Zhao, Y.; Lajus, A.; et al. Genome sequence of luminous piezophile Photobacterium phosphoreum ANT-2200. Genome Announc. 2014, 2, e00096-14. [Google Scholar] [CrossRef]
- Zhang, S.D.; Petersen, N.; Zhang, W.J.; Cargou, S.; Ruan, J.; Murat, D.; Santini, C.-L.; Song, T.; Kato, T.; Notareschi, P.; et al. Swimming behaviour and magnetotaxis function of the marine bacterium strain MO-1. Environ. Microbiol. Rep. 2014, 6, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, I.; Goodall-Copestake, W.; Thorne, M.A.; Schlitt, T.; Ávila-Jiménez, M.L.; Pearce, D.A. Extremophiles in an Antarctic marine ecosystem. Microorganisms 2016, 4, 8. [Google Scholar] [CrossRef]
- Gonçalves, V.N.; Oliveira, F.S.; Carvalho, C.R.; Schaefer, C.E.; Rosa, C.A.; Rosa, L.H. Antarctic rocks from continental Antarctica as source of potential human opportunistic fungi. Extremophiles 2017, 21, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Wu, G.; Zhan, T.; Shao, Z.; Liu, Z. Characterization of a cold-adapted and salt-tolerant esterase from a psychrotrophic bacterium Psychrobacter pacificensis. Extremophiles 2013, 17, 809–819. [Google Scholar] [CrossRef]
- Lin, A.; Wu, G.; Gu, Q.; Zhu, T.; Li, D. New eremophilane-type sesquiterpenes from an Antarctic deep-sea-derived fungus, Penicillium sp. PR19 N-1. Arch. Pharm. Res. 2014, 37, 839–844. [Google Scholar] [CrossRef]
- Boysen, J.M.; Saeed, N.; Hillmann, F. Natural products in the predatory defense of the filamentous fungal pathogen Aspergillus fumigatus. Beilstein J. Org. Chem. 2021, 17, 1814–1827. [Google Scholar] [CrossRef]
- Cen, Q.W.; Wang, Z.Y.; Tang, Z.X.; Zhang, Y.; Chen, T.; Xue, D.W.; Xu, M.-F.; Bai, X.-L.; Zhou, T.; Shi, L.E. Initial purification of antimicrobial fermentation metabolites from Paecilomyces cicadae and its antimicrobial mechanism. LWT 2021, 148, 111785. [Google Scholar] [CrossRef]
- Anwar, A.; Kanwal, Q.; Sadiqa, A.; Razaq, T.; Khan, I.H.; Javaid, A.; Khan, S.; Tag-Eldin, E.; Ouladsmane, M. Synthesis and antimicrobial analysis of high surface area strontium-substituted calcium phosphate nanostructures for bone regeneration. Int. J. Mol. Sci. 2023, 24, 14527. [Google Scholar] [CrossRef]
- Guo, Y.; Zhou, H.; Qin, L.; Pang, Z.; Qin, T.; Ren, H.; Pan, Z.; Zhou, J. Frequency, antimicrobial resistance, and genetic diversity of Klebsiella pneumoniae in food samples. PLoS ONE 2016, 11, e0153561. [Google Scholar] [CrossRef]
- Mo, Y.; Tan, W.C.; Cooper, B.S. Antibiotic duration for common bacterial infections—A systematic review. JAC Antimicrob. Resist. 2025, 7, dlae215. [Google Scholar] [CrossRef]
- Zucconi, L.; Canini, F.; Temporiti, M.E.; Tosi, S. Extracellular enzymes and bioactive compounds from Antarctic terrestrial fungi for bioprospecting. Int. J. Environ. Res. Public Health 2020, 17, 6459. [Google Scholar] [CrossRef] [PubMed]
- Willems, T.; De Mol, M.L.; De Bruycker, A.; De Maeseneire, S.L.; Soetaert, W.K. Alkaloids from marine fungi: Promising antimicrobials. Antibiotics 2020, 9, 340. [Google Scholar] [CrossRef]
- Liao, J.; Wang, L.; Ding, S.; Tian, G.; Hu, H.; Wang, Q.; Yin, W. Molybdenum-based antimicrobial nanomaterials: A comprehensive review. Nano Today 2023, 50, 101875. [Google Scholar] [CrossRef]
- Christiansen, J.V.; Isbrandt, T.; Petersen, C.; Sondergaard, T.E.; Nielsen, M.R.; Pedersen, T.B.; Sørensen, J.L.; Larsen, T.O.; Frisvad, J.C. Fungal quinones: Diversity, producers, and applications of quinones from Aspergillus, Penicillium, Talaromyces, Fusarium, and Arthrinium. Appl. Microbiol. Biotechnol. 2021, 105, 8157–8193. [Google Scholar] [CrossRef]
- Dahlem Junior, M.A.; Nguema Edzang, R.W.; Catto, A.L.; Raimundo, J.M. Quinones as an efficient molecular scaffold in the antibacterial/antifungal or antitumoral arsenal. Int. J. Mol. Sci. 2022, 23, 14108. [Google Scholar] [CrossRef] [PubMed]
- Rogozhin, E.A.; Sadykova, V.S.; Baranova, A.A.; Vasilchenko, A.S.; Lushpa, V.A.; Mineev, K.S.; Georgieva, M.L.; Kul’ko, A.B.; Krasheninnikov, M.E.; Lyundup, A.V.; et al. A novel lipopeptaibol emericellipsin A with antimicrobial and antitumor activity produced by the extremophilic fungus Emericellopsis alkalina. Molecules 2018, 23, 2785. [Google Scholar] [CrossRef]
- Baranova, A.A.; Alferova, V.A.; Korshun, V.A.; Tyurin, A.P. Antibiotics from extremophilic micromycetes. Russ. J. Bioorganic Chem. 2020, 46, 903–971. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhao, S.; Yu, Y.; Chen, Z.; Shen, H.; Zhou, L. Beauvericin K, a new antifungal beauvericin analogue from a marine-derived Fusarium sp. Nat. Prod. Commun. 2016, 11, 1825–1826. [Google Scholar]
- Liu, M.; Sun, W.; Wang, J.; He, Y.; Zhang, J.; Li, F.; Qi, C.; Zhu, H.; Xue, Y.; Hu, Z.; et al. Bioactive secondary metabolites from the marine-associated fungus Aspergillus terreus. Bioorg. Chem. 2018, 80, 525–530. [Google Scholar] [CrossRef]
- Scharf, D.H.; Heinekamp, T.; Brakhage, A.A. Gliotoxin—A fungal virulence factor and its role in Aspergillus fumigatus pathogenicity. Curr. Pharm. Design 2012, 18, 4859–4873. [Google Scholar]
- Singh, S.P.; Qureshi, A.; Hassan, W. Mechanisms of action by antimicrobial agents: A review. McGill J. Med. 2021, 19. [Google Scholar] [CrossRef]
- Denning, D.W. Echinocandins: A new class of antifungal. J. Antimicrob. Chemother. 2002, 49, 889–891. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Ahuja, S.; Garg, S. Fungal secondary metabolites: Biological activity and potential applications. In Recent Trends in Mycological Research: Volume 1: Agricultural and Medical Perspective; Springer: Cham, Switzerland, 2021; pp. 159–188. [Google Scholar]
- Waditee-Sirisattha, R.; Kageyama, H.; Takabe, T. Halophilic microorganism resources and their applications in industrial and environmental biotechnology. AIMS Microbiol. 2016, 2, 42–54. [Google Scholar] [CrossRef]
- Kour, D.; Kaur, T.; Devi, R.; Rana, K.L.; Yadav, N.; Rastegari, A.A.; Yadav, A.N. Biotechnological applications of beneficial microbiomes for evergreen agriculture and human health. In Trends of Microbial Biotechnology for Sustainable Agriculture and Biomedicine Systems: Perspectives for Human Health; Rastegari, A.A., Yadav, A.N., Yadav, N., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 255–279. [Google Scholar]
- Yadav, A.N.; Kour, D.; Kaur, T.; Devi, R.; Guleria, G.; Rana, K.L.; Yadav, N.; Rastegari, A.A. Microbial biotechnology for sustainable biomedicine systems: Current research and future challenges. In Trends of Microbial Biotechnology for Sustainable Agriculture and Biomedicine Systems: Perspectives for Human Health; Rastegari, A.A., Yadav, A.N., Yadav, N., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 281–292. [Google Scholar]
- Arifeen, M.Z.U.; Ma, Y.-N.; Xue, Y.-R.; Liu, C. Deep-Sea Fungi Could Be the New Arsenal for Bioactive Molecules. Mar. Drugs 2019, 18, 9. [Google Scholar] [CrossRef]
- Petersen, L.E.; Kellermann, M.Y.; Schupp, P.J. Secondary Metabolites of Marine Microbes: From Natural Products Chemistry to Chemical Ecology. In YOUMARES 9—The Oceans: Our Research, Our Future: Proceedings of the 2018 Conference for YOUng MArine RESearchers in Oldenburg, Germany; Jungblut, S., Liebich, V., Bode-Dalby, M., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 159–180. [Google Scholar]
- Yadav, A.N.; Kumar, R.; Kumar, S.; Kumar, V.; Sugitha, T.; Singh, B.; Chauhan, V.; Dhaliwal, H.S.; Saxena, A.K. Beneficial microbiomes: Biodiversity and potential biotechnological applications for sustainable agriculture and human health. J. Appl. Biol. Biotechnol. 2017, 5, 45–57. [Google Scholar] [CrossRef]
- Yadav, A.N.; Saxena, A.K. Biodiversity and biotechnological applications of halophilic microbes for sustainable agriculture. J. Appl. Biol. Biotechnol. 2018, 6, 48–55. [Google Scholar] [CrossRef]
- Gómez-Silva, B. Lithobiontic life: “Atacama rocks are well and alive”. Antonie Van Leeuwenhoek 2018, 111, 1333–1343. [Google Scholar] [CrossRef]
- Okoro, C.K.; Brown, R.; Jones, A.L.; Andrews, B.A.; Asenjo, J.A.; Goodfellow, M.; Bull, A.T. Diversity of culturable actinomycetes in hyper-arid soils of the Atacama Desert, Chile. Antonie Van Leeuwenhoek 2009, 95, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Rosa, L.H.; Zani, C.L.; Cantrell, C.L.; Duke, S.O.; Van Dijck, P.; Desideri, A.; Rosa, C.A. Fungi in Antarctica: Diversity, ecology, effects of climate change, and bioprospection for bioactive compounds. In Fungi of Antarctica: Diversity, Ecology and Biotechnological Applications; Springer: Berlin/Heidelberg, Germany, 2019; pp. 1–17. [Google Scholar]
- Bakermans, C.; Bergholz, P.W.; Ayala-del-Río, H.; Tiedje, J. Genomic insights into cold adaptation of permafrost bacteria. In Permafrost Soils; Springer: Berlin/Heidelberg, Germany, 2009; pp. 159–168. [Google Scholar]
- Bowman, J.P. Genomic analysis of psychrophilic prokaryotes. In Psychrophiles: From Biodiversity to Biotechnology; Springer: Berlin, Germany, 2008; pp. 265–284. [Google Scholar]
- Unell, M. Physiological, Genetic and Proteomic Characterization of Arthrobacter Chlorophenolicus During Growth on Different Phenolic Substrates or Temperatures; Swedish University of Agricultural Sciences: Uppsala, Sweden, 2008; No. 2008: 26. [Google Scholar]
- Gocheva, Y.; Krumova, E.; Slokoska, L.; Miteva, J.; Angelova, M. Cell response of Antarctic and temperate strains of Penicillium spp. to different growth temperatures. Mycol. Res. 2006, 110, 1347–1354. [Google Scholar] [CrossRef]
- Ruisi, S.; Barreca, D.; Selbmann, L.; Zucconi, L.; Onofri, S. Fungi in Antarctica. Rev. Environ. Sci. Biotechnol. 2007, 6, 127–141. [Google Scholar] [CrossRef]
- Tosi, S.; Kostadinova, N.; Krumova, E.; Pashova, S.; Dishliiska, V.; Spassova, B.; Vassilev, S.; Angelova, M. Antioxidant enzyme activity of filamentous fungi isolated from Livingston Island, Maritime Antarctica. Polar Biol. 2010, 33, 1227–1237. [Google Scholar] [CrossRef]
- Feller, G.; Gerday, C. Psychrophilic enzymes: Hot topics in cold adaptation. Nat. Rev. Microbiol. 2003, 1, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Collins, T.; Roulling, F.; Piette, F.; Marx, J.C.; Feller, G.; Gerday, C.; D’Amico, S. Fundamentals of cold-adapted enzymes. In Psychrophiles: From Biodiversity to Biotechnology; Margesin, R., Schinner, F., Gerday, C., Marx, J.C., Eds.; Springer: Berlin, Germany, 2007; pp. 211–227. [Google Scholar]
- Krumova, E.; Abrashev, R.; Dishliyska, V.; Stoyancheva, G.; Kostadinova, N.; Miteva-Staleva, J.; Spasova, B.; Angelova, M. Cold-active catalase from the psychrotolerant fungus Penicillium griseofulvum. J. Biol. Microbiol. 2021, 61, 782–794. [Google Scholar] [CrossRef]
- Giordano, D. Bioactive Molecules from Extreme Environments. Mar. Drugs 2020, 18, 640. [Google Scholar] [CrossRef]
- Gittel, A.; Bárta, J.; Kohoutová, I.; Mikutta, R.; Owens, S.; Gilbert, J.; Urich, T. Distinct microbial communities associated with buried soils in the Siberian tundra. ISME J. 2014, 8, 841–853. [Google Scholar] [CrossRef] [PubMed]
- Deslippe, J.R.; Hartmann, M.; Grayston, S.J.; Simard, S.W.; Mohn, W.W. Stable isotope probing implicates a species of Cortinarius in carbon transfer through ectomycorrhizal fungal mycelial networks in Arctic tundra. New Phytol. 2016, 210, 383–390. [Google Scholar] [CrossRef]
- Fujiyoshi, M.; Yoshitake, S.; Watanabe, K.; Murota, K.; Tsuchiya, Y.; Uchida, M.; Nakatsubo, T. Successional changes in ectomycorrhizal fungi associated with the polar willow Salix polaris in a deglaciated area in the High Arctic, Svalbard. Polar Biol. 2011, 34, 667–673. [Google Scholar] [CrossRef]
- Arenz, B.E.; Blanchette, R.A. Distribution and abundance of soil fungi in Antarctica at sites on the Peninsula, Ross Sea Region, and McMurdo Dry Valleys. Soil Biol. Biochem. 2011, 43, 308–315. [Google Scholar] [CrossRef]
- Park, C.H.; Kim, K.M.; Elvebakk, A.; Kim, O.S.; Jeong, G.; Hong, S.G. Algal and fungal diversity in Antarctic lichens. J. Eukaryot. Microbiol. 2015, 62, 196–205. [Google Scholar] [CrossRef]
- Nagano, Y.; Nagahama, T.; Abe, F. Cold-adapted yeasts in deep-sea environments. In Cold-Adapted Yeasts; Buzzini, P., Margesin, R., Eds.; Springer: Berlin, Germany, 2013; pp. 149–171. [Google Scholar]
- Singh, P.; Raghukumar, C.; Verma, P.; Shouche, Y. Phylogenetic diversity of culturable fungi from the deep-sea sediments of the Central Indian Basin and their growth characteristics. Fungal Divers. 2010, 40, 89–102. [Google Scholar] [CrossRef]
- Singh, P.; Raghukumar, C.; Meena, R.M.; Verma, P.; Shouche, Y. Fungal diversity in deep-sea sediments revealed by culture-dependent and culture-independent approaches. Fungal Ecol. 2012, 5, 543–553. [Google Scholar] [CrossRef]
- Che Zain, M.S.; Lee, S.Y.; Nasir, N.M.; Fakurazi, S.; Shaari, K. Metabolite characterization and correlations with antioxidant and wound healing properties of oil palm (Elaeis guineensis Jacq.) leaflets via 1H-NMR-based metabolomics approach. Molecules 2020, 25, 5636. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2019, 36, 122–173. [Google Scholar] [CrossRef]
- Jansson, J.K.; Tas, N. The microbial ecology of permafrost. Nat. Rev. Microbiol. 2014, 12, 414–425. [Google Scholar] [CrossRef]
- Bakermans, C.; Skidmore, M.L.; Douglas, S.; McKay, C.P. Molecular characterization of bacteria from permafrost of the Taylor Valley, Antarctica. FEMS Microbiol. Ecol. 2014, 89, 331–346. [Google Scholar] [CrossRef]
- De Maayer, P.; Anderson, D.; Cary, C.; Cowan, D.A. Some like it cold: Understanding the survival strategies of psychrophiles. EMBO Rep. 2014, 15, 508–517. [Google Scholar] [CrossRef]
- Hu, W.; Zhang, Q.; Tian, T.; Cheng, G.; An, L.; Feng, H. The microbial diversity, distribution, and ecology of permafrost in China: A review. Extremophiles 2015, 19, 693–705. [Google Scholar] [CrossRef]
- Xing, C.; Chen, D.; Xie, C.; Liu, Q.; Zhong, T.; Shao, Z.; Liu, G.; Luo, L.; Yang, X. Anti-food allergic compounds from Penicillium griseofulvum MCCC 3A00225, a deep-sea-derived fungus. Mar. Drugs 2021, 19, 224. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, G.; Osman, S.; Kukkadapu, R.; Engelhard, M.; Vaishampayan, P.A.; Andersen, G.L.; Sani, R.K. Microbial and mineralogical characterizations of soils collected from the deep biosphere of the former Homestake gold mine, South Dakota. Microb. Ecol. 2010, 60, 539–550. [Google Scholar] [CrossRef]
- Urbieta, M.S.; Donati, E.R.; Chan, K.G.; Shahar, S.; Sin, L.L.; Goh, K.M. Thermophiles in the genomic era: Biodiversity, science, and applications. Biotechnol. Adv. 2015, 33, 633–647. [Google Scholar] [CrossRef]
- Bajpai, P. Developments and Applications of Enzymes from Thermophilic Microorganisms; Elsevier: Amsterdam, The Netherlands, 2023. [Google Scholar]
- Hidese, R.; Fukuda, W.; Niitsu, M.; Fujiwara, S. Identification of branched-chain polyamines in hyperthermophiles. In Polyamines; Alcazar, R., Tiburcio, A.F., Eds.; Humana Press: New York, NY, USA, 2018; pp. 81–94. [Google Scholar]
- Goh, K.M.; Kahar, U.M.; Chai, Y.Y.; Chong, C.S.; Chai, K.P.; Ranjani, V.; Illias, R.M.; Chan, K.G. Recent discoveries and applications of Anoxybacillus. Appl. Microbiol. Biotechnol. 2013, 97, 1475–1488. [Google Scholar] [CrossRef] [PubMed]
- Zeldes, B.M.; Keller, M.W.; Loder, A.J.; Straub, C.T.; Adams, M.W.W.; Kelly, R.M. Extremely thermophilic microorganisms as metabolic engineering platforms for production of fuels and industrial chemicals. Front. Microbiol. 2015, 6, 1209. [Google Scholar] [CrossRef] [PubMed]
- Gessler, N.N.; Egorova, A.S.; Belozerskaya, T.A. Melanin pigments of fungi under extreme environmental conditions. Appl. Biochem. Microbiol. 2014, 50, 105–113. [Google Scholar] [CrossRef]
- Conley, C.A.; Ishkhanova, G.; McKay, C.P.; Cullings, K. A preliminary survey of non-lichenized fungi cultured from the hyperarid Atacama Desert of Chile. Astrobiology 2006, 6, 521–526. [Google Scholar] [CrossRef]
- Santiago, I.F.; Goncalves, V.N.; Gomez-Silva, B.; Galetovic, A.; Rosa, L.H. Fungal diversity in the Atacama Desert. Antonie Van Leeuwenhoek 2018, 111, 1345–1360. [Google Scholar] [CrossRef]
- Grishkan, I.; Nevo, E. Spatiotemporal distribution of soil microfungi in the Makhtesh Ramon area, central Negev desert, Israel. Fungal Ecol. 2010, 3, 326–337. [Google Scholar] [CrossRef]
- Vargas Castillo, R.; Beck, A. Photobiont selectivity and specificity in Caloplaca species in a fog-induced community in the Atacama Desert, northern Chile. Fungal Biol. 2012, 116, 665–676. [Google Scholar] [CrossRef]
- Gonçalves, V.N.; Cantrell, C.L.; Wedge, D.E.; Ferreira, M.C.; Soares, M.A.; Jacob, M.R.; Rosa, L.H. Fungi associated with rocks of the A tacama D esert: Taxonomy, distribution, diversity, ecology and bioprospection for bioactive compounds. Environ. Microbiol. 2016, 18, 232–245. [Google Scholar] [CrossRef]
- Martinelli, L.; Zalar, P.; Gunde-Cimerman, N.; Azua-Bustos, A.; Sterflinger, K.; Piñar, G. Aspergillus atacamensis and A. salisburgensis: Two new halophilic species from hypersaline/arid habitats with a Phialosimplex-like morphology. Extremophiles 2017, 21, 755–773. [Google Scholar] [CrossRef]
- Symanczik, S.; Błaszkowski, J.; Chwat, G.; Boller, T.; Wiemken, A.; Al-Yahyaei, N.; Al-Yahya’ei, M.N. Three new species of arbuscular mycorrhizal fungi discovered at one location in a desert of Oman: Diversispora omaniana, Septoglomus nakheelum and Rhizophagus arabicus. Mycology 2014, 106, 243–259. [Google Scholar] [CrossRef]
- Zalar, P.; de Hoog, G.S.; Schroers, H.J.; Frank, J.M.; Gunde-Cimerman, N. Taxonomy and phylogeny of the xerophilic genus Wallemia (Wallemiomycetes and Wallemiales, cl. et ord. nov.). Antonie Van Leeuwenhoek 2005, 87, 311–328. [Google Scholar] [CrossRef] [PubMed]
- Jancic, S.; Frisvad, J.C.; Kocev, D.; Gostincar, C.; Dzeroski, S.; Gunde-Cimerman, N. Production of secondary metabolites in extreme environments: Food- and airborne Wallemia spp. produce toxic metabolites at hypersaline conditions. PLoS ONE 2016, 11, e0169116. [Google Scholar] [CrossRef]
- Peng, X.P.; Wang, Y.; Liu, P.P.; Hong, K.; Chen, H.; Yin, X.; Zhu, W.-M. Aromatic compounds from the halotolerant fungal strain of Wallemia sebi PXP-89 in a hypersaline medium. Arch. Pharm. Res. 2011, 34, 907–912. [Google Scholar] [CrossRef]
- Jiang, W.; Ye, P.; Chen, C.T.A.; Wang, K.; Liu, P.; He, S.; Wu, B. Two novel hepatocellular carcinoma cycle inhibitory cyclodepsipeptides from a hydrothermal vent crab-associated fungus Aspergillus clavatus C2WU. Mar. Drugs 2013, 11, 4761–4772. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.S.; Niu, X.M.; Wang, Y.L.; Guo, J.P.; Pan, W.Z.; Huang, X.W.; Zhang, K.Q. Isolation of putative biosynthetic intermediates of prenylated indole alkaloids from a thermophilic fungus Talaromyces thermophilus. Org. Lett. 2010, 12, 4356–4359. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.P.; Tan, J.L.; Wang, Y.L.; Wu, H.Y.; Zhang, C.P.; Niu, X.-M.; Pan, W.Z.; Huang, X.W.; Zhang, K.-Q. Isolation of talathermophilins from the thermophilic fungus Talaromyces thermophilus YM3-4. J. Nat. Prod. 2011, 74, 2278–2281. [Google Scholar] [CrossRef]
- Azua-Bustos, A.; Urrejola, C.; Vicuña, R. Life at the dry edge: Microorganisms of the Atacama Desert. FEBS Lett. 2012, 586, 2939–2945. [Google Scholar] [CrossRef]
- Cordero, R.R.; Damiani, A.; Jorquera, J.; Sepulveda, E.; Caballero, M.; Fernandez, S.; Feron, S.; Llanillo, P.J.; Carrasco, J.; Laroze, D.; et al. Ultraviolet radiation in the Atacama Desert. Antonie Van Leeuwenhoek 2018, 111, 1301–1313. [Google Scholar] [CrossRef]
- Bull, A.T.; Asenjo, J.A.; Goodfellow, M.; Gómez-Silva, B. The Atacama Desert: Technical resources and the growing importance of novel microbial diversity. Annu. Rev. Microbiol. 2016, 70, 215–234. [Google Scholar] [CrossRef]
- Marchetta, A. Assessment of fungal diversity present in Arctic and Antarctic lakes and selection of Heavy Metal tolerant fungal isolates. Ph.D. Thesis, Università degli Studi di Messina, Messina, Italy, 2023. [Google Scholar]
- Navarro-Gonzalez, R.; Rainey, F.A.; Molina, P.; Bagaley, D.R.; Hollen, B.J.; de la Rosa, J.; Small, A.M.; Quinn, R.C.; Grunthaner, F.J.; Cáceres, L.; et al. Mars-like soils in the Atacama Desert, Chile, and the dry limit of microbial life. Science 2003, 302, 1018–1021. [Google Scholar] [CrossRef]
- Schulze-Makuch, D.; Wagner, D.; Kounaves, S.P.; Mangelsdorf, K.; Devine, K.G.; de Vera, J.P.; Zamorano, P.; Schmitt-Kopplin, P.; Grossart, H.-P.; Parro, V.; et al. Transitory microbial habitat in the hyperarid Atacama Desert. Proc. Natl. Acad. Sci. USA 2018, 115, 2670–2675. [Google Scholar] [CrossRef] [PubMed]
- Azua-Bustos, A.; Fairen, A.G.; Gonzalez-Silva, C.; Ascaso, C.; Carrizo, D.; Fernandez-Martínez, M.A.; Fernandez-Sampedro, M.; Garcia-Descalzo, L.; García-Villadangos, M.; Martin-Redondo, M.P.; et al. Unprecedented rains decimate surface microbial communities in the hyperarid core of the Atacama Desert. Sci. Rep. 2018, 8, 16706. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Quatrini, R. Acidophile microbiology in space and time. In Acidophiles: Life in Extremely Acidic Environments; Quatrini, R., Johnson, D.B., Eds.; Caister Academic Press: Norfolk, UK, 2016; pp. 3–16. [Google Scholar]
- Druschel, G.K.; Baker, B.J.; Gihring, T.M.; Banfield, J.F. Acid mine drainage biogeochemistry at Iron Mountain, California. Geochem. Trans. 2004, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Mirete, S.; Morgante, V.; Gonzalez-Pastor, J.E. Acidophiles: Diversity and mechanisms of adaptation to acidic environments. In Adaption of Microbial Life to Environmental Extremes; Stan-Lotter, H., Fendrihan, S., Eds.; Springer: Cham, Switzerland, 2017; pp. 227–251. [Google Scholar]
- Denef, V.J.; Mueller, R.S.; Banfield, J.F. AMD biofilms: Using model communities to study microbial evolution and ecological complexity in nature. ISME J. 2010, 4, 599–610. [Google Scholar] [CrossRef]
- Konings, W.N.; Albers, S.V.; Koning, S.; Driessen, A.J. The cell membrane plays a crucial role in survival of bacteria and archaea in extreme environments. Anton. Leeuw. 2002, 81, 61–72. [Google Scholar] [CrossRef]
- Falteisek, L.; Cepicka, I. Microbiology of diverse acidic and non-acidic microhabitats within a sulfidic ore mine. Extremophiles 2012, 16, 911–922. [Google Scholar] [CrossRef]
- Stierle, D.B.; Stierle, A.A.; Patacini, B.; McIntyre, K.; Girtsman, T.; Bolstad, E. Berkeleyones and related meroterpenes from a deep water acid mine waste fungus that inhibit the production of interleukin 1-b from induced inflammasomes. J. Nat. Prod. 2011, 74, 2273–2277. [Google Scholar] [CrossRef]
- Stierle, D.B.; Stierle, A.A.; Girtsman, T.; McIntyre, K.; Nichols, J. Caspase-1 and-3 inhibiting drimane sesquiterpenoids from the extremophilic fungus Penicillium solitum. J. Nat. Prod. 2012, 75, 262–266. [Google Scholar] [CrossRef]
- Díaz-Cardenas, C.; Cantillo, A.; Rojas, L.Y.; Sandoval, T.; Fiorentino, S.; Robles, J.; Ramos, F.A.; Zambrano, M.M.; Baena, S. Microbial diversity of saline environments: Searching for cytotoxic activities. AMB Express 2017, 7, 223. [Google Scholar] [CrossRef]
- Enache, M.; Teodosiu, G.; Itoh, T.; Kamekura, M.; Stan-Lotter, H. Halophilic microorganisms from manmade and natural hypersaline environments: Physiology, ecology, and biotechnological potential. In Adaption of Microbial Life to Environmental Extremes; Stan-Lotter, H., Fendrihan, S., Eds.; Springer: Cham, Switzerland, 2017; pp. 201–226. [Google Scholar]
- Oren, A. Bioenergetic aspects of halophilism. Microbiol. Mol. Biol. Rev. 1999, 63, 334–348. [Google Scholar] [CrossRef]
- Gunde-Cimerman, N.; Zalar, P. Extremely halotolerant and halophilic fungi inhabit brine in solar salterns around the globe. Food Technol. Biotechnol. 2014, 52, 170–179. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).