Metabolic Engineering for the Biosynthesis of Pentalenene in the Rapidly Growing Bacterium Vibrio natriegens
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Reagents
2.2. Culture Medium
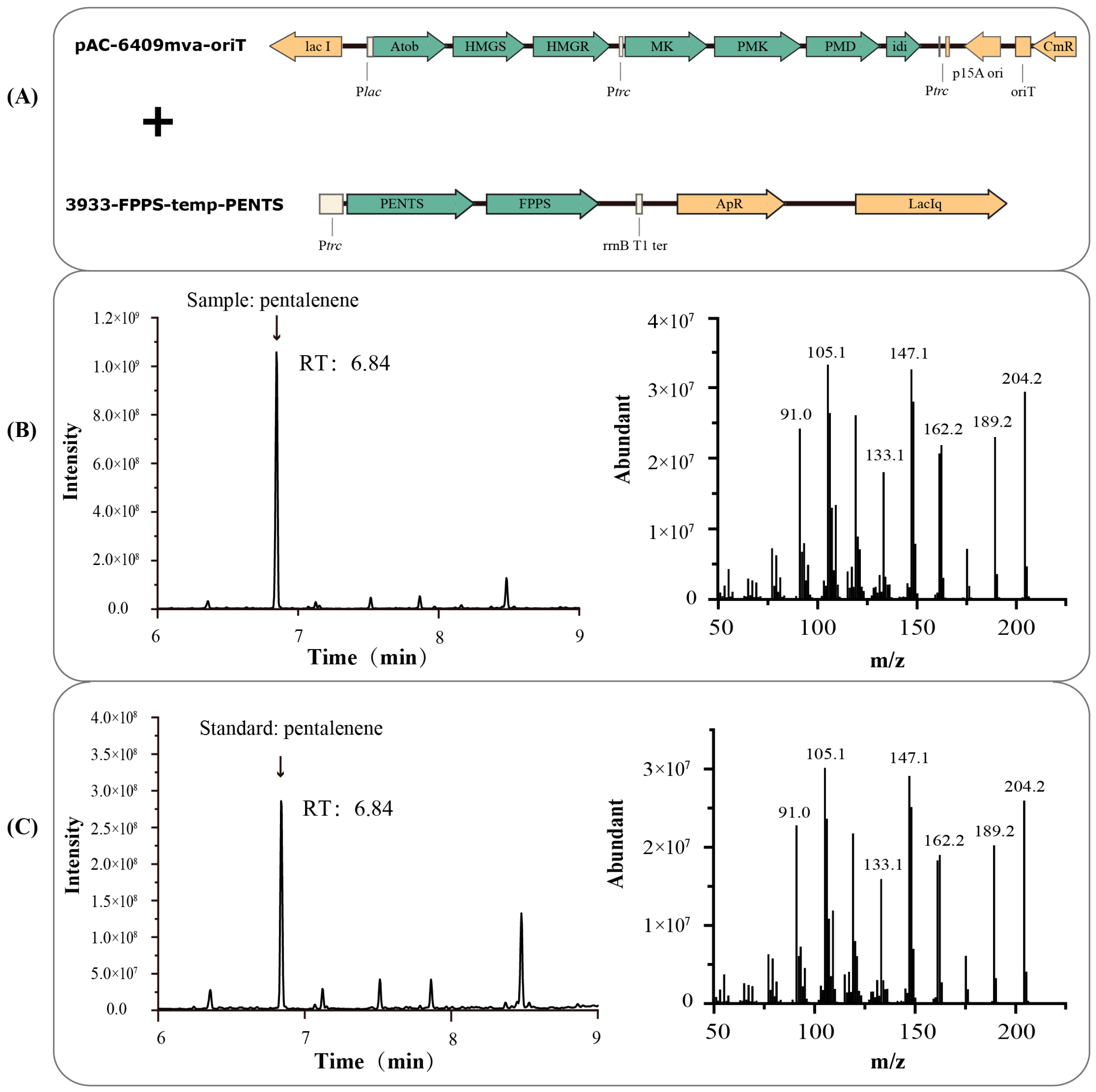
2.3. Plasmid and Strain Construction
2.4. Bacterial Conjugation
2.5. Shake Fermentation
2.6. Identification and Quantitative Analysis of Products
2.7. Optimization of Expression Conditions for Pentalenene Production by V. natriegens
2.8. Transcriptomics Analysis Methods
3. Results and Analysis
3.1. Production and Characterization of Pentalenene in V. natriegens
3.2. Optimization of Culture Conditions and Downstream Modules to Enhance Pentalenene Production
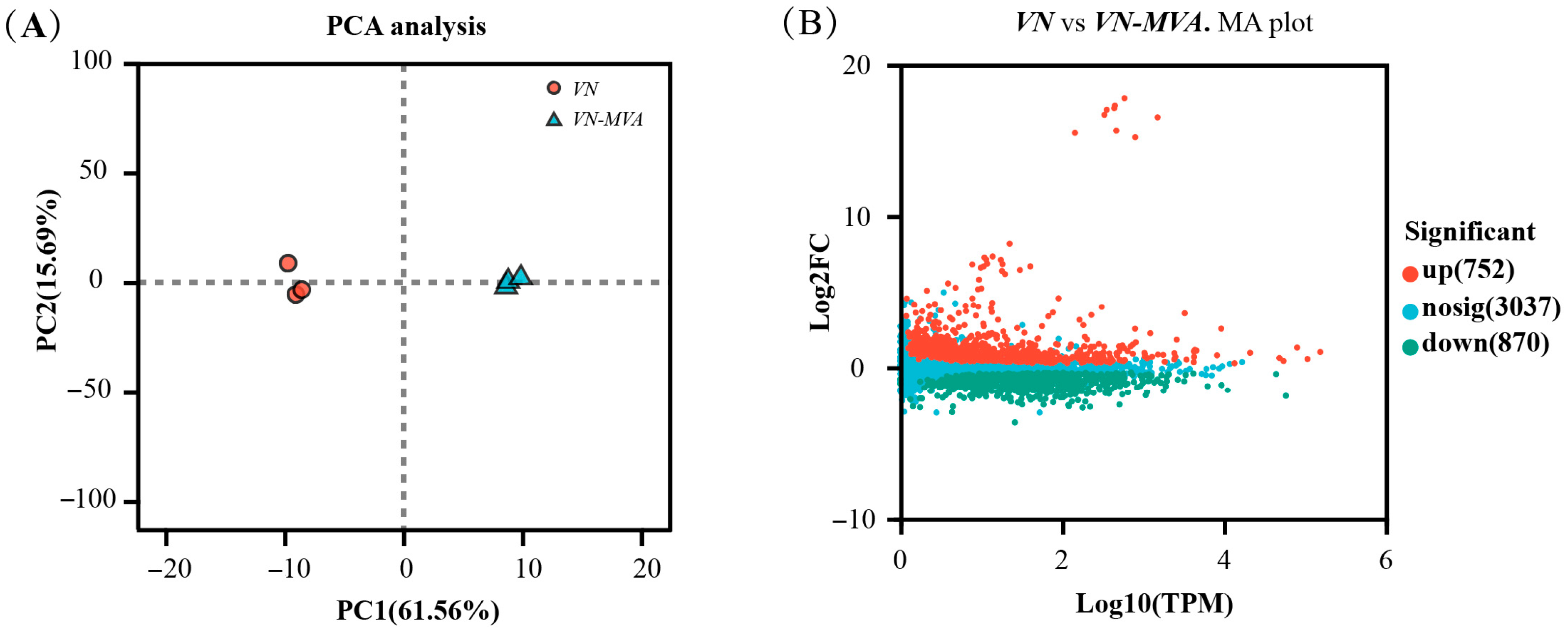
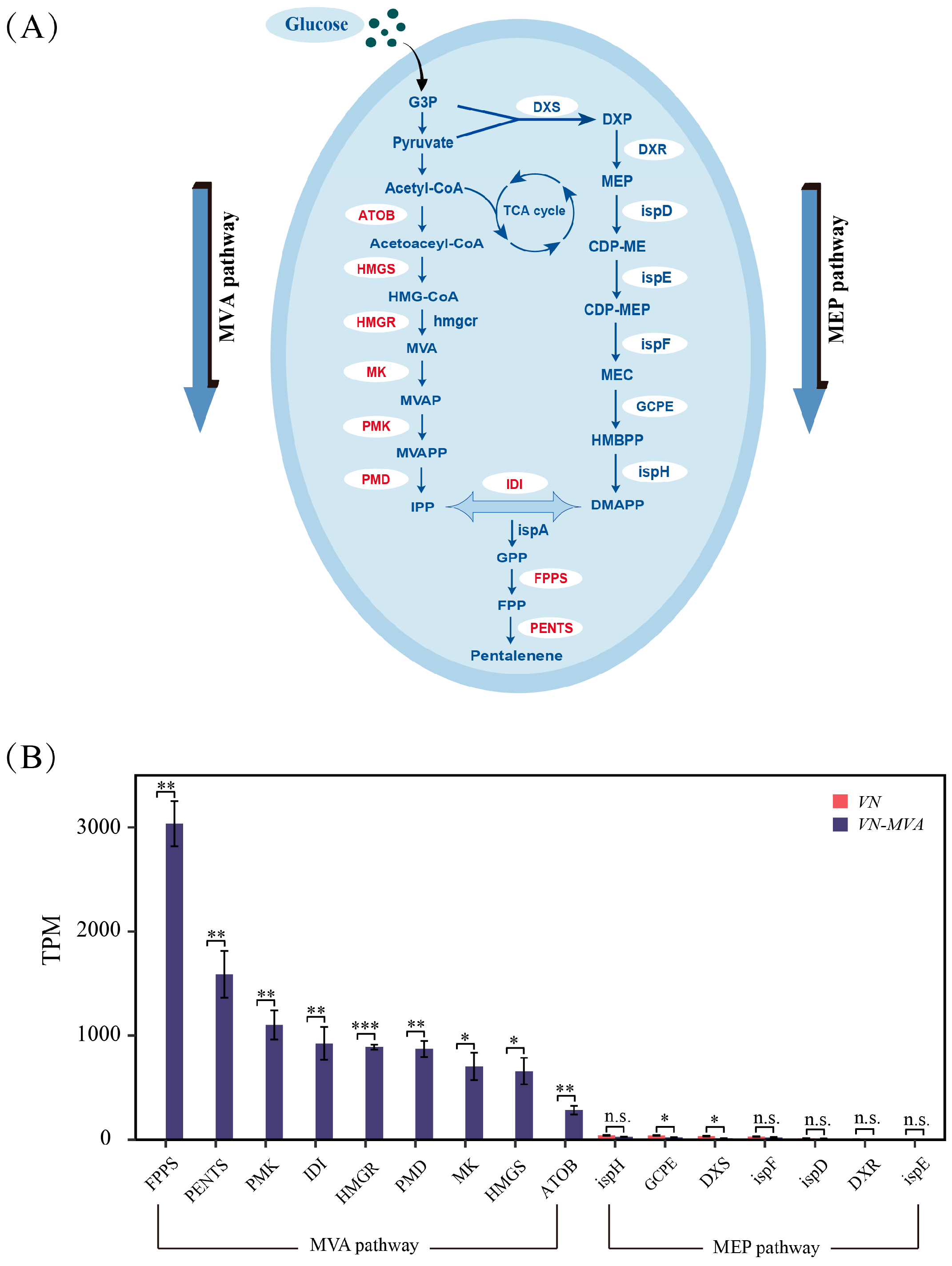
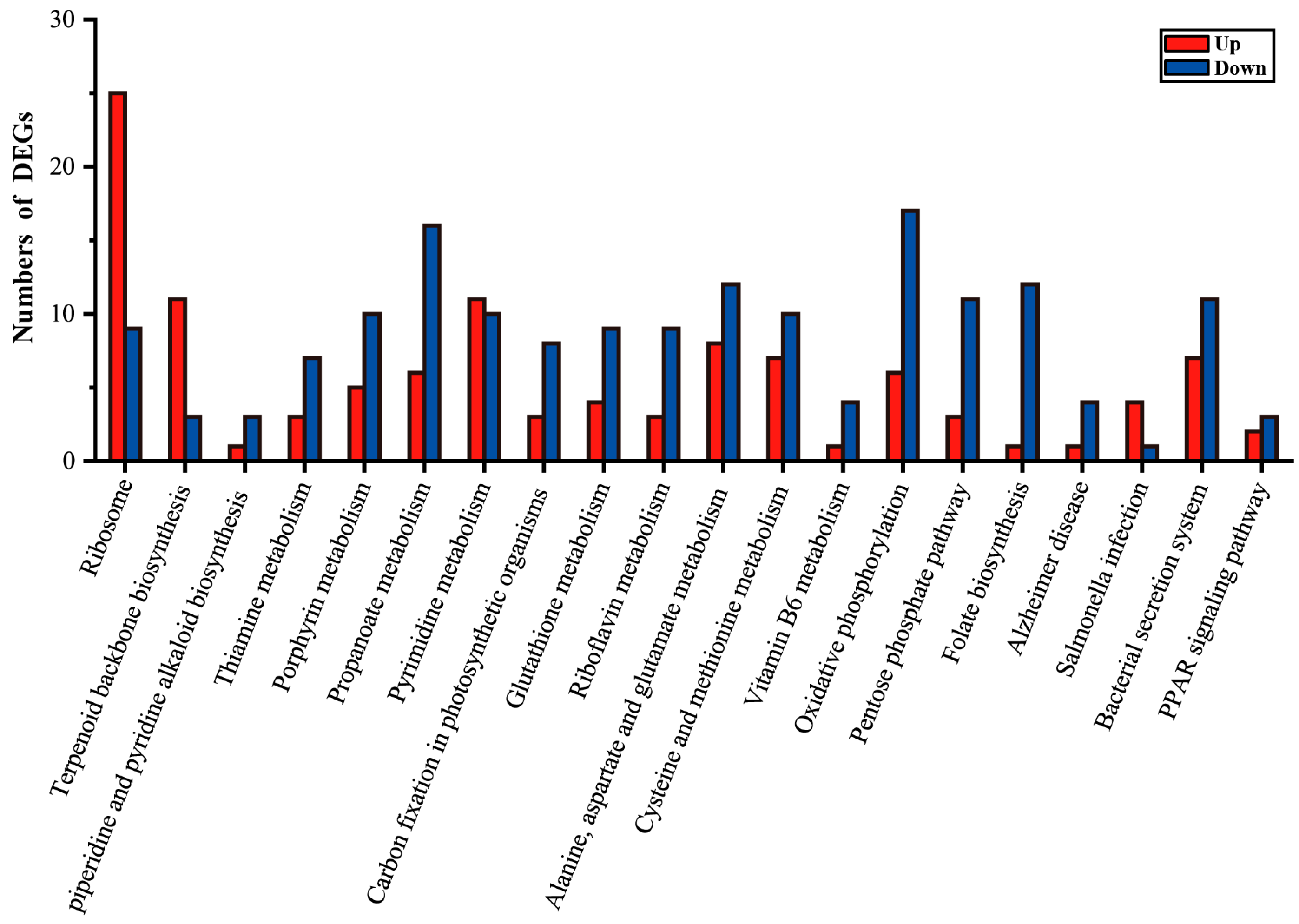
3.3. Transcriptome-Based Analysis of the Key Regulatory Factors of Pentalenene Synthesis Efficiency
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schleicher, L.; Muras, V.; Claussen, B.; Pfannstiel, J.; Blombach, B.; Dibrov, P.; Fritz, G.; Steuber, J. Vibrio natriegens as Host for Expression of Multisubunit Membrane Protein Complexes. Front. Microbiol. 2018, 9, 2537. [Google Scholar] [CrossRef]
- Fuchs, H.; Ullrich, S.R.; Hedrich, S. Vibrio natriegens as a superior host for the production of c-type cytochromes and difficult-to-express redox proteins. Sci. Rep. 2024, 14, 6093. [Google Scholar] [CrossRef]
- Biener, R.; Horn, T.; Komitakis, A.; Schendel, I.; König, L.; Hauenstein, A.; Ludl, A.; Speidel, A.; Schmid, S.; Weißer, J.; et al. High-cell-density cultivation of Vibrio natriegens in a low-chloride chemically defined medium. Appl. Microbiol. Biotechnol. 2023, 107, 7043–7054. [Google Scholar] [CrossRef]
- Ellis, G.A.; Tschirhart, T.; Spangler, J.; Walper, S.A.; Medintz, I.L.; Vora, G.J. Exploiting the Feedstock Flexibility of the Emergent Synthetic Biology Chassis Vibrio natriegens for Engineered Natural Product Production. Mar. Drugs 2019, 17, 679. [Google Scholar] [CrossRef] [PubMed]
- Mojica, N.; Kersten, F.; Montserrat-Canals, M.; Iii, G.R.H.; Tislevoll, A.M.; Cordara, G.; Teter, K.; Krengel, U. Using Vibrio natriegens for High-Yield Production of Challenging Expression Targets and for Protein Perdeuteration. Biochemistry 2024, 63, 587–598. [Google Scholar] [CrossRef]
- Liu, X.; Han, X.; Peng, Y.; Tan, C.; Wang, J.; Xue, H.; Xu, P.; Tao, F. Rapid production of l-DOPA by Vibrio natriegens, an emerging next-generation whole-cell catalysis chassis. Microb. Biotechnol. 2022, 15, 1610–1621. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.D.; Tschirhart, T.; Compton, J.; Hennessa, T.M.; VanArsdale, E.; Wang, Z. Rapid, high-titer biosynthesis of melanin using the marine bacterium Vibrio natriegens. Front. Bioeng. Biotechnol. 2023, 11, 1239756. [Google Scholar] [CrossRef]
- Seto, H.; Yonehara, H. Studies on the biosynthesis of pentalenolactone. III. Isolation of a biosynthetic intermediate hydrocarbon, pentalenene. J. Antibiot. 1980, 33, 92–93. [Google Scholar] [CrossRef]
- Tarasova, E.V.; Luchnikova, N.A.; Grishko, V.V.; Ivshina, I.B. Actinomycetes as Producers of Biologically Active Terpenoids: Current Trends and Patents. Pharmaceuticals 2023, 16, 872. [Google Scholar] [CrossRef]
- Huang, Y.; Ye, Z.; Wan, X.; Yao, G.; Duan, J.; Liu, J.; Yao, M.; Sun, X.; Deng, Z.; Shen, K.; et al. Systematic Mining and Evaluation of the Sesquiterpene Skeletons as High Energy Aviation Fuel Molecules. Adv. Sci. 2023, 10, e2300889. [Google Scholar] [CrossRef]
- Liu, H.; Fang, S.; Zhao, L.; Men, X.; Zhang, H. A Single Active-Site Mutagenesis Confers Enhanced Activity and/or Changed Product Distribution to a Pentalenene Synthase from Streptomyces sp. PSKA01. Bioengineering 2023, 10, 392. [Google Scholar] [CrossRef]
- Cheng, T.; Wang, L.; Sun, C.; Xie, C. Optimizing the downstream MVA pathway using a combination optimization strategy to increase lycopene yield in Escherichia coli. Microb. Cell Factories 2022, 21, 121. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Cheng, P.; Sun, J.; Zhang, Y.; Zheng, Y.; Jiang, X.-R.; Rao, X. Engineering a mevalonate pathway in Halomonas bluephagenesis for the production of lycopene. Front. Microbiol. 2023, 13, 1100745. [Google Scholar] [CrossRef]
- Liu, C.-L.; Tian, T.; Alonso-Gutierrez, J.; Garabedian, B.; Wang, S.; Baidoo, E.E.K.; Benites, V.; Chen, Y.; Petzold, C.J.; Adams, P.D.; et al. Renewable production of high density jet fuel precursor sesquiterpenes from Escherichia coli. Biotechnol. Biofuels 2018, 11, 285. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Nie, Q.; Ren, M.; Feng, H.; Jiang, X.; Zheng, Y.; Liu, M.; Zhang, H.; Xian, M. Metabolic engineering of Escherichia colifor the biosynthesis of alpha-pinene. Biotechnol. Biofuels 2013, 6, 60. [Google Scholar] [CrossRef]
- Bao, S.-H.; Jiang, H.; Zhu, L.-Y.; Yao, G.; Han, P.-G.; Wan, X.-K.; Wang, K.; Song, T.-Y.; Liu, C.-J.; Wang, S.; et al. A dynamic and multilocus metabolic regulation strategy using quorum-sensing-controlled bacterial small RNA. Cell Rep. 2021, 36, 109413. [Google Scholar] [CrossRef] [PubMed]
- Strand, T.A.; Lale, R.; Degnes, K.F.; Lando, M.; Valla, S. A New and Improved Host-Independent Plasmid System for RK2-Based Conjugal Transfer. PLoS ONE 2014, 9, e90372. [Google Scholar] [CrossRef]
- Hoffart, E.; Grenz, S.; Lange, J.; Nitschel, R.; Müller, F.; Schwentner, A.; Feith, A.; Lenfers-Lücker, M.; Takors, R.; Blombach, B. High Substrate Uptake Rates Empower Vibrio natriegens as Production Host for Industrial Biotechnology. Appl. Environ. Microbiol. 2017, 83, e01614-17. [Google Scholar] [CrossRef]
- Levin, J.Z.; Yassour, M.; Adiconis, X.; Nusbaum, C.; Thompson, D.A.; Friedman, N.; Gnirke, A.; Regev, A. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods 2010, 7, 709–715. [Google Scholar] [CrossRef]
- Parkhomchuk, D.; Borodina, T.; Amstislavskiy, V.; Banaru, M.; Hallen, L.; Krobitsch, S.; Lehrach, H.; Soldatov, A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009, 37, e123. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Zhu, H.; Hu, L.; Rozhkova, T.; Wang, X.; Li, C. Spectrophotometric analysis of bioactive metabolites and fermentation optimisation of Streptomyces sp. HU2014 with antifungal potential against Rhizoctonia solani. Biotechnol. Biotechnol. Equip. 2023, 37, 231–242. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, M.; Li, Y.; Cui, Y.; Zhang, Y.; Wang, Z.; Wang, M.; Huang, Y. Application of response surface methodology to optimize the production of antimicrobial metabolites by Micromonospora Y15. Biotechnol. Biotechnol. Equip. 2017, 31, 1016–1025. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, W.; Yang, Q. Transcriptome Analysis Reveals the Regulation of Aureobasidium pullulans under Different pH Stress. Int. J. Mol. Sci. 2023, 24, 16103. [Google Scholar] [CrossRef] [PubMed]
- Daniel, R.M.; Danson, M.J. Temperature and the catalytic activity of enzymes: A fresh understanding. FEBS Lett. 2013, 587, 2738–2743. [Google Scholar] [CrossRef]
- Wendering, P.; Nikoloski, Z. Model-driven insights into the effects of temperature on metabolism. Biotechnol. Adv. 2023, 67, 108203. [Google Scholar] [CrossRef]
- Hoffmann, K.H. Metabolic and Enzyme Adaptation to Temperature. In Environmental Physiology and Biochemistry of Insects; Hoffmann, K.H., Ed.; Springer: Berlin/Heidelberg, Germany, 1985; pp. 1–32. [Google Scholar] [CrossRef]
- Yang, J.; Guo, L. Biosynthesis of β-carotene in engineered E. coli using the MEP and MVA pathways. Microb. Cell Factories 2014, 13, 160. [Google Scholar] [CrossRef]
- Yang, C.; Gao, X.; Jiang, Y.; Sun, B.; Gao, F.; Yang, S. Synergy between methylerythritol phosphate pathway and mevalonate pathway for isoprene production in Escherichia coli. Metab. Eng. 2016, 37, 79–91. [Google Scholar] [CrossRef]
- Xu, X.; Liu, Y.; Luan, J.; Liu, R.; Wang, Y.; Liu, Y.; Xu, A.; Zhou, B.; Han, F.; Shang, W. Effect of downregulated citrate synthase on oxidative phosphorylation signaling pathway in HEI-OC1 cells. Proteome Sci. 2022, 20, 14. [Google Scholar] [CrossRef]
- Chang, Y.; Zhang, B.; Du, M.; Geng, Z.; Wei, J.; Guan, R.; An, S.; Zhao, W. The vital hormone 20-hydroxyecdysone controls ATP production by upregulating binding of trehalase 1 with ATP synthase subunit α in Helicoverpa armigera. J. Biol. Chem. 2022, 298, 101565. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; He, H.; Li, H.; Lu, W.; Guo, T.; Kong, J. The global regulator CodY responds to oxidative stress by the regulation of glutathione biosynthesis in Streptococcus thermophilus. J. Dairy Sci. 2017, 100, 8768–8775. [Google Scholar] [CrossRef]
- Ting, K.K.Y.; Floro, E.; Dow, R.; Jongstra-Bilen, J.; Cybulsky, M.I.; Rocheleau, J.V. Measuring the rate of NADPH consumption by glutathione reductase in the cytosol and mitochondria. PLoS ONE 2024, 19, e0309886. [Google Scholar] [CrossRef] [PubMed]
- Watermann, P.; Arend, C.; Dringen, R. G6PDi-1 is a Potent Inhibitor of G6PDH and of Pentose Phosphate pathway-dependent Metabolic Processes in Cultured Primary Astrocytes. Neurochem. Res. 2023, 48, 3177–3189. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.H.Y.; Ho, T.C.S.; Parle, D.R.; Leeper, F.J. Furan-based inhibitors of pyruvate dehydrogenase: SAR study, biochemical evaluation and computational analysis. Org. Biomol. Chem. 2023, 21, 1755–1763. [Google Scholar] [CrossRef]
- van Rossum, H.M.; Kozak, B.U.; Pronk, J.T.; van Maris, A.J. Engineering cytosolic acetyl-coenzyme A supply in Saccharomyces cerevisiae: Pathway stoichiometry, free-energy conservation and redox-cofactor balancing. Metab. Eng. 2016, 36, 99–115. [Google Scholar] [CrossRef]
- Lu, S.; Zhou, C.; Guo, X.; Du, Z.; Cheng, Y.; Wang, Z.; He, X. Enhancing fluxes through the mevalonate pathway in Saccharomyces cerevisiae by engineering the HMGR and β-alanine metabolism. Microb. Biotechnol. 2022, 15, 2292–2306. [Google Scholar] [CrossRef]
- Hoff, J.; Daniel, B.; Stukenberg, D.; Thuronyi, B.W.; Waldminghaus, T.; Fritz, G. Vibrio natriegens: An ultrafast-growing marine bacterium as emerging synthetic biology chassis. Environ. Microbiol. 2020, 22, 4394–4408. [Google Scholar] [CrossRef]

| Pathway | Gene Name | Annotation | log2 (FC) | Up or Down |
|---|---|---|---|---|
| Oxidative phosphorylation | NDH | NADH dehydrogenase | −1.13 * | Down |
| petB | cytochrome B | −0.74 * | Down | |
| petC | cytochrome C | −0.41 * | Down | |
| cyoB | cytochrome o ubiquinol oxidase subunit I | −1.69 * | Down | |
| cyoC | cytochrome o ubiquinol oxidase subunit III | −2.27 * | Down | |
| cyoD | Cytochrome o ubiquinol oxidase | −2.14 * | Down | |
| atpA | ATP F0F1 synthase subunit alpha | −0.52 * | Down | |
| atpC | ATP synthase F0F1 subunit epsilon | −0.93 * | Down | |
| atpD | ATP synthase F0F1 subunit beta | −0.75 * | Down | |
| atpG | ATP F0F1 synthase subunit gamma | −0.88 * | Down | |
| Glutathione metabolism | GSR | glutathione reductase | 0.64 * | Up |
| Pentose phosphate pathway | G6PDH | glucose-6-phosphate dehydrogenase | −0.87 * | Down |
| Thiamine metabolism | DXS | 1-deoxy-D-xylulose-5-phosphate synthase | −0.96 * | Down |
| thiH | thiamine biosynthesis protein ThiH | −1.63 * | Down | |
| thiM | hydroxyethylthiazole kinase | −1.61 * | Down | |
| tenA | hypothetical protein | −1.68 * | Down | |
| thiI | tRNA s(4)U8 sulfurtransferase | −0.64 * | Down | |
| thiG | thiazole synthase | −0.93 * | Down | |
| thiK | thiamine kinase | −0.62 * | Down |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, L.; Lin, R.; Jiang, H.; Yao, G.; Liu, J.; Han, P.; Wan, X.; Chen, C.; Zhang, Y.; Bao, S.; et al. Metabolic Engineering for the Biosynthesis of Pentalenene in the Rapidly Growing Bacterium Vibrio natriegens. Fermentation 2025, 11, 249. https://doi.org/10.3390/fermentation11050249
Hu L, Lin R, Jiang H, Yao G, Liu J, Han P, Wan X, Chen C, Zhang Y, Bao S, et al. Metabolic Engineering for the Biosynthesis of Pentalenene in the Rapidly Growing Bacterium Vibrio natriegens. Fermentation. 2025; 11(5):249. https://doi.org/10.3390/fermentation11050249
Chicago/Turabian StyleHu, Lujun, Rui Lin, Hui Jiang, Ge Yao, Jiajia Liu, Penggang Han, Xiukun Wan, Chang Chen, Yunfei Zhang, Shaoheng Bao, and et al. 2025. "Metabolic Engineering for the Biosynthesis of Pentalenene in the Rapidly Growing Bacterium Vibrio natriegens" Fermentation 11, no. 5: 249. https://doi.org/10.3390/fermentation11050249
APA StyleHu, L., Lin, R., Jiang, H., Yao, G., Liu, J., Han, P., Wan, X., Chen, C., Zhang, Y., Bao, S., & Wang, F. (2025). Metabolic Engineering for the Biosynthesis of Pentalenene in the Rapidly Growing Bacterium Vibrio natriegens. Fermentation, 11(5), 249. https://doi.org/10.3390/fermentation11050249





