Abstract
The exploration of microbial genetic resources for the production of fFen-flavor Baijiu has not only enriched the microbial library for baijiu production but has also laid the foundation for process improvement and strain optimization in baijiu brewing. In this study, a total of 177 fungal isolates were screened, including Saccharomyces cerevisiae, non-Saccharomyces cerevisiae, molds, and some pathogenic bacteria. Among them, Saccharomyces cerevisiae was the most abundant with 119 isolates, playing a major role in the fermentation of baijiu production. A total of 148 bacterial isolates were obtained from the fermentation mash samples, showing greater diversity compared to fungi. Bacillus species were the most abundant, with 94 isolates. Bacillus licheniformis, in particular, can produce a rich enzymatic system and flavor precursors, making it an important contributor to the sensory quality of baijiu. Lactic acid bacteria were the second most abundant, with 16 isolates. Additionally, five pathogenic fungal species were identified, including Candida pelliculosa, Candida lusitaniae, Fusarium oxysporum, Fusarium solani and Talaromyces marneffei. Six pathogenic bacterial species were also isolated, namely Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus xylosus, Moraxella osloensis, Actinomyces meyeri and Stenotrophomonas maltophilia. Finally, two strains of high acetate ethyl ester-producing yeast and lactate-degrading bacteria with good tolerance to temperature, pH, and ethanol concentration were identified as Saccharomyces cerevisiae and Bacillus licheniformis, respectively.
1. Introduction
Fen-flavor baijiu is one of the basic aroma types of Chinese baijiu, represented by Shanxi Fen-flavor baijiu. It utilizes sorghum as the brewing substrate and undergoes traditional pit fermentation, steaming and solid-state fermentation processes, adhering to the fundamental characteristics of ancient baijiu production methods, and is considered the origin of Chinese baijiu. Fen-flavor baijiu exhibits distinct flavor characteristics of purity, mellowness, sweetness, and a refreshing aftertaste. These style features are closely associated with the rich microbial communities in the brewing environment. Micro-organisms play a crucial role in the Fen-flavor baijiu production process. The diverse micro-organisms collectively create a complex and reliable micro-ecological habitat. Brewing micro-organisms can independently or synergistically utilize the nutrients in the raw materials within this microenvironment to produce ethanol and various unique flavor compounds.
One of the quality evaluation criteria of Fen-flavor baijiu is the flavor of the baijiu body. The production of high-quality Fen-flavor baijiu is inseparable from the regulation of flavor substances in baijiu. The flavor substance has a close relationship with micro-organisms, and the regulation of flavor cannot be separated from the understanding and regulation of micro-organisms in the fermentation process. Zheng [1] et al. showed that the yeasts in the Fen-flavor daqu mainly include: Saccharomycopsis fibuligera, Pichia anomala, Pichia kudriavzevii and Saccharomyces cerevisiae. The molds mainly include Absidia corymbifera, Aspergillus, Mucor circinelloides, Pseudocercospora phrymae, and Rhizopus stolonifer, with Absidia corymbifera being the dominant fungus among filamentous fungi. The bacteria found in the Fen-flavor daqu mainly belong to the genera Bacillus, Lactobacillus, Lactococcus, Streptococcus, Leuconostoc, Staphylococcus, and Acetobacter [2,3]. Wang [4] et al. detected a total of 204 species of prokaryotic micro-organisms in the fermented mash after three days, with 10 dominant genera identified, including Lactobacillus, Leuconostoc, Staphylococcus, Gluconobacter, Acetobacter, Petrimonas, Clostridium, Ruminococcus, Methanobacterium and Methanobrevibacter. Based on these studies, it can be seen that the micro-organisms involved in the fermentation process of Fen-flavor baijiu are very complex, and at the same time, the flavor substances produced by these micro-organisms according to their independent or synergistic effects are also very complex.
The flavor substances of Fen-flavor baijiu are mainly divided into acids, esters, alcohols, carbonyl compounds and off-odours [5], of which the main aroma of esters is ethyl acetate and ethyl lactate [6]. Fen-flavor baijiu as a kind of light-flavour baijiu; the absolute content of ethyl acetate and ethyl lactate and the proportionality between them have a great influence on the quality and style characteristics of Fen-flavor baijiu. Generally the content ratio of ethyl acetate and ethyl lactate is about 1: (0.6~0.8). However, in the production of baijiu, there is a common problem of a high content of ethyl lactate, resulting in the disproportion between ethyl lactate and the main aroma, which seriously affects the quality of baijiu. There are two forms of ester formation in baijiu: one is the esterification of alcohols and acids; the other is the metabolism of ester-producing yeast, especially ethyl acetate, which is generally believed to be produced in baijiu by the fermentation of ester-producing yeast [7]. In recent years, more and more attention has been paid to the mechanism and regulation of ethyl lactate production in baijiu. Ren JY et al. [8] increased the content of ethyl lactate by genetic modification of production value yeast, and Du Set al [9] regulated the content of ethyl lactate by repeated inoculation of granulocytes. There is a lack of research on reducing the content of ethyl lactate, mainly because of its stable nature, which is not easy to decompose. It can be done mostly by reducing the precursor substance of ethyl lactate lactic acid to reduce the content of ethyl lactate in baijiu [10].
This study attempts to deepen the understanding of micro-organisms in the fermentation process of Fen-flavor baijiu through new techniques, as well as to discover micro-organisms related to the regulation of flavor substances. Culturomics is a method for isolation of micro-organisms using multiple culture conditions, and identification of bacterial species using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF-MS) and 16S rRNA gene sequencing [11]. Culturomics is a novel approach to the study of complex microbial ecosystems, with the potential to detect low abundance taxa; to provide pure cultures of strains for polyphasic taxonomic characterization of new species; and to allow for the study of interactions between different strains present in a given microbiota, among other advantages [12]. Culturomics can make up for the shortcomings of traditional microbial isolation methods and macrogenomics techniques. Currently, culturomics is mostly applied to the study of intestinal micro-organisms, and is less applied to the micro-organisms of baijiu [13].
Therefore, the aim of this study was to apply culture genomics technology to mine the microbial germplasm resources of Fen-flavor baijiu brewing, and to screen out functional strains with degradation of lactic acid and high production of ethyl acetate from the point of view of regulating the absolute content and proportionality of ethyl acetate and lactic acid ethyl ester, the main aromatic components of Fen-flavor baijiu.
2. Materials and Method
2.1. Reagents and Materials
Sampling of fermented grains of Baijiu: fermented grains from a Fen-flavor baijiu factory in Shanxi. Take 4 pairs, 6 pairs, 15 pairs, and 24 pairs of time-fermented grains as experimental samples.
Commercialized culture medium: LB culture medium was purchased from Beijing Aoboxing Biotechnology Co., Ltd., Beijing, China; Gaoshi No. 1 medium, R2A medium, DG18 medium, and PDA medium were purchased from Qingdao Haibo Biotechnology Co., Ltd., Qingdao, China; MRS medium, Rose Bengal medium, and YPD medium were purchased from Solebao Biotechnology Co., Ltd., Beijing, China.
Ester production medium: glucose 8%, yeast extract 1%, and peptone 2%.
Sodium lactate medium (g/L): sodium lactate 20.0, yeast extract 5.0, ammonium sulfate 5.0, potassium dihydrogen phosphate 1.0, magnesium sulfate heptahydrate 0.3, agar powder 20.0, pH 6.8, 121 °C sterilization for 20 min.
Sodium lactate fermentation medium (g/L): sodium lactate 20.0, yeast extract 5.0, peptone 5.0, ammonium sulfate 5.0, potassium dihydrogen phosphate 1.0, potassium dihydrogen phosphate 1.0, sodium chloride 0.5, magnesium sulfate heptahydrate 0.5, pH 6.8, 121 °C sterilization for 20 min.
2.2. Instruments and Equipment
The instruments and equipment used in this experiment mainly include an intelligent constant temperature oscillation incubator (Shanghai Zhichu Instrument Co., Ltd., Shanghai, China), a super clean workbench (Suzhou Antai Air Technology Co., Ltd., Suzhou, China), a high-pressure steam sterilization pot (Shanghai Boxun Industrial Co., Ltd., Shanghai, China), MALDI-TOF-MS (Hexin CMI-3000, Guangzhou Hexin Instrument Co., Ltd., Guangzhou, China), a high-performance liquid chromatography (Agilent 1260 Infinity II system, Agilent Technology Co., Ltd.,Santa Clara, CA USA) gas chromatograph GC-2010 (SHIMADZU, Kyoto, Japan), etc.
2.3. Experimental Methods
2.3.1. Setting of Cultivation Conditions
Microbial growth requires nutrients and a proper environment including a carbon source, a nitrogen source, inorganic salts, growth factors and water, as well as an environment including temperature, pH, aerobic conditions and an interface. Based on the literature on the nutritional requirements and growth conditions of fermented grains, different solid plates were selected and designed for isolation and cultivation, including a common commercial medium and the original medium. For the microbial culturomics in Baijiu fermented grain samples, the designed isolation and culture conditions are shown in Table 1.

Table 1.
Isolation and cultivation conditions of micro-organisms in fermented grains.
2.3.2. Isolation and Purification of Fermented Grains Strains
Fungal isolation and purification: take 5 g of fermented grains and 45 mL of sterile physiological saline, add sterile glass beads, and shake in a constant temperature incubator at a speed of 180 r/min and 28 °C for 30 min. For gradient dilutions, 1 mL of oscillating medium was added to 9 mL of saline. Then, 200 μL of the dilutions in the 10−2, 10−3, and 10−4 gradients were taken and applied to different media (0.1 g/L chloramphenicol was added to the media to prevent bacterial contamination) for plate coating, and each gradient was applied two to three times in parallel. Cultivate at 28 °C for 48 h. Select single colonies, and observe the characteristics of single colonies, such as colony size, color, coloration, etc. Select colonies with different morphologies to be isolated and purified by streaking on the medium two to three times until single colonies appear.
Bacterial isolation and purification: Take 5 g of fermented grains and 45 mL of sterile physiological saline, add sterile glass beads, and shake in a constant temperature incubator at a speed of 180 r/min and 37 °C for 30 min. For gradient dilutions, 1 mL of oscillating medium was added to 9 mL of saline. Then, 200 μL of the dilutions in the 10−2, 10−3, and 10−4 gradients were taken and applied to different media for plate coating, and each gradient was applied two to three times in parallel. Cultivate at 37 °C for 48 h. Select single colonies, and observe the characteristics of single colonies, such as colony size, color, coloration, etc. Select colonies with different morphologies to be isolated and purified by streaking on the medium two to three times until single colonies appear.
2.3.3. Screening of Functional Strains
The yeast strain isolated and purified from the fermented grains was activated and cultured for 24 h, with an OD value of 0.8. Then, it was inoculated onto an ester producing medium and incubated for 4 days under 30 °C oscillation. Then add 80 mL of alcohol (60%) to the fermentation broth and distill 100 mL of the distillate. Filter the distillate through a microporous filter membrane and determine the content of ethyl acetate using gas chromatography. Chromatographic conditions: gas chromatograph GC-2010 (SHIMADZU, Kyoto, Japan); FID detector; chromatographic column: CP-WAX strongly polar polyethylene glycol chromatographic column; detector temperature: 260 °C, injection port temperature: 240 °C; the internal standard is n-amyl acetate; temperature rise program: hold at 50 °C for 4 min, then raise to 100 °C at 10 °C/min, and then raise to 240 °C at 40 °C/min for 1 min; air flow rate 300 mL/min, hydrogen flow rate 30 mL/min, nitrogen flow rate 20 mL/min; split ratio: 30:1.
Using traditional plate dilution separation and purification methods, lactic acid degrading bacteria were isolated and purified from fermented grains in a sodium lactate medium. The isolated and purified strains were inoculated with 2 mL of 80 mL of sodium lactate fermentation medium and incubated at 37 °C for 7 days. The fermentation broth was centrifuged for 10 min at 4 °C and 8000 r/min conditions, and then a microfiber filtration membrane (0.22 μm filter with inorganic membrane, dilute the filtrate 20 times, and then use high-performance liquid chromatography (injection volume of 10μL)) was used to determine the remaining amount of lactic acid. Select the strain with low lactic acid content in the fermentation broth as the target strain for the next step of screening. Chromatographic conditions: column: waters C18 (250mm× 4.6mm, 5 µm); mobile phase: methanol: 20 mmol/L KH2PO4 = 3:97; injection volume is 10 μL; flow rate: 1.5 mL/min; detector wavelength: 210 nm; column temperature: 25 °C.
Lactic acid utilization rate = (initial lactic acid amount − remaining lactic acid amount) ÷ initial lactic acid amount × 100%.
2.3.4. Study on the Tolerance of Strains
Alcohol tolerance test: fill 43 mL, 45 mL, 46 mL, 47 mL, 48 mL, and 49 mL of liquid culture media in a 150 mL triangular flask, sterilize, and then add 1 mL, 2 mL, 3 mL, 4 mL, 5 mL, and 6 mL of anhydrous ethanol accordingly. Make the alcohol content of the culture medium 2%, 4%, 6%, 8%, 10%, and 12%, respectively. The strains were accessed and incubated at 30 °C and 120 rpm with oscillation for 24 h. The OD values at 600 nm were determined.
Temperature tolerance test: fill 150 mL triangular flasks with an appropriate amount of liquid medium, access the strains placed in 30 °C, 34 °C, 37 °C, 40 °C, respectively, and 120 rpm vibration culture for 24 h, and determine the OD value of the 600 nm wavelength, to examine the heat resistance ability.
Acid resistance test: the pH of the medium was adjusted to 2.0, 3.0, 4.0, 5.0 and 6.0 with 1 mol/L hydrochloric acid, and the strains were cultured at 30 °C and 120 rpm for 24 h. The OD values at 600 nm were measured to investigate the acid resistance of the strains.
2.4. Morphological Identification of Strains and MALDI-TOF-MS Identification
The screened functional strains were characterized morphologically, and the excavated strains were identified using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF-MS) [14] and each strain was identified at least three times in parallel. The identification scoring criteria are shown in Figure 1.
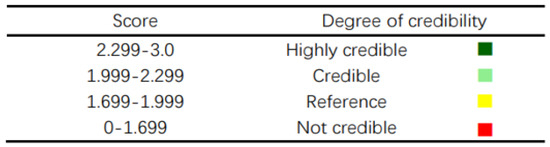
Figure 1.
Scoring criteria.
2.5. Molecular Biology Analysis of Strains
Send the cultivated strain solution to Shanxi Bocui Biotechnology Co., Ltd., Taiyuan, China, for sequencing. The sequencing results were compared with the blast sequence in NCBI. Select representative strain sequences from the database and use MEGA5.1 software to construct a phylogenetic tree using the neighbour-joining method.
2.6. Strain Conservation
Pure cultures of micro-organisms that had been isolated and cultured and identified by MALDI-TOF-MS were suspended and dispersed into sterile cryopreservation tubes containing strain cryopreservation solution (30% glycerol + 70% liquid bacterial suspension), and the strains were numbered and placed into an ultra-low-temperature refrigerator at −80 °C for long-term cryopreservation.
3. Result and Analysis
3.1. Results of Fungal Isolation
In this experiment, the isolation and screening of fungi from fermented grain samples yielded 177 pure cultures (Table 2), and the identification of the strains revealed that Saccharomyces cerevisiae was the most isolated with 119 strains. Some non-saccharomycetes such as Pichia kudriavzevii, Candida parapsilosis, and Pichia fermentans were isolated and 44 strains were obtained. On the other hand, fewer strains of molds were isolated only 8 strains, which belonged to Penicillium oxalicum and Aspergillus fumigatus, respectively. The microbial population and its number in the brewing process of Fen-flavor baijiu are mainly affected by the surrounding environment, such as temperature, water, oxygen, etc. When the acidity and alcohol content increase and the brewing environment is lacking in oxygen, the molds will soon wither away [15]. Two strains of Candida lusitaniae were also isolated, as well as one strain each of Candida pelliculosa, Fusarium oxysporum, Fusarium solani, and Talaromyces marneffei.

Table 2.
Isolation and Identification Results of Fungi in Fermented Grains (177 Strains in Total).
3.2. Results of Bacterial Isolation
A total of 148 bacterial strains were obtained from the bacterial isolation and screening of the fermented grain samples in this experiment (Table 3). Compared with fungi, the bacterial species were more abundant. The genus Bacillus was the most abundant, with 94 strains; 76 strains of Bacillus licheniformis, 7 strains of Bacillus subtilis, 5 strains of Bacillus pumilus, 3 strains of Bacillus amyloliquefaciens, 2 strains of Bacillus cereus, and 1 strain of Bacillus simplex were also isolated. This shows that Bacillus licheniformis is the dominant bacterium for Fen-flavor baijiu spirits. In this experiment, 16 different species of lactic acid bacteria were isolated and screened as 4 strains of Lactobacillus plantarum, 6 strains of Lactobacillus brevis, 3 strains of Pediococcus pentosaceus, 1 strain of Lactobacillus paracasei, 1 strain of Lactobacillus fermentum and 1 strain of Lactobacillus beijerinck. Some Staphylococcus epidermidis, Staphylococcus aureus, Staphylococcus xylosus, Moraxella osloensis, Actinomyces meyeri and Stenotrophomonas maltophilia were also isolated.

Table 3.
Isolation and identification results of bacteria in fermented grains (148 strains in total).
3.3. Screening Results of Functional Strains
3.3.1. Screening and Characterization of Ethyl Acetate-Producing Yeasts
Screening for High-Yielding Ethyl Acetate Yeasts
A total of 23 yeast strains were isolated and purified from the fermented grains using a solid ester-producing medium. After the activation and inoculation of the 23 yeast strains obtained from the isolation and purification into the ester-producing culture solution, the fermentation products were processed by centrifugation and filtration through a microporous filter membrane and then tested for the yield of ethyl acetate by gas chromatography. The results of the screening are shown in Table 4. The results showed that nine strains of bacteria had the ability to produce ethyl acetate, namely J2, J3, J6, J11, J12, J14, J17, J20, and J23. Four of the strains, J2, J6, J11, and J14, had higher ester production capacities, which were all greater than 0.5 g/L. Strain J6 had the highest ethyl acetate yield, which was 1.701 ± 0.11 g/L, followed by strain J2, with an ethyl acetate yield of 1.198 ± 0.36 g/L.

Table 4.
Identification results of ethyl acetate production.
Studies on the Tolerance of High-Yielding Ethyl Acetate Yeasts
Fermented grains are the main carrier substrate for baijiu fermentation, and their microenvironment is characterized by high ethanol concentration, high temperature and low pH. Therefore, the screened high-yielding ethyl acetate yeasts need to be examined for acid, temperature and alcohol resistance to achieve the selection of suitable strains for this purpose. As can be seen from Figure 2A–C, low pH, high alcohol content, and high temperature inhibited the growth of J2, J6, J11, J14. Except for J6, which was less tolerant, the growth trends of the three plants were similar, but the ethyl acetate production capacity of J2 (1.198 ± 0.36 g/L) was much higher than that of J11 (0.641 ± 0.05 g/L) and J14 (0.575 ± 0.24 g/L). J2 was selected for the subsequent study.
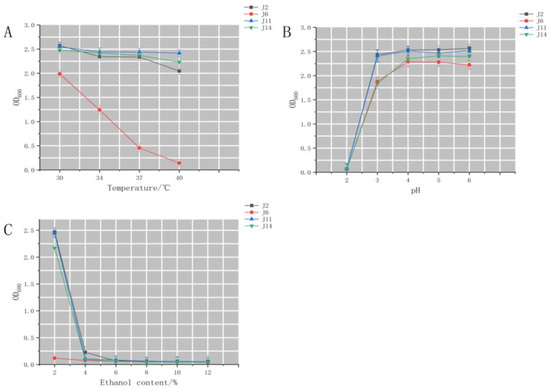
Figure 2.
Effect of environmental change on yeast growth (A) Effect of temperature on yeast growth (B) Effect of pH on yeast growth (C) Effect of alcohol by volume on yeast.
Colony Morphology and MALDI-TOF-MS Identification of High-Yield Ethyl Acetate Yeast
From Figure 3A of the J2 colony, it can be seen that the J2 colony is creamy white, with neat edges, uniform colony texture, and positive and negative sides, and the surface of the colony is moist, smooth, sticky and easy to pick up, which is in line with the basic morphological characteristics of yeast colonies. The strain was identified by MALDI-TOF-MS, and the preliminary identification result of the strain was Saccharomyces cerevisiae, as shown in Figure 3B,C.
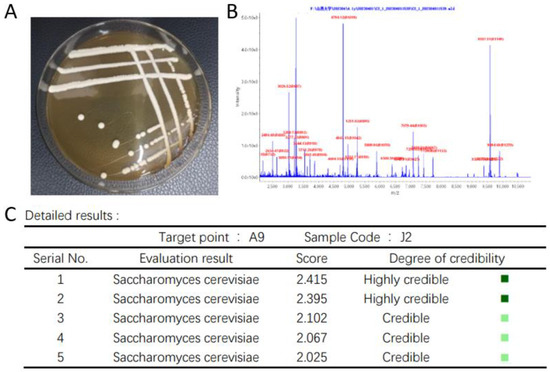
Figure 3.
Colony morphology and biological mass spectrometry identification of strain J2 (A) Colony morphology of strain J2 (B) Biomass spectrometry of strain J2 (C) Biomass spectrometry identification results of strain J2.
Molecular Biological Validation of High-Yielding Ethyl Acetate Yeasts
J2 was subjected to 26srDNA sequencing, and the results obtained from sequencing were subjected to blast sequence comparison in NCBI. Then, the phylogenetic tree of yeast strains related to the J2 gene sequence of Saccharomyces cerevisiae strains was plotted using MEGA-X software. As shown in Figure 4, the phylogenetic tree in the figure showed that strain J2 and Saccharomyces cerevisiae clustered on a branch and were close relatives of each other. Combined with the analysis of colony shapes and MALDI-TOF-MS measurement results, strain J2 was identified as belonging to Saccharomyces cerevisiae.

Figure 4.
Adjacent phylogenetic tree of strain J2 26s rDNA gene sequence and its phylogenetic relationship.
3.3.2. Screening and Characterization of Lactic Acid Degrading Bacteria
Screening of Lactic Acid Degrading Bacteria
A total of 14 strains of bacteria were isolated and purified from the fermented grains using a sodium lactate solid medium. After the activation of the 14 strains of bacteria obtained from the isolation and purification, they were inoculated into the liquid fermentation of sodium lactate, and the fermentation products were processed by centrifugation and filtration through microporous filtration membranes to determine the lactic acid content by using high-performance liquid chromatography (HPLC), and to calculate their lactic acid degradation rate. The final results of the assay are shown in Figure 5, which indicate that these 14 strains have the ability to degrade lactic acid, among which the degradation rates of R1, R3, R8, and R12 exceeded 80%, and the lactic acid degradation rates of R1 and R12 reached almost 100%. Therefore, these four strains were selected for the subsequent tolerance study.
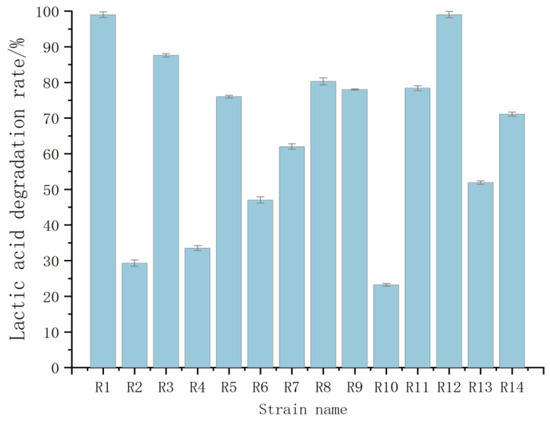
Figure 5.
Lactic acid degradation rates of different strains.
Tolerance Studies of Lactic Acid Degrading Bacteria
The same pH, alcohol concentration, and temperature tolerance experiments were performed on the four bacteria mentioned above with lactic acid degradation rates greater than 80%. From Figure 6A–C, it can be seen that low pH, high alcohol content, and low temperature all inhibited the growth of R1, R3, R8, and R12. However, overall, R12 always maintained high activity regardless of changes in environmental conditions, so R12 was selected for subsequent studies.
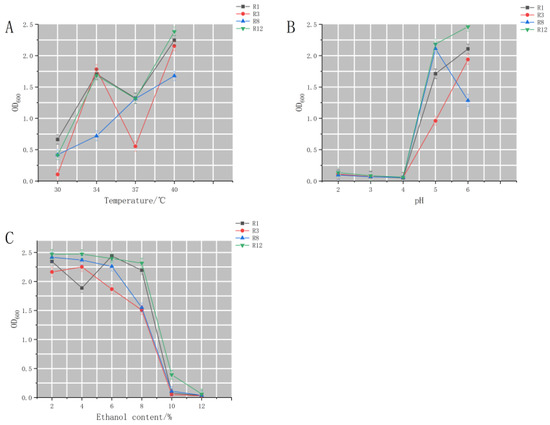
Figure 6.
Effect of environmental change on the growth of lactic acid degrading bacteria (A) Effect of temperature on the growth of lactic acid degrading bacteria (B) Effect of pH on the growth of lactic acid degrading bacteria (C) Effect of alcohol by volume on the growth of lactic acid degrading bacteria.
Colony Morphology and MALDI-TOF-MS Identification of Lactic Acid Degrading Bacteria
From Figure 7A, the single colony of R12 was small and irregularly shaped with untidy edges, and the strain was identified by MALDI-TOF-MS. From Figure 7B,C, the preliminary results of the strain were identified as Bacillus licheniformis.
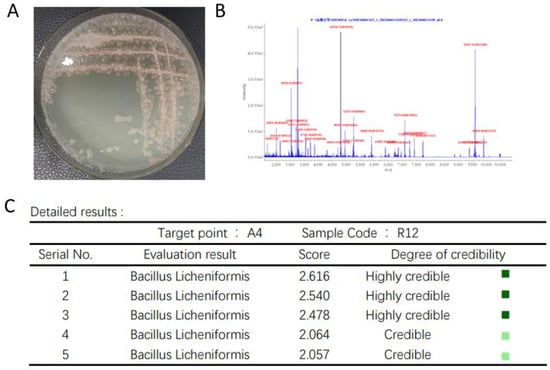
Figure 7.
Colony morphology and biological mass spectrometry identification of strain R12 (A) Colony morphology of strain R12 (B) Biomass spectrometry of strain R12 (C) Biomass spectrometry identification results of strain R12.
Molecular Biological Validation of Lactic Acid Degrading Bacteria
R12 was subjected to 16S rRNA sequencing, and the results obtained from sequencing were subjected to blast sequence comparison in NCBI. Then, the phylogenetic tree of the strains related to the R12 gene sequence of strain R12 was drawn using MEGA-X software. As shown in Figure 8, the phylogenetic tree in the figure showed that strains R12 and Bacillus licheniformis clustered on a branch and were close relatives of each other. Combined with the analysis of colony shapes and MALDI-TOF-MS measurement results, strain J2 was identified as belonging to Bacillus licheniformis.
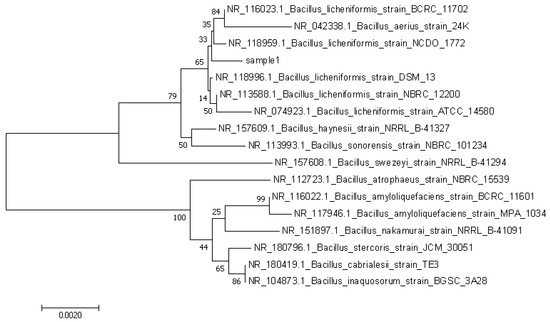
Figure 8.
Adjacent phylogenetic tree of the phylogenetic relationship between the 16S rRNA gene sequence of strain R12.
4. Conclusions and Discussion
Different micro-organisms all play vital roles in the brewing process of Fen-flavor baijiu, and together they create a complex and complete micro-ecological environment for the brewing of Fen-flavor baijiu. In this experiment, 177 pure fungal cultures and 148 pure bacteria cultures were screened from fermented grain samples using culturomics.
The screened fungi included mainly Saccharomyces cerevisiae, non-Saccharomyces cerevisiae, molds and some pathogenic bacteria. Among them, S. cerevisiae was isolated the most, with 119 strains. Saccharomyces cerevisiae plays the main fermentation role in the production of baijiu, which is important for the flavor and quality of baijiu [16]. Yeasts can be divided into two categories according to their function in the fermentation process: Saccharomyces cerevisiae, which mainly produce alcohol, and non-Saccharomyces cerevisiae, which ferment less efficiently but also make an important contribution to flavor. They are essential to the process of producing flavor substances in baijiu. For example, in the production of baijiu, the production of the ester aroma class of yeast is mostly non-Saccharomyces cerevisiae such as Candida parapsilosis, Pichia kudriavzevii, and so on [17]. Li [18] and others isolated and screened Pichia kudriavzevii obtained from fermented grains during the fermentation of baijiu with different degrees of esterification capacity for acids and alcohols, which can give the baijiu a more full-bodied aroma and contribute greatly to the sensory quality of the baijiu. More and more studies are now proving that mixed fermentations of non-Saccharomyces cerevisiae with Saccharomyces cerevisiae can produce unique styles and types of products. P. kudriavzevii has strong acid tolerance and lactic acid decomposition, as demonstrated by Deng [19] et al. Co-cultivation of P. kudriavzevii with S. cerevisiae can greatly improve lactic acid consumption and restore the ethanol metabolism of S. cerevisiae.
The main molds screened included Mycobacterium, Rhizoctonia, Aspergillus, Ploughshares, and Penicillium. Molds have a significant impact on improving the quality of baijiu fermentation because they can produce a variety of enzymes during the growth phase, break down fermentation ingredients, and produce aroma and flavor components in baijiu [20]. On the other hand, molds are also able to produce a variety of metabolites, for example, P. variotii is able to produce acetic acid, phenylethanol, linoleic acid, methylbutanol, phenylacetic acid, and other substances, which are flavor substances and flavor precursors in foods, and have an important effect on the flavor and texture of fermented foods.
Isolated cultures have produced more species of bacteria than fungi. Bacteria mainly include Bacillus, Lactobacillus and Staphylococcus. The genus Bacillus was the most isolated, with 94 strains. Bacillus cohn is widely distributed in the process of baijiu brewing and is rich in diversity, such as Bacillus licheniformis, Bacillus amyloliquefaciens, Bacillus subtilis, Bacillus cereus, and Bacillus methylotrophicus [21]. Bacillus cohn occupies an important proportion in the micro-organisms of baijiu brewing because it has a good high-temperature tolerance.
There are a large number of lactic acid bacteria in nature, so a large number of lactic acid bacteria are enriched in the open production of baijiu. The lactic acid bacteria mainly include Weisseria, Lactobacillus, Leuconostoc, etc. [22]. Lactic acid bacteria can be produced through the metabolism of carbohydrates and lactic acid, acetic acid and other small molecules of organic acid directly or as a precursor of flavor substances indirectly involved in the formation of the flavor of baijiu; and lactic acid bacteria can also be used to inhibit the growth of stray bacteria by lowering the pH value of the environment as well as the production of bacterial inhibitory substances, which reduces the occurrence of spoilage [23]. Antagonistic substances produced by Lactobacillus plantarum were found to be effective in inhibiting the growth of a wide range of gram-positive and gram-negative bacteria in food products and prolonging the shelf-life of food products [24]. Lactobacillus casei is a frequently used active probiotic, such as a unique mutant strain of Lactobacillus casei shirota that ferments Yakult, whose main role is to stimulate metabolism and control the microenvironment of the human digestive tract [25]. Pediococcus pentosaceus has also been noted to have the ability to produce bacteriocins, which have been shown to greatly limit the growth of foodborne pathogens [26].
Staphylococcus spp. are widely distributed in nature and can cause serious infections in humans and animals. Staphylococcus aureus is a common pathogen in baijiu, and it causes a variety of superficial and invasive infections, such as osteomyelitis, pneumonia, endocarditis and sepsis [27]. However, some studies have shown that yellow water, a fermentation by-product of baijiu, has high antibacterial activity and good antibacterial properties against Staphylococcus aureus [28]. Staphylococcus epidermidis is a common commensal organism on the skin and mucous membranes of humans and mammals, which usually does not cause disease, but forms a strong biofilm on the surface of the infected area, which enhances its resistance to antimicrobial drugs, thus causing a variety of infectious diseases [29].
Ethyl acetate and ethyl lactate are the main aroma components of Fen-flavor baijiu, and their absolute content and the proportional relationship affect the quality of Fen-flavor baijiu. At present, in the process of baijiu production, the content of ethyl lactate is too high, which leads to the imbalance of ethyl lactate and ethyl acetate, which seriously affects the quality of baijiu. Therefore, it is of great significance to improve the content of ethyl acetate and reduce ethyl lactate in the process of baijiu production to improve the quality of baijiu and promote the sustainable development of the baijiu industry.
The functional strains isolated from the fermentation process of Fen-flavor baijiu in this study provide potential strains for the regulation of the absolute contents of ethyl acetate and ethyl lactate as well as the proportional relationship in Fen-flavor baijiu. They were identified as Saccharomyces cerevisiae and Bacillus licheniformis, which are important functional bacteria in baijiu brewing.
As a parthenogenetic anaerobic eukaryotic organism, Saccharomyces cerevisiae can ferment glucose into ethanol and carbon dioxide under anaerobic conditions, which is widely used in the fields of food, medicine and brewing [30]. Saccharomyces cerevisiae is the main bacterium of alcoholic fermentation in the process of baijiu production, and its fermentation ability directly affects the raw material yield. The by-products of alcoholic fermentation, such as esters and alcohols and other substances, affect the style of the product, which plays a pivotal role in the production of baijiu. Moreover, Saccharomyces cerevisiae is highly tolerant to ethanol, and it has been shown that Saccharomyces cerevisiae accumulates high levels of ergosterol and has altered plasma membrane ATPase when the alcohol concentration is high, which improves the strain’s ability to survive alcohol stress [31]. Some researchers have also selected and bred high alcohol yielding Saccharomyces cerevisiae [32], high ethanol tolerance Saccharomyces cerevisiae [31], etc. by changing the culture conditions, with a view to providing benefits and process improvement for the baijiu industry. Bacillus licheniformis is an important flavor contributing bacterium that affects the taste of baijiu by producing C4 chemicals such as pyrazine compounds, volatile acids, aromatics and phenolic compounds.
It has been shown in the literature that a moderate reduction of lactic acid enhances the stability and functionality of Jiang-flavor Baijiu communities [33], but this study has not yet explored the changes of these two strains during the production of Fen-flavor baijiu, and the subsequent effects of the inoculum amount as well as the proportion of inoculum on the quality of Fen-flavor baijiu in the fermentation process. The effects of the two functional strains of bacteria on ethyl acetate and ethyl lactate of Fen-flavor baijiu, as well as on the fermentation flora, will be observed subsequently from the experimental simulation point of view.
Author Contributions
J.Z.: Writing—review and editing, Supervision, Funding acquisition. T.Z. and Z.H.: Conceptualization, Writing—original draft, Formal analysis. T.B. and L.D.: Writing—review and editing, Resources, Funding acquisition, Supervision, Investigation. B.B.: Writing—review and editing, Supervision, Investigation. M.C. and Y.Y.: Writing—review and editing, Supervision, Funding acquisition, Project administration. S.F.: Investigation, Funding acquisition, Writing—review and editing, Formal analysis. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the General Youth Fund Project of Shanxi Province (20210302124511), Xinghuacun College Open Project Foundation (XCSXU-KF-202306, XCSXU-KF-202313, XCSXU-KF-202317, XCSXU-KF-202328).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available within the article.
Acknowledgments
We would like to express our sincere gratitude to the funding agency for their financial support. Also, we are indebted to all the participants who dedicated their time and efforts in this research. Their contributions have been crucial to the success of this study.
Conflicts of Interest
Author Min Chen was employed by the Shanxi Food Research Institute. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Zheng, X.-W.; Yan, Z.; Han, B.-Z.; Zwietering, M.H.; Samson, R.A.; Boekhout, T.; Nout, M.J.R. Complex microbiota of a Chinese “Fen” liquor fermentation starter (Fen-Daqu), revealed by culture-dependent and culture-independent methods. Food Microbiol. 2012, 31, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Y.; Gao, Y.B.; Fan, Q.W.; Xu, Y. Characterization and comparison of microbial community of different typical Chinese liquor Daqus by PCR-DGGE. Lett. Appl. Microbiol. 2011, 53, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.-W.; Yan, Z.; Nout, M.J.R.; Smid, E.J.; Zwietering, M.H.; Boekhout, T.; Han, J.-S.; Han, B.-Z. Microbiota dynamics related to environmental conditions during the fermentative production of Fen-Daqu, a Chinese industrial fermentation starter. Int. J. Food Microbiol. 2014, 182–183, 57–62. [Google Scholar] [CrossRef]
- Wang, X.; Du, H.; Xu, Y. Source tracking of prokaryotic communities in fermented grain of Chinese strong-flavor liquor. Int. J. Food Microbiol. 2017, 244, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Zhang, K.; Zou, W.; Hou, Y. Three main flavour types of Chinese Baijiu: Characteristics, research, and perspectives. J. Inst. Brew. 2021, 127, 317–326. [Google Scholar] [CrossRef]
- Niu, Y.; Yao, Z.; Xiao, Q.; Xiao, Z.; Ma, N.; Zhu, J. Characterization of the key aroma compounds in different light aroma type Chinese liquors by GC-olfactometry, GC-FPD, quantitative measurements, and aroma recombination. Food Chem. 2017, 233, 204–215. [Google Scholar] [CrossRef]
- Fan, G.; Sun, B.; Xu, D.; Teng, C.; Fu, Z.; Du, Y.; Li, X. Isolation and Identification of High-Yield Ethyl Acetate-Producing Yeast from Gujinggong Daqu and Its Fermentation Characteristics. J. Am. Soc. Brew. Chem. 2018, 76, 117–124. [Google Scholar] [CrossRef]
- Ren, J.-Y.; Liu, G.; Chen, Y.-F.; Jiang, S.; Ma, Y.-R.; Zheng, P.; Guo, X.-W.; Xiao, D.-G. Enhanced Production of Ethyl Lactate in Saccharomyces cerevisiae by Genetic Modification. J. Agric. Food Chem. 2020, 68, 13863–13870. [Google Scholar] [CrossRef]
- Du, S.; Yao, L.; Zhong, B.; Qin, J.; He, S.; Liu, Y.; Wu, Z. Enhancing synthesis of ethyl lactate in rice baijiu fermentation by adding recovered granular cells. J. Biosci. Bioeng. 2024, 137, 388–395. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.; Jian, H.; Xu, X.; Xi, Y. Biocycle Fermentation Based on Lactic Acid Bacteria and Yeast for the Production of Natural Ethyl Lactate. ACS Omega 2019, 4, 16009–16015. [Google Scholar] [CrossRef]
- Lagier, J.-C.; Dubourg, G.; Million, M.; Cadoret, F.; Bilen, M.; Fenollar, F.; Levasseur, A.; Rolain, J.-M.; Fournier, P.-E.; Raoult, D. Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 2018, 16, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Fournier, P.-E.; Lagier, J.-C.; Dubourg, G.; Raoult, D. From culturomics to taxonomogenomics: A need to change the taxonomy of prokaryotes in clinical microbiology. Anaerobe 2015, 36, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Naud, S.; Khelaifia, S.; Mbogning Fonkou, M.D.; Dione, N.; Lagier, J.C.; Raoult, D. Proof of Concept of Culturomics Use of Time of Care. Front. Cell. Infect. Microbiol. 2020, 10, 524769. [Google Scholar] [CrossRef] [PubMed]
- Schwake, D.O.; Sandrin, T.; Zhang, L.; Abbaszadegan, M. Strain-Level Characterization of Legionella Environmental Isolates via MALDI-TOF-MS. Microorganisms 2022, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Huan, W.; Yongguang, H.; Yunli, H. Microbiome diversity and evolution in stacking fermentation during different rounds of Jiang-flavoured Baijiu brewing. LWT 2021, 143, 111119. [Google Scholar]
- Urso, R.; Rantsiou, K.; Dolci, P.; Rolle, L.; Comi, G.; Cocolin, L. Yeast biodiversity and dynamics during sweet wine production as determined by molecular methods. FEMS Yeast Res. 2008, 8, 1053–1062. [Google Scholar] [CrossRef]
- Cristian, V. The impact of non-Saccharomyces yeasts in the production of alcoholic beverages. Appl. Microbiol. Biotechnol. 2016, 100, 9861–9874. [Google Scholar]
- Li, R.-R.; Xu, M.; Zheng, J.; Liu, Y.-J.; Sun, C.-H.; Wang, H.; Guo, X.-W.; Xiao, D.-G.; Wu, X.-L.; Chen, Y.-F. Application Potential of Baijiu Non-Saccharomyces Yeast in Winemaking Through Sequential Fermentation With Saccharomyces cerevisiae. Front. Microbiol. 2022, 13, 902597. [Google Scholar] [CrossRef]
- Nan, D.; Hai, D.; Yan, X. Cooperative Response of Pichia kudriavzevii and Saccharomyces cerevisiae to Lactic Acid Stress in Baijiu Fermentation. J. Agric. Food Chem. 2020, 68, 4903–4911. [Google Scholar]
- Liu, S.; Yang, L.; Zhou, Y.; He, S.; Li, J.; Sun, H.; Yao, S.; Xu, S. Effect of mixed moulds starters on volatile flavor compounds in rice wine. LWT 2019, 112, 108215. [Google Scholar] [CrossRef]
- Wu, X.; Jiang, Q.; Wang, Z.; Xu, Y.; Chen, W.; Sun, J.; Liu, Y. Diversity, enzyme production and antibacterial activity of Bacillus strains isolated from sesame-flavored liquor Daqu. Arch. Microbiol. 2021, 203, 5831–5839. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Xiong, Y.; Xie, Y.; Zhang, H.; Jiang, K.; Pang, X.N.; Huang, M. Metabolic characteristics of lactic acid bacteria and interaction with yeast isolated from light-flavor Baijiu fermentation. Food Biosci. 2022, 50, 102102. [Google Scholar] [CrossRef]
- Jiang, F.-G.; Cheng, H.-J.; Liu, D.; Wei, C.; An, W.-J.; Wang, Y.-F.; Sun, H.-T.; Song, E.-L. Treatment of Whole-Plant Corn Silage with Lactic Acid Bacteria and Organic Acid Enhances Quality by Elevating Acid Content, Reducing pH, and Inhibiting Undesirable Microorganisms. Front. Microbiol. 2020, 11, 593088. [Google Scholar] [CrossRef] [PubMed]
- Heredia-Castro, P.Y.; Méndez-Romero, J.I.; Hernández-Mendoza, A.; Acedo-Félix, E.; González-Córdova, A.F.; Vallejo-Cordoba, B. Antimicrobial activity and partial characterization of bacteriocin-like inhibitory substances produced by Lactobacillus spp. isolated from artisanal Mexican cheese. J. Dairy Sci. 2015, 98, 8285–8293. [Google Scholar] [CrossRef] [PubMed]
- Harbige, L.S.; Pinto, E.; Allgrove, J.; Thomas, L.V. Immune Response of Healthy Adults to the Ingested Probiotic Lactobacillus casei Shirota. Scand. J. Immunol. 2016, 84, 353–364. [Google Scholar] [CrossRef]
- Wanderley Porto, M.C.; de Souza de Azevedo, P.O.; Lourenço, F.R.; Converti, A.; Vitolo, M.; de Souza Oliveira, R.P. Effect of Polydextrose on the Growth of Pediococcus pentosaceus as Well as Lactic Acid and Bacteriocin-like Inhibitory Substances (BLIS) Production. Microorganisms 2022, 10, 1898. [Google Scholar] [CrossRef]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef]
- Kang, J.; Sun, Y.; Huang, X.; Ye, L.; Chen, Y.; Chen, X.; Zheng, X.; Han, B.-Z. Unraveling the microbial compositions, metabolic functions, and antibacterial properties of Huangshui, a byproduct of Baijiu fermentation. Food Res. Int. 2022, 157, 111320. [Google Scholar] [CrossRef]
- Michael, O. Molecular basis of Staphylococcus epidermidis infections. Semin. Immunopathol. 2012, 34, 201–214. [Google Scholar]
- Huang, S.; Liu, T.; Peng, B.; Geng, A. Enhanced ethanol production from industrial lignocellulose hydrolysates by a hydrolysate-cofermenting Saccharomyces cerevisiae strain. Bioprocess. Biosyst. Eng. 2019, 42, 883–896. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, Y.; Du, G.; Zhou, J.; Chen, J. Exogenous ergosterol protects Saccharomyces cerevisiae from D-limonene stress. J. Appl. Microbiol. 2013, 114, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Fumio, M.; Chikara, F.; Takashi, K.; Jun, I. Engineering strategy of yeast metabolism for higher alcohol production. Microb. Cell Factories 2011, 10, 70. [Google Scholar]
- Wei, J.; Nie, Y.; Du, H.; Xu, Y. Reduced lactic acid strengthens microbial community stability and function during Jiang-flavour Baijiu fermentation. Food Biosci. 2024, 59, 103935. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).