A Novel Strategy for Further Enhancing Superior Properties of Thermophilic Endoglucanase from Acidomyces richmondensis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Cultivation Conditions
2.2. Rational Mining of Candidate Modification Sites
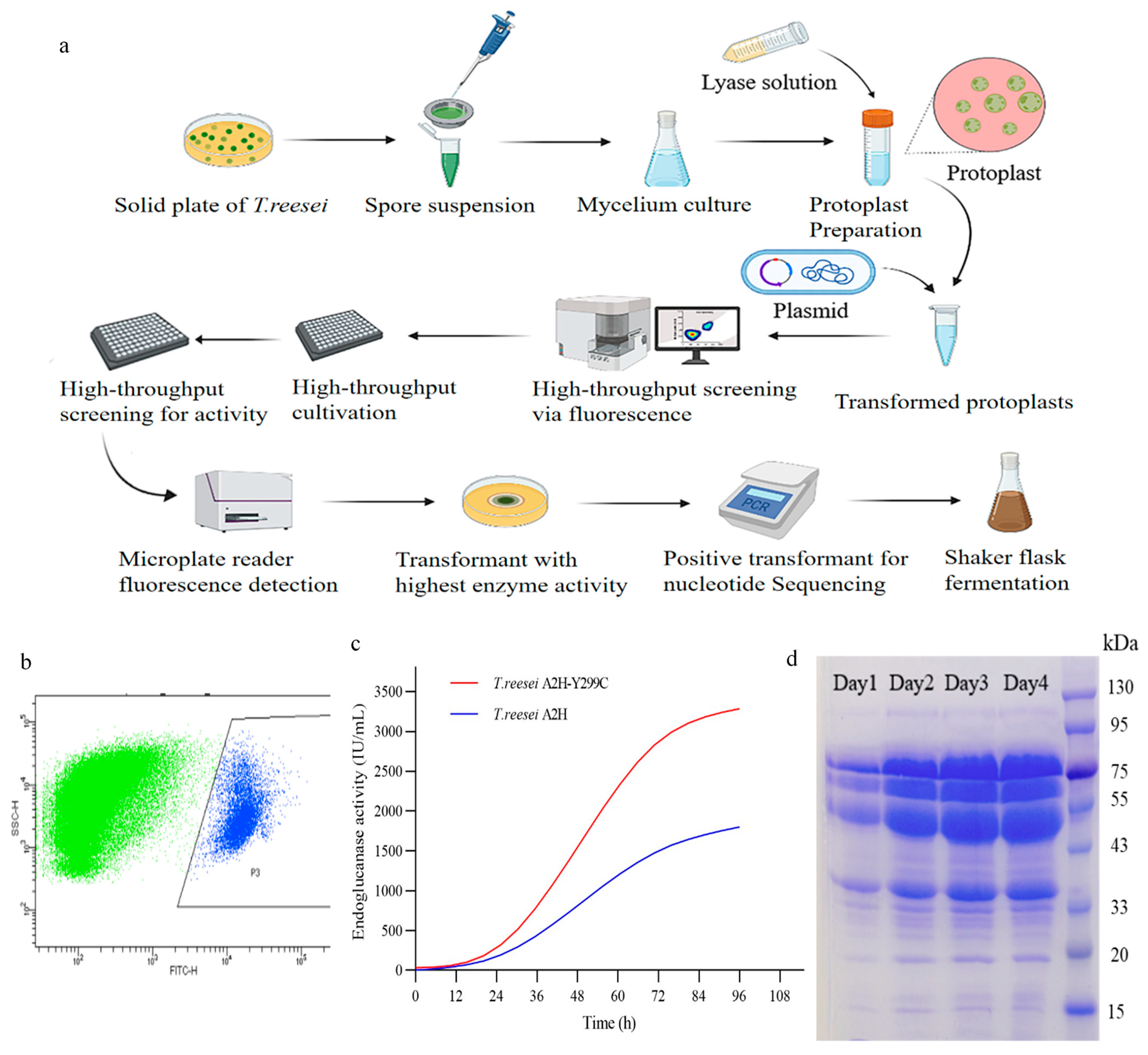
2.3. Mutant Library and High-Throughput Screening
2.4. High-Throughput Screening for Activity
2.5. Nucleotide Sequencing and Sequence Analysis
2.6. Determination of Endoglucanase Activity
2.7. Thermostability Assay of EG
2.8. SDS-PAGE Analysis
2.9. B Factor Values Analysis
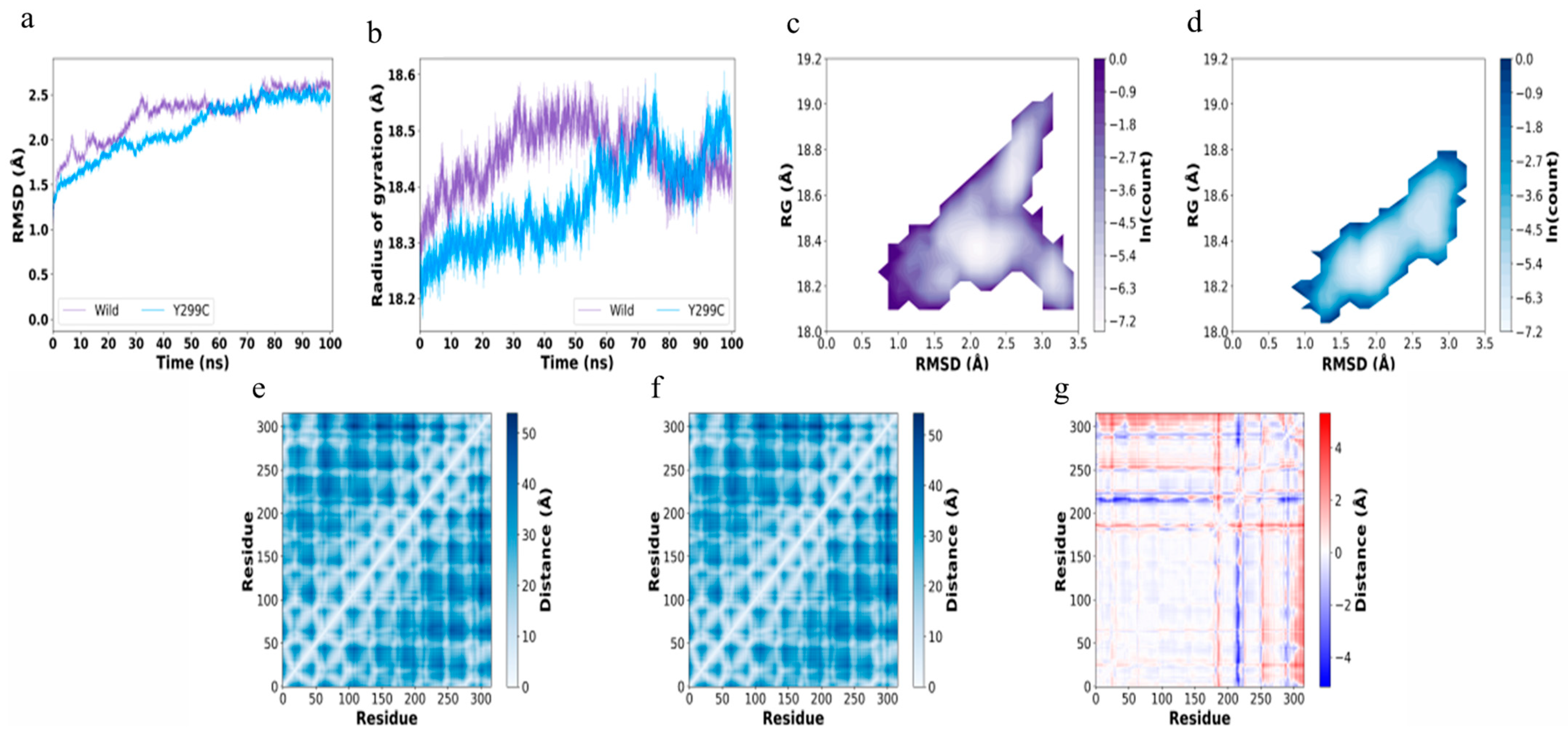
2.10. Molecular Dynamics (MD) Simulations
3. Results and Discussion
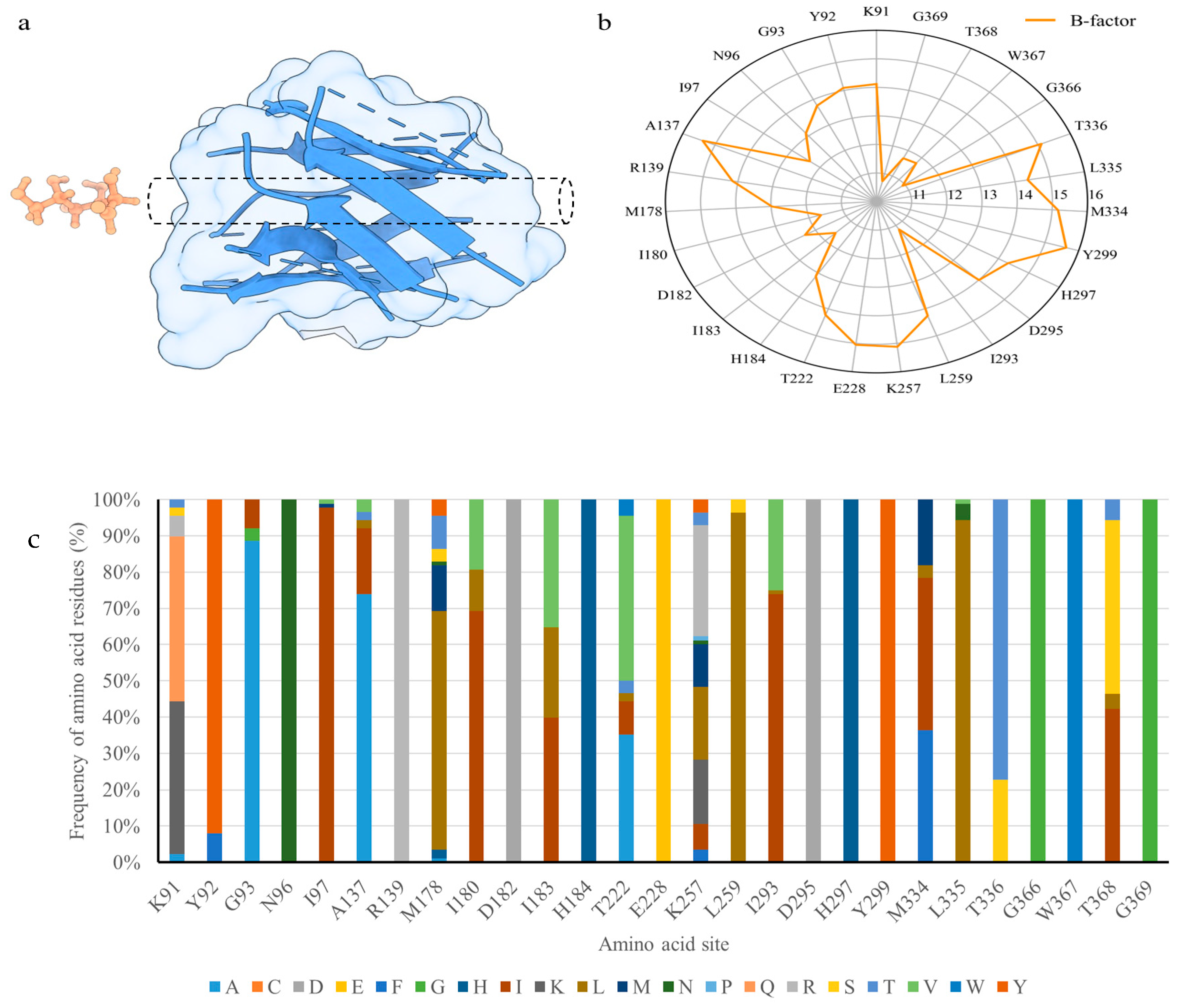
3.1. Three-Dimensional Model of Cel5A from Acidomyces Richmondensis
3.2. Selection of Mutational Target Sites through B-Factor Analysis
3.3. High-Throughput Screening of Transformants with High Enzymatic Activity
3.4. Improvement of Enzyme Activity and Thermostability in the Mutant EG
3.5. Mechanisms for Stabilizing EG Mutations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Qaiser, H.; Kaleem, A.; Abdullah, R.; Iqtedar, M.; Hoessli, D.C. Overview of lignocellulolytic enzyme systems with special reference to valorization of lignocellulosic biomass. Protein. Pept. Lett. 2021, 28, 1349–1364. [Google Scholar] [CrossRef] [PubMed]
- Marriott, P.E.; Gómez, L.D.; McQueen-Mason, S.J. Unlocking the potential of lignocellulosic biomass through plant science. New. Phytol. 2016, 209, 1366–1381. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Qu, Y. Integrated engineering of enzymes and microorganisms for improving the efficiency of industrial lignocellulose deconstruction. Eng. Microbiol. 2021, 1, 100005. [Google Scholar] [CrossRef]
- Davies, G.J.; Dodson, G.G.; Hubbard, R.E.; Tolley, S.P.; Dauter, Z.; Wilson, K.S.; Hjort, C.; Mikelsen, J.M.; Rasmussen, G.; Schülein, M. Structure and function of endoglucanase V. Nature 1993, 365, 362–364. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic. Acids Res. 2014, 42, 490–495. [Google Scholar] [CrossRef]
- Aspeborg, H.; Coutinho, P.M.; Wang, Y.; Brumer, H.; Henrissat, B. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol. Biol. 2013, 12, 186. [Google Scholar] [CrossRef]
- Tseng, C.W.; Ko, T.P.; Guo, R.T.; Huang, J.W.; Wang, H.C.; Huang, C.H.; Cheng, Y.S.; Wang, A.H.J.; Liu, J.R. Substrate binding of a GH5 endoglucanase from the ruminal fungus Piromyces rhizinflata. Acta Crystallogr. Sect. F 2011, 67, 1189–1194. [Google Scholar] [CrossRef]
- Leggio, L.L.; Larsen, S. The 1.62 Å structure of Thermoascus aurantiacus endoglucanase: Completing the structural picture of subfamilies in glycol-side hydrolase family 5. FEBS Lett. 2002, 523, 103–108. [Google Scholar] [CrossRef]
- Lee, T.M.; Farrow, M.F.; Arnold, F.H.; Mayo, S.L. A structural study of Hypocrea jecorina Cel5A. Protein. Sci. 2011, 20, 1935–1940. [Google Scholar] [CrossRef]
- Liu, G.; Li, Q.; Shang, N.; Huang, J.W.; Ko, T.P.; Liu, W.; Zheng, Y.; Han, X.; Chen, Y.; Chen, C.C.; et al. Functional and structural analyses of a 1,4-β-endoglucanase from Ganoderma lucidum. Enzyme. Microb. Technol. 2016, 86, 67–74. [Google Scholar] [CrossRef]
- Yan, J.; Liu, W.; Li, Y.; Lai, H.L.; Zheng, Y.; Huang, J.W.; Chen, C.C.; Chen, Y.; Jin, J.; Li, H.; et al. Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-β-endoglucanase. Biochem. Biophys. Res. Commun. 2016, 475, 8–12. [Google Scholar] [CrossRef]
- Wierenga, R. The TIM-barrel fold: A versatile framework for efficient enzymes. FEBS Lett. 2001, 492, 193–198. [Google Scholar] [CrossRef] [PubMed]
- You, S.; Tu, T.; Zhang, L.J.; Wang, Y.; Huang, H.Q.; Ma, R.; Shi, P.J.; Bai, Y.G.; Su, X.Y.; Lin, Z.M.; et al. Improvement of the thermostability and catalytic efficiency of a highly active β-glucanase from Talaromyces leycettanus JCM 12802 by optimizing residual charge-charge interactions. Biotechnol Biofuels. 2016, 9, 124. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.B. Cellulases and biofuels. Curr. Opin. Biotechnol. 2009, 20, 295–299. [Google Scholar] [CrossRef]
- Celestino, K.R.S.; Cunha, R.B.; Felix, C.R. Characterization of a β-glucanase produced by Rhizopus microsporus var. microsporus, and its potential for application in the brewing industry. BMC Biochem. 2006, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Liu, Y.; Liu, M.; Wang, S.; Wang, Q. Improving the thermostability of a thermostable endoglucanase from Chaetomium thermophilum by engineering the conserved non-catalytic residue and N-glycosylation site. Int. J. Biol. Macromol. 2020, 164, 3361–3368. [Google Scholar] [CrossRef] [PubMed]
- Juturu, V.; Wu, J.C. Microbial cellulases: Engineering, production and applications. Renew. Sustain. Energy. Rev. 2014, 33, 188–203. [Google Scholar] [CrossRef]
- Martínez, A.C.; Balcázar, L.E.; Dantán, G.E.; Folch, M.J.L. Celulasas fúngicas: Aspectos biológicos y aplicaciones en la industria energética. Rev. Latinoam. Microbiol. 2008, 50, 119–131. [Google Scholar]
- Zheng, F.; Vermaas, V.J.; Zheng, J.; Wang, Y.; Tu, T.; Wang, X.; Xie, X.; Yao, B.; Beckham, G.T.; Luo, H. Activity and thermostability of GH5 endoglucanase chimeras from mesophilic and thermophilic parents. Appl. Environ. Microbiol. 2019, 85, e02079-18. [Google Scholar] [CrossRef]
- Ferrè, F.; Clote, P. DiANNA: A web server for disulfide connectivity prediction. Nucleic Acids Res. 2005, 33, W230–W232. [Google Scholar] [CrossRef]
- Pace, C.N.; Fu, H.; Fryar, K.L.; Landua, J.; Trevino, S.R.; Shirley, B.A.; Hendricks, M.M.; Limura, S.; Gajiwala, K.; Scholtz, J.M.; et al. Contribution of hydrophobic interactions to protein stability. J. Mol. Biol. 2011, 408, 514–528. [Google Scholar] [CrossRef]
- Tong, L.; Zheng, J.; Wang, X.; Wang, X.; Huang, H.; Yang, H.; Tu, T.; Wang, Y.; Bai, Y.; Luo, H.; et al. Improvement of thermostability and catalytic efciency of glucoamylase from Talaromyces leycettanus JCM12802 via site-directed mutagenesis to enhance industrial saccharifcation applications. Biotechnol. Biofuels 2021, 14, 202. [Google Scholar] [CrossRef]
- Gao, L.; Guo, Q.; Xu, R.; Dong, H.; Zhou, C.; Yu, Z.; Zhang, Z.; Wang, L.; Chen, X.; Wu, X. Loop optimization of Trichoderma reesei endoglucanases for balancing the activity–stability trade-off through cross-strategy between machine learning and the B-factor analysis. GCB Bioenergy 2022, 15, 128–142. [Google Scholar] [CrossRef]
- Li, F.; Xie, Y.; Gao, X.; Shan, M.; Sun, C.; Niu, Y.D.; Shan, A. Screening of cellulose degradation bacteria from Min pigs and optimization of its cellulase production. Electron. J. Biotechn. 2020, 48, 29–35. [Google Scholar] [CrossRef]
- Gomi, K.; Iimura, Y.; Hara, S. Integrative transformation of Aspergillus oryzaewith a plasmid containing the Aspergillus nidulans argB gene. Agr. Biol. Chem. 1987, 51, 2549–2555. [Google Scholar]
- Karplus, P.A.; Schulz, G.E. Prediction of chain flexibility in proteins. Naturwissenschaften 1985, 72, 212–213. [Google Scholar] [CrossRef]
- You, S.; Chen, C.C.; Tu, T.; Wang, X.; Ma, R.; Cai, H.Y.; Guo, R.T.; Luo, H.Y.; Yao, B. Insight into the functional roles of Glu175 in the hyperthermostable xylanase XYL10C-ΔN through structural analysis and site-saturation mutagenesis. Biotechnol. Biofuels 2018, 11, 159. [Google Scholar] [CrossRef]
- Guo, Y.; Tu, T.; Zheng, J.; Bai, Y.; Huang, H.; Su, X.; Wang, Y.; Wang, Y.; Yao, B.; Luo, H. Improvement of BsAPA Aspartic Protease Thermostability via Autocatalysis-Resistant Mutation. J. Agric. Food Chem. 2019, 67, 10505–10512. [Google Scholar] [CrossRef]
- Parthasarathy, S.; Murthy, M.R.N. Protein thermal stability: Insights from atomic displacement parameters (B values). Protein. Eng. 2000, 13, 9–13. [Google Scholar] [CrossRef]
- Reetz, M.T.; Carballeira, J.D.; Vogel, A. Iterative saturation mutagenesis on the basis of B factors as a strategy for increasing protein thermostability. Angew. Chem. Int. Ed. 2006, 45, 7745–7751. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, Y.; Song, X.; Hong, J.; Zhang, Y.; Yao, L. Improving Trichoderma reesei Cel7B thermostability by targeting the weak spots. J. Chem. Inf. Model. 2014, 54, 2826–2833. [Google Scholar] [CrossRef]
- Li, F.; Liu, Q.; Li, X.; Zhang, C.; Li, J.; Sun, W.; Liu, D.; Xiao, D.; Tian, C. Construction of a new thermophilic fungus Myceliophthora thermophila platform for enzyme production using a versatile 2A peptide strategy combined with efficient CRISPR-Cas9 system. Biotechnol. Lett. 2020, 42, 1181–1191. [Google Scholar] [CrossRef]
- Kurahashi, R.; Tanaka, S.; Takano, K. Activity-stability trade-off in random mutant proteins. J. Biosci. Bioeng. 2019, 128, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, K.S. Defying the activity–stability trade-off in enzymes: Taking advantage of entropy to enhance activity and thermostability. Crit. Rev. Biotechnol. 2017, 37, 309–322. [Google Scholar] [CrossRef]
- Li, G.; Zhou, X.; Li, Z.; Liu, Y.; Liu, D.; Miao, Y.; Wan, Q.; Zhang, R. Significantly improving the thermostability of a hyperthermophilic GH10 family xylanase XynAF1 by semi-rational design. Appl. Microbiol. Biotechnol. 2021, 105, 4561–4576. [Google Scholar] [CrossRef]
| Strains | ArCel5A | ArCel5A-Y299C |
|---|---|---|
| Conditions | Residual activity | |
| 30 min at 70 °C | 76.9 ± 1.1% | 93.8 ± 1.4% |
| 10 min at 80 °C | 71.2 ± 0.92% | 91.5 ± 1.2% |
| Y299A | Y299H | Y299D | Y299R | Y299F | |
| ΔΔG | −2.83 | −0.8 | −0.75 | −0.56 | −0.6 |
| Y299C | Y299G | Y299E | Y299K | Y299L | |
| ΔΔG | 0.34 | −3.47 | −0.83 | −1.2 | −0.92 |
| Y299M | Y299Q | Y299S | Y299Y | Y299T | |
| ΔΔG | −0.56 | −0.99 | −1.68 | 0 | −0.64 |
| Y299I | Y299W | Y299P | Y299V | ||
| ΔΔG | −1.12 | −0.63 | −2.55 | −0.73 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Zhang, Z.; Li, Y.; Yuan, J.; Dong, H.; Bao, T.; Wu, X.; Gu, L.; Zhang, J.; Gao, L. A Novel Strategy for Further Enhancing Superior Properties of Thermophilic Endoglucanase from Acidomyces richmondensis. Fermentation 2024, 10, 27. https://doi.org/10.3390/fermentation10010027
Wang S, Zhang Z, Li Y, Yuan J, Dong H, Bao T, Wu X, Gu L, Zhang J, Gao L. A Novel Strategy for Further Enhancing Superior Properties of Thermophilic Endoglucanase from Acidomyces richmondensis. Fermentation. 2024; 10(1):27. https://doi.org/10.3390/fermentation10010027
Chicago/Turabian StyleWang, Shengjie, Zherui Zhang, Yi Li, Jie Yuan, Haofan Dong, Tongtong Bao, Xin Wu, Lingfang Gu, Jian Zhang, and Le Gao. 2024. "A Novel Strategy for Further Enhancing Superior Properties of Thermophilic Endoglucanase from Acidomyces richmondensis" Fermentation 10, no. 1: 27. https://doi.org/10.3390/fermentation10010027
APA StyleWang, S., Zhang, Z., Li, Y., Yuan, J., Dong, H., Bao, T., Wu, X., Gu, L., Zhang, J., & Gao, L. (2024). A Novel Strategy for Further Enhancing Superior Properties of Thermophilic Endoglucanase from Acidomyces richmondensis. Fermentation, 10(1), 27. https://doi.org/10.3390/fermentation10010027







