vtRNA2-1/nc886 Produces a Small RNA That Contributes to Its Tumor Suppression Action through the microRNA Pathway in Prostate Cancer
Abstract
1. Introduction
2. Results
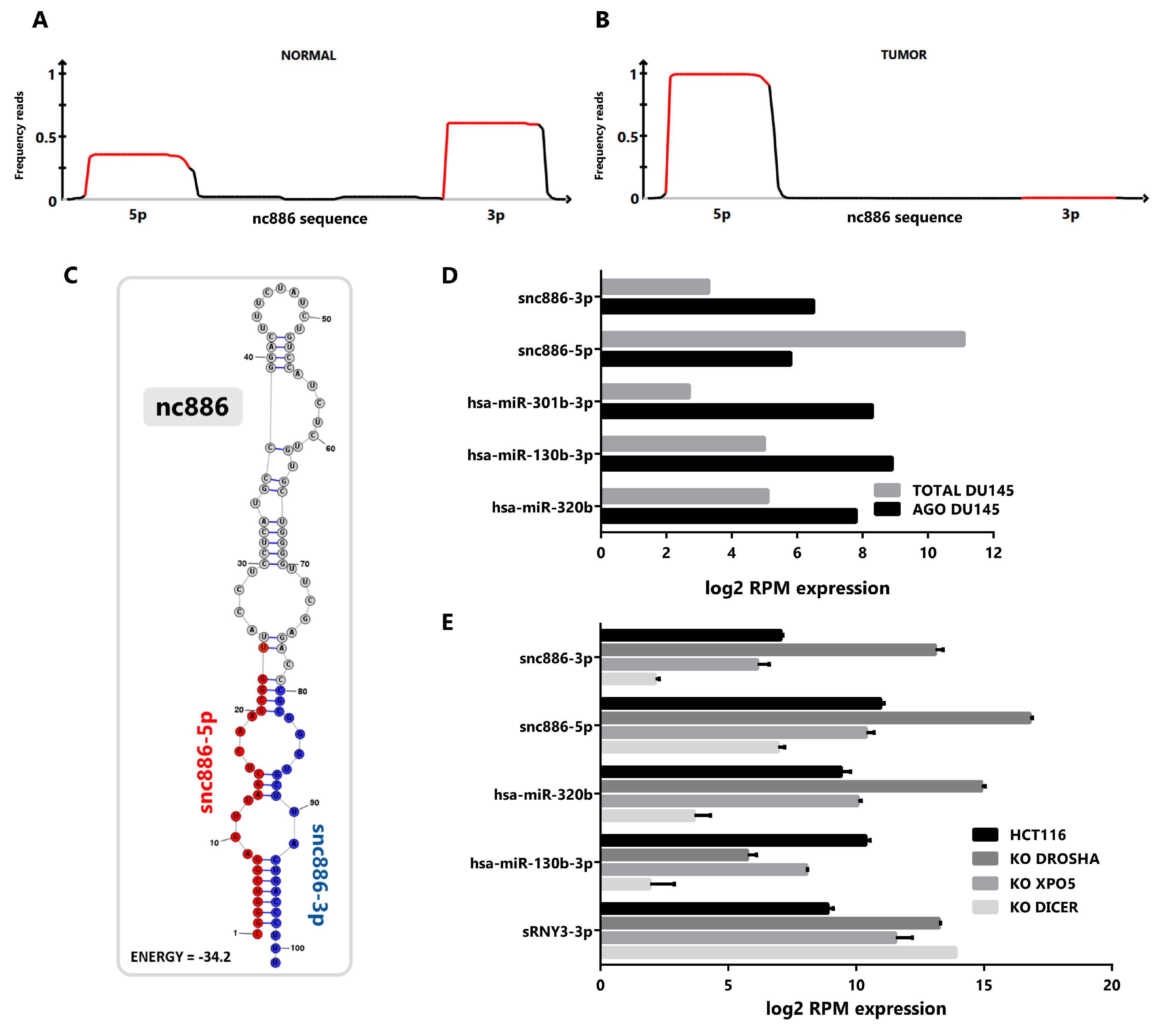
2.1. vtRNA2-1/nc886 Produces Small RNAs with microRNA-Like Features
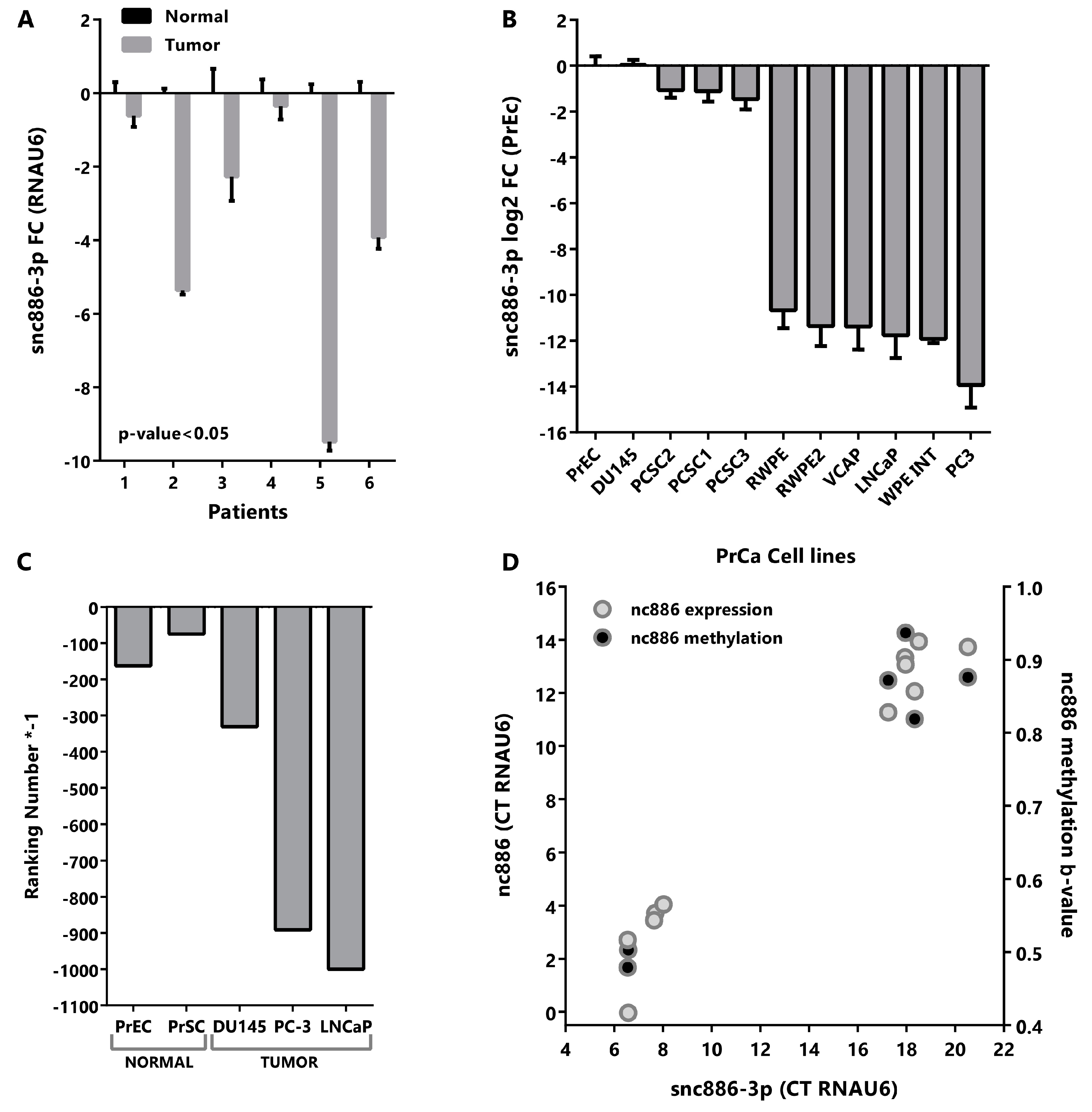
2.2. Snc886-3p Has the Expression Profile of a Tumor Suppressor in Prostate Cells
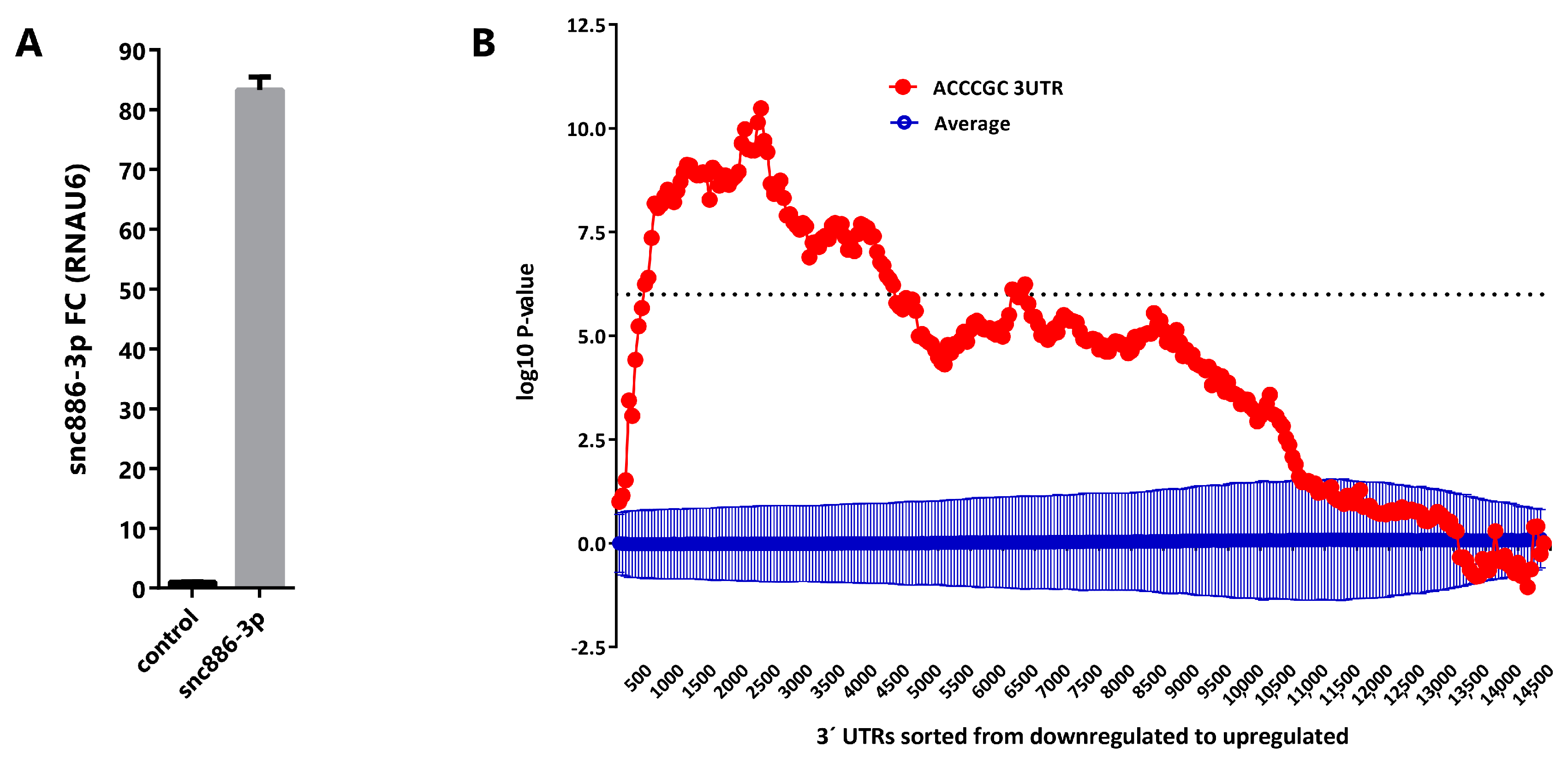
2.3. Snc886-3p Modulates Transcripts Affecting Cell Cycle and Apoptosis
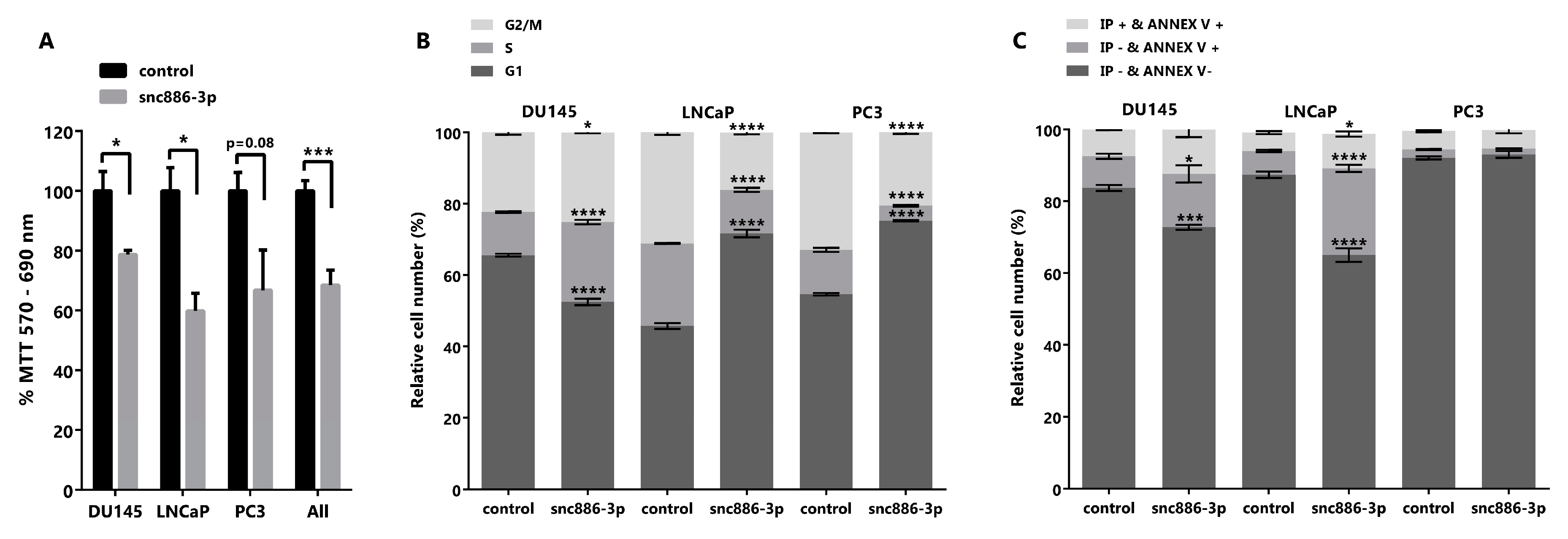
2.4. Snc886-3p Causes a Decrease in Cell Viability
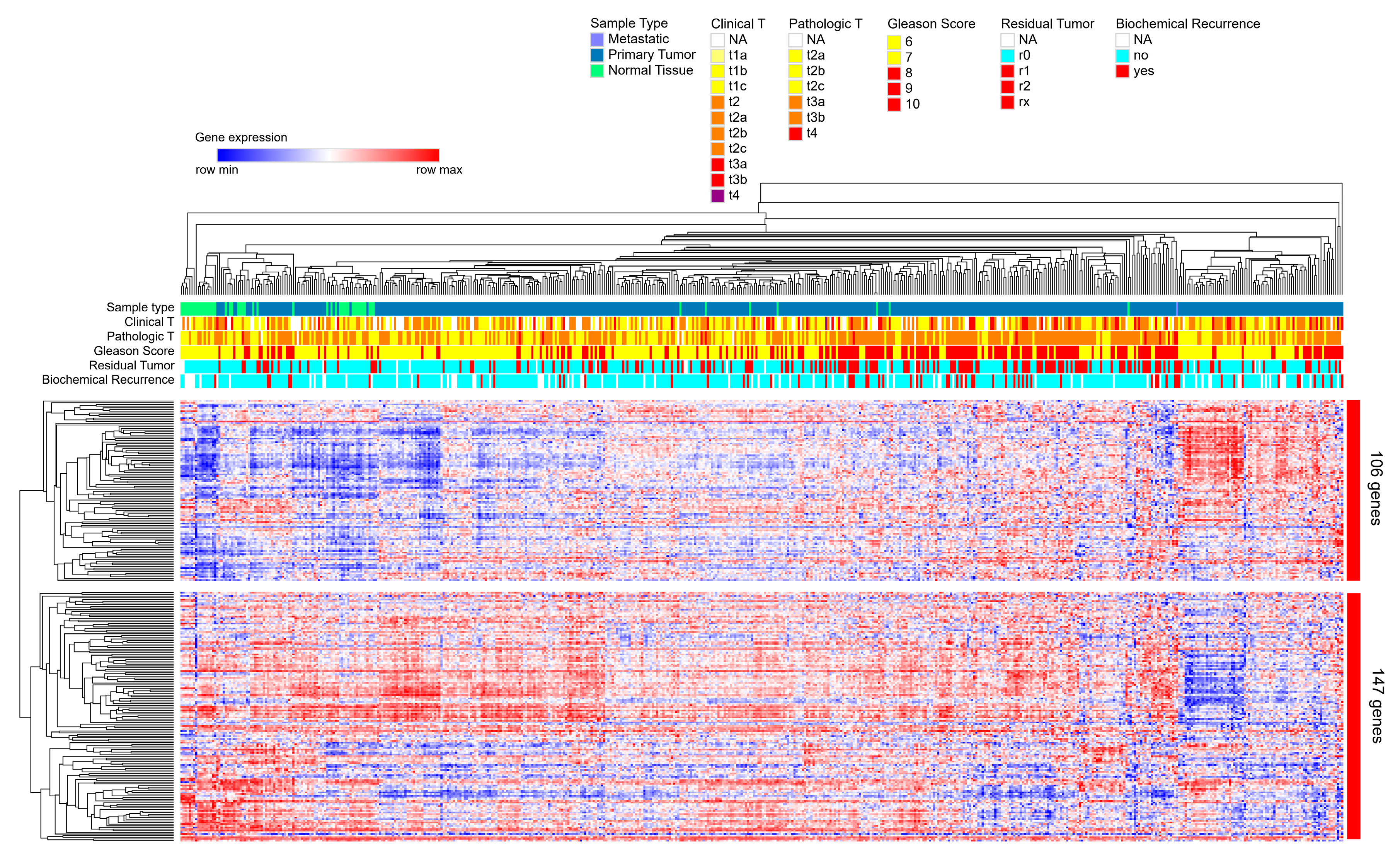
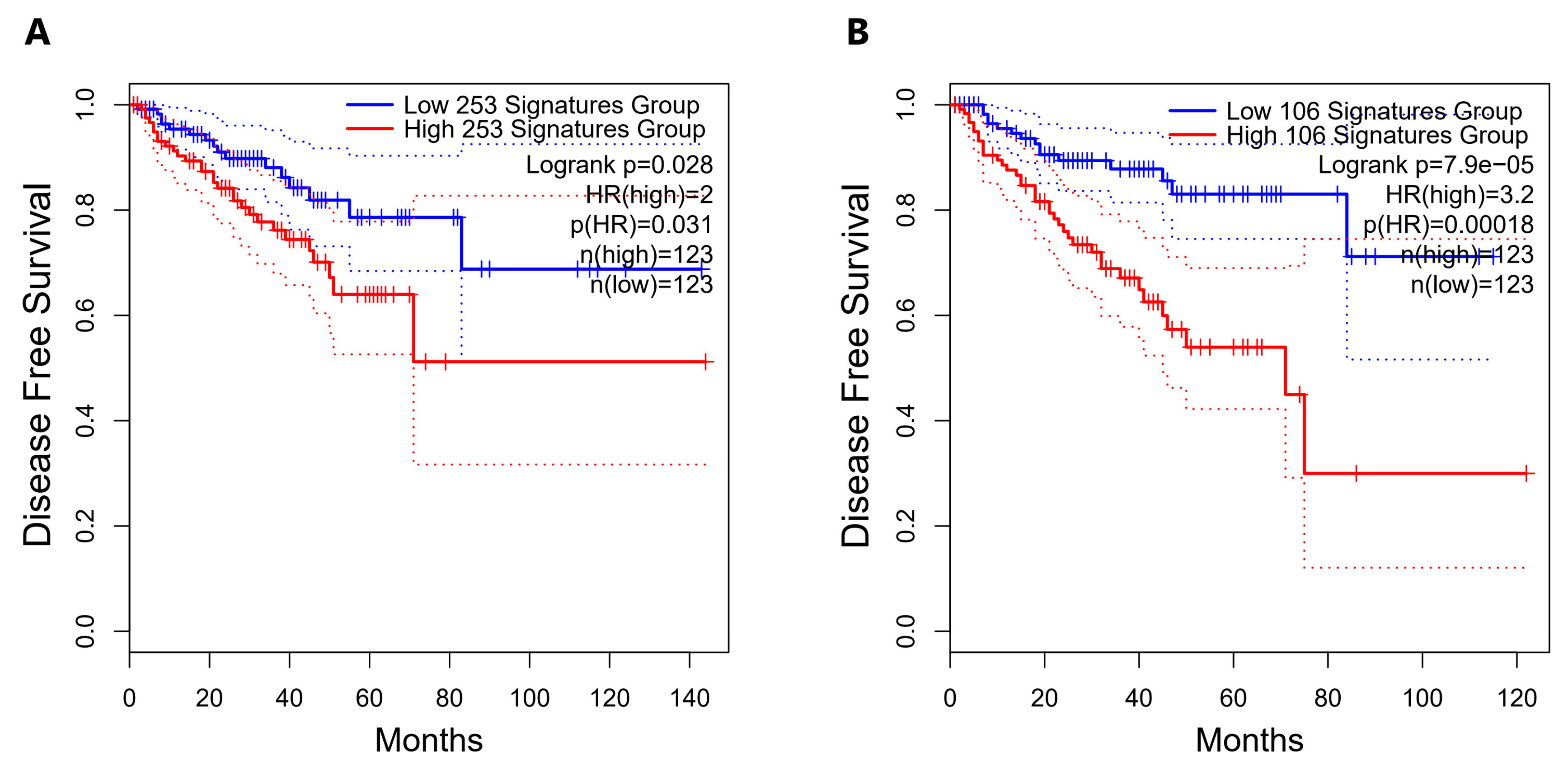
2.5. Direct Candidate Target Genes of snc886-3p Are Associated with Clinical Worse Prognosis in Prostate Cancer Patients
3. Discussion
4. Materials and Methods
4.1. Human Specimens
4.2. Cell Lines
4.3. Cell Transfection
4.4. RNA Extraction Reverse Transcription and Quantitative Real Time PCR
4.5. Microarray Experiments
4.6. Cytotoxicity Assay
4.7. Flow Cytometry for DNA Content Analysis
4.8. Annexin V Alexa Fluor 488/PI Apoptosis Detection Assay
4.9. Dataset Analysis
4.9.1. Analysis of microRNA Microarray Datasets
4.9.2. Analysis of Small RNA Transcriptomic Datasets
4.9.3. Analysis of Methylation Microarray Datasets
4.9.4. Heatmap of Hierarchical Clusterization of 253 snc886-3p Candidate Direct Target Genes
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ncRNA | non-coding RNA |
| vtRNA | vault RNA |
| 3′-UTR | three prime untranslated region |
| PrCa | Prostate Cancer |
| PSA | Prostate-Specific Antigen |
| nc886 | non-coding RNA 886 (vtRNA2-1) |
| Pre-miR-886 | hsa-mir-886 precursor of microRNAs hsa-miR-886-3p and 5p |
| snc886s | small non-coding RNA derived from nc886 |
| snc886-3p | small non-coding RNA derived from nc886 at 3′ region |
| snc886-5p | small non-coding RNA derived from nc886 at 5′ region |
| svtRNA2-1 | small non-coding RNA derived from vtRNA2-1/nc886 |
| PAR-CLIP | photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation |
| PRAD-TCGA | Prostate Adenocarcinoma—The Cancer Genome Atlas |
| SRA | Sequence Read Archive |
| GEO | Gene Expression Omnibus |
| DEG | Differentially Expressed Gene |
| MFE | Maximum Free Energy |
| RISC | RNA-induced silencing complex |
| qRT-PCR | quantitative Reverse Transcription Polymerase Chain Reaction |
| TSS200nt | 200nt region upstream to the transcription start site |
| GSEA | Gene Set Enrichment Analysis |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide |
References
- Ceder, Y. Non-coding RNAs in prostate cancer: From discovery to clinical applications. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2016; Volume 886, pp. 155–170. [Google Scholar]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- Kohaar, I.; Petrovics, G.; Srivastava, S. A Rich Array of Prostate Cancer Molecular Biomarkers: Opportunities and Challenges. Int. J. Mol. Sci. 2019, 20, 1813. [Google Scholar] [CrossRef] [PubMed]
- Bijnsdorp, I.V.; Van Royen, M.E.; Verhaegh, G.W.; Martens-Uzunova, E.S. The Non-Coding Transcriptome of Prostate Cancer: Implications for Clinical Practice. Mol. Diagn. Ther. 2017, 21, 385–400. [Google Scholar] [CrossRef] [PubMed]
- Bolton, E.M.; Tuzova, A.V.; Walsh, A.L.; Lynch, T.; Perry, A.S. Noncoding RNAs in prostate cancer: The long and the short of it. Clin. Cancer Res. 2014, 20, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Mouraviev, V.; Lee, B.; Patel, V.; Albala, D.; Johansen, T.E.B.; Partin, A.; Ross, A.; Perera, R.J. Clinical prospects of long noncoding RNAs as novel biomarkers and therapeutic targets in prostate cancer. Prostate Cancer Prostatic Dis. 2016, 19, 14–20. [Google Scholar] [CrossRef]
- Griffiths-Jones, S. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A Mammalian microRNA Expression Atlas Based on Small RNA Library Sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef]
- Stadler, P.F.; Chen, J.J.-L.; Hackermüller, J.; Hoffmann, S.; Horn, F.; Khaitovich, P.; Kretzschmar, A.K.; Mosig, A.; Prohaska, S.J.; Qi, X.; et al. Evolution of Vault RNAs. Mol. Boil. Evol. 2009, 26, 1975–1991. [Google Scholar] [CrossRef]
- Kedersha, N.L.; Rome, L.H.; Krotoski, D.M.; Domingo, C.; Bronner-Fraser, M. Isolation and characterization of a novel ribonucleoprotein particle: Large structures contain a single species of small RNA. J. Cell Boil. 1986, 103, 699–709. [Google Scholar] [CrossRef]
- Van Zon, A.; Mossink, M.H.; Scheper, R.J.; Sonneveld, P.; Wiemer, E.A.C. The vault complex. Cell. Mol. Life Sci. 2003, 60, 1828–1837. [Google Scholar] [CrossRef]
- Lee, K.; Kunkeaw, N.; Jeon, S.H.; Lee, I.; Johnson, B.H.; Kang, G.-Y.; Bang, J.Y.; Park, H.S.; Leelayuwat, C.; Lee, Y.S. Precursor miR-886, a novel noncoding RNA repressed in cancer, associates with PKR and modulates its activity. RNA 2011, 17, 1076–1089. [Google Scholar] [CrossRef]
- Jeon, S.H.; Johnson, B.H.; Lee, Y.S. A Tumor Surveillance Model: A Non-Coding RNA Senses Neoplastic Cells and Its Protein Partner Signals Cell Death. Int. J. Mol. Sci. 2012, 13, 13134–13139. [Google Scholar] [CrossRef]
- Jeon, S.H.; Lee, K.; Lee, K.S.; Kunkeaw, N.; Johnson, B.H.; Holthauzen, L.M.F.; Gong, B.; Leelayuwat, C.; Lee, Y.S. Characterization of the direct physical interaction of nc886, a cellular non-coding RNA, and PKR. FEBS Lett. 2012, 586, 3477–3484. [Google Scholar] [CrossRef]
- Golec, E.; Lind, L.; Qayyum, M.; Blom, A.M.; King, B.C. The Noncoding RNA nc886 Regulates PKR Signaling and Cytokine Production in Human Cells. J. Immunol. 2019, 202, 131–141. [Google Scholar] [CrossRef]
- Fort, R.S.; Mathó, C.; Geraldo, M.V.; Ottati, M.C.; Yamashita, A.S.; Saito, K.C.; Leite, K.R.M.; Méndez, M.; Maedo, N.; Méndez, L.; et al. Nc886 is epigenetically repressed in prostate cancer and acts as a tumor suppressor through the inhibition of cell growth. BMC Cancer 2018, 18, 127. [Google Scholar] [CrossRef]
- Cloonan, N.; Wani, S.; Xu, Q.; Gu, J.; Lea, K.; Heater, S.; Barbacioru, C.; Steptoe, A.L.; Martin, H.C.; Nourbakhsh, E.; et al. MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Boil. 2011, 12, R126. [Google Scholar] [CrossRef]
- Treppendahl, M.B.; Qiu, X.; Søgaard, A.; Yang, X.; Nandrup-Bus, C.; Hother, C.; Andersen, M.K.; Kjeldsen, L.; Möllgaard, L.; Hellström-Lindberg, E.; et al. Allelic methylation levels of the noncoding VTRNA2-1 located on chromosome 5q31.1 predict outcome in AML. Blood 2012, 119, 206–216. [Google Scholar] [CrossRef]
- Miñones-Moyano, E.; Friedländer, M.R.; Pallares, J.; Kagerbauer, B.; Porta, S.; Escaramís, G.; Ferrer, I.; Estivill, X.; Marti, E. Upregulation of a small vault RNA (svtRNA2-1a) is an early event in Parkinson disease and induces neuronal dysfunction. RNA Boil. 2013, 10, 1093–1106. [Google Scholar] [CrossRef]
- Jung, K.; Fendler, A.; Stephan, C.; Honey, R.J.; Stewart, R.J.; Pace, K.T.; Erbersdobler, A.; Samaan, S.; Yousef, G.M. miRNAs can predict prostate cancer biochemical relapse and are involved in tumor progression. Int. J. Oncol. 2011, 39, 1183–1192. [Google Scholar] [CrossRef]
- Nordentoft, I.K.; Birkenkamp-Demtroder, K.; Agerbæk, M.; Theodorescu, D.; Ostenfeld, M.S.; Hartmann, A.; Borre, M.; Ørntoft, T.F.; Dyrskjøt, L. miRNAs associated with chemo-sensitivity in cell lines and in advanced bladder cancer. BMC Med. Genom. 2012, 5, 40. [Google Scholar] [CrossRef]
- Tahiri, A.; Leivonen, S.K.; Lüders, T.; Steinfeld, I.; Aure, M.R.; Geisler, J.; Mäkelä, R.; Nord, S.; Riis, M.L.H.; Yakhini, Z.; et al. Deregulation of cancer-related miRNAs is a common event in both benign and malignant human breast tumors. Carcinogenesis 2014, 35, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.-F.; Zou, J.; Bao, Z.-J.; Dong, J. miR-93 suppresses proliferation and colony formation of human colon cancer stem cells. World J. Gastroenterol. 2011, 17, 4711–4717. [Google Scholar] [CrossRef]
- Gao, W.; Shen, H.; Liu, L.; Xu, J.; Xu, J.; Shu, Y. MiR-21 overexpression in human primary squamous cell lung carcinoma is associated with poor patient prognosis. J. Cancer Res. Clin. Oncol. 2011, 137, 557–566. [Google Scholar] [CrossRef]
- Cao, J.; Song, Y.; Bi, N.; Shen, J.; Liu, W.; Fan, J.; Sun, G.; Tong, T.; He, J.; Shi, Y.; et al. DNA Methylation-Mediated Repression of miR-886-3p Predicts Poor Outcome of Human Small Cell Lung Cancer. Cancer Res. 2013, 73, 3326–3335. [Google Scholar] [CrossRef]
- Bi, N.; Cao, J.; Song, Y.; Shen, J.; Liu, W.; Fan, J.; He, J.; Shi, Y.; Zhang, X.; Lu, N.; et al. A MicroRNA Signature Predicts Survival in Early Stage Small-Cell Lung Cancer Treated with Surgery and Adjuvant Chemotherapy. PLoS ONE 2014, 9, e91388. [Google Scholar] [CrossRef]
- Shen, J.; Zhou, W.; Bi, N.; Song, Y.-M.; Zhang, F.-Q.; Zhan, Q.-M.; Wang, L.-H. MicroRNA-886-3P functions as a tumor suppressor in small cell lung cancer. Cancer Boil. Ther. 2018, 19, 1185–1192. [Google Scholar] [CrossRef]
- Xiong, Y.; Zhang, L.; Holloway, A.K.; Wu, X.; Su, L.; Kebebew, E. MiR-886-3p Regulates Cell Proliferation and Migration, and Is Dysregulated in Familial Non-Medullary Thyroid Cancer. PLoS ONE 2011, 6, e24717. [Google Scholar] [CrossRef]
- Dettmer, M.S.; Perren, A.; Moch, H.; Komminoth, P.; Nikiforov, Y.E.; Nikiforova, M.N. MicroRNA profile of poorly differentiated thyroid carcinomas: New diagnostic and prognostic insights. J. Mol. Endocrinol. 2014, 52, 181–189. [Google Scholar] [CrossRef]
- Pillai, M.M.; Yang, X.; Balakrishnan, I.; Bemis, L.; Torok-Storb, B. MiR-886-3p Down Regulates CXCL12 (SDF1) Expression in Human Marrow Stromal Cells. PLoS ONE 2010, 5, e14304. [Google Scholar] [CrossRef]
- Mahishi, L.H.; Hart, R.P.; Lynch, D.R.; Ratan, R.R. miR-886-3p levels are elevated in Friedreich ataxia. J. Neurosci. 2012, 32, 9369–9373. [Google Scholar] [CrossRef]
- McDonald, A.C.; Vira, M.; Walter, V.; Shen, J.; Raman, J.D.; Sanda, M.G.; Patil, D.; Taioli, E. Circulating microRNAs in plasma among men with low-grade and high-grade prostate cancer at prostate biopsy. Prostate 2019, 79, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Chen, D.; Su, Z.; Li, Y.; Yu, W.; Zhang, Q.; Yang, L.; Li, C.; Yang, S.; Ni, L.; et al. miR-886-3p upregulation in clear cell renal cell carcinoma regulates cell migration, proliferation and apoptosis by targeting PITX1. Int. J. Mol. Med. 2014, 34, 1409–1416. [Google Scholar] [CrossRef] [PubMed]
- Schou, J.V.; Rossi, S.; Jensen, B.V.; Nielsen, D.L.; Pfeiffer, P.; Høgdall, E.; Yilmaz, M.; Tejpar, S.; Delorenzi, M.; Kruhøffer, M.; et al. miR-345 in Metastatic Colorectal Cancer: A Non-Invasive Biomarker for Clinical Outcome in Non-KRAS Mutant Patients Treated with 3rd Line Cetuximab and Irinotecan. PLoS ONE 2014, 9, e99886. [Google Scholar] [CrossRef]
- Okumura, T.; Kojima, H.; Miwa, T.; Sekine, S.; Hashimoto, I.; Hojo, S.; Nagata, T.; Shimada, Y. The expression of microRNA 574-3p as a predictor of postoperative outcome in patients with esophageal squamous cell carcinoma. World J. Surg. Oncol. 2016, 14, 228. [Google Scholar] [CrossRef]
- Kunkeaw, N.; Lee, Y.-S.; Im, W.R.; Jang, J.J.; Song, M.-J.; Yang, B.; Park, J.-L.; Kim, S.-Y.; Ku, Y.; Kim, Y.; et al. Mechanism mediated by a noncoding RNA, nc886, in the cytotoxicity of a DNA-reactive compound. Proc. Natl. Acad. Sci. USA 2019, 116, 8289–8294. [Google Scholar] [CrossRef]
- Wyman, S.K.; Knouf, E.C.; Parkin, R.K.; Fritz, B.R.; Lin, D.W.; Dennis, L.M.; Krouse, M.A.; Webster, P.J.; Tewari, M. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res. 2011, 21, 1450–1461. [Google Scholar] [CrossRef]
- Marshall, E.A.; Sage, A.P.; Ng, K.W.; Martinez, V.D.; Firmino, N.S.; Bennewith, K.L.; Lam, W.L. Small non-coding RNA transcriptome of the NCI-60 cell line panel. Sci. Data 2017, 4, 170157. [Google Scholar] [CrossRef]
- Takayama, K.-I.; Misawa, A.; Suzuki, T.; Takagi, K.; Hayashizaki, Y.; Fujimura, T.; Homma, Y.; Takahashi, S.; Urano, T.; Inoue, S. TET2 repression by androgen hormone regulates global hydroxymethylation status and prostate cancer progression. Nat. Commun. 2015, 6, 8219. [Google Scholar] [CrossRef]
- Kodama, Y.; Shumway, M.; Leinonen, R. International Nucleotide Sequence Database Collaboration The Sequence Read Archive: Explosive growth of sequencing data. Nucleic Acids Res. 2011, 40, D54–D56. [Google Scholar] [CrossRef]
- Clough, E.; Barrett, T. The Gene Expression Omnibus Database. In Breast Cancer; Springer Science and Business Media LLC: New York, NY, USA, 2016; Volume 1418, pp. 93–110. [Google Scholar]
- Berezikov, E.; Robine, N.; Samsonova, A.; Westholm, J.O.; Naqvi, A.; Hung, J.H.; Okamura, K.; Dai, Q.; Bortolamiol-Becet, D.; Martin, R.; et al. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 2011, 21, 203–215. [Google Scholar] [CrossRef]
- Park, J.-E.; Heo, I.; Tian, Y.; Simanshu, D.K.; Chang, H.; Jee, D.; Patel, D.J.; Kim, V.N. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 2011, 475, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Bellaousov, S.; Reuter, J.S.; Seetin, M.G.; Mathews, D.H. RNAstructure: Web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 2013, 41, W471–W474. [Google Scholar] [CrossRef] [PubMed]
- Calderon, B.M.; Conn, G.L. Human noncoding RNA 886 (nc886) adopts two structurally distinct conformers that are functionally opposing regulators of PKR. RNA 2017, 23, 557–566. [Google Scholar] [PubMed]
- Calderon, B.M.; Conn, G.L. A human cellular noncoding RNA activates the antiviral protein 2′–5′-oligoadenylate synthetase 1. J. Boil. Chem. 2018, 293, 16115–16124. [Google Scholar] [CrossRef]
- Starega-Roslan, J.; Koscianska, E.; Kozlowski, P.; Krzyzosiak, W.J. The role of the precursor structure in the biogenesis of microRNA. Cell. Mol. Life Sci. 2011, 68, 2859–2871. [Google Scholar] [CrossRef] [PubMed]
- Finnegan, E.F.; Pasquinelli, A.E. MicroRNA biogenesis: Regulating the regulators. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 51–68. [Google Scholar] [CrossRef]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Boil. 2009, 10, 126–139. [Google Scholar] [CrossRef]
- Hamilton, M.P.; Rajapakshe, K.I.; Bader, D.A.; Cerne, J.Z.; Smith, E.A.; Coarfa, C.; Hartig, S.M.; McGuire, S.E. The Landscape of microRNA Targeting in Prostate Cancer Defined by AGO-PAR-CLIP. Neoplasia 2016, 18, 356–370. [Google Scholar] [CrossRef]
- Kim, Y.-K.; Kim, B.; Kim, V.N. Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, E1881–E1889. [Google Scholar] [CrossRef]
- Chen, C.-J.; Heard, E. Small RNAs derived from structural non-coding RNAs. Methods 2013, 63, 76–84. [Google Scholar] [CrossRef]
- Van Dongen, S.; Abreu-Goodger, C.; Enright, A.J. Detecting microRNA binding and siRNA off-target effects from expression data. Nat. Methods 2008, 5, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Mahadevappa, R.; Neves, H.; Yuen, S.M.; Bai, Y.; McCrudden, C.M.; Yuen, H.F.; Wen, Q.; Zhang, S.D.; Kwok, H.F. The prognostic significance of Cdc6 and Cdt1 in breast cancer. Sci. Rep. 2017, 7, 985. [Google Scholar] [CrossRef]
- Li, C.; Zhou, X.; Wang, Y.; Jing, S.; Yang, C.; Sun, G.; Liu, Q.; Cheng, Y.; Wang, L. miR-210 regulates esophageal cancer cell proliferation by inducing G2/M phase cell cycle arrest through targeting PLK1. Mol. Med. Rep. 2014, 10, 2099–2104. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Shaoqing, Y.; Ruxin, Z.; Guojun, L.; Zhiqiang, Y.; Hua, H.; Shudong, Y.; Jie, Z. Microarray Analysis of Differentially Expressed microRNAs in Allergic Rhinitis. Am. J. Rhinol. Allergy 2011, 25, e242–e246. [Google Scholar] [CrossRef] [PubMed]
- Amort, M.; Nachbauer, B.; Tuzlak, S.; Kieser, A.; Schepers, A.; Villunger, A.; Polacek, N. Expression of the vault RNA protects cells from undergoing apoptosis. Nat. Commun. 2015, 6, 7030. [Google Scholar] [CrossRef]
- Persson, H.; Kvist, A.; Vallon-Christersson, J.; Medstrand, P.; Borg, A.; Rovira, C. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nature 2009, 11, 1268–1271. [Google Scholar] [CrossRef]
- Hussain, S.; Sajini, A.A.; Blanco, S.; Dietmann, S.; Lombard, P.; Sugimoto, Y.; Paramor, M.; Gleeson, J.G.; Odom, D.T.; Ule, J.; et al. NSun2-Mediated Cytosine-5 Methylation of Vault Noncoding RNA Determines Its Processing into Regulatory Small RNAs. Cell Rep. 2013, 4, 255–261. [Google Scholar] [CrossRef]
- Vermeulen, A.; Behlen, L.; Reynolds, A.; Wolfson, A.; Marshall, W.S.; Karpilow, J.; Khvorova, A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA 2005, 11, 674–682. [Google Scholar] [CrossRef]
- Starega-Roslan, J.; Krol, J.; Koscianska, E.; Kozlowski, P.; Szlachcic, W.J.; Sobczak, K.; Krzyzosiak, W.J. Structural basis of microRNA length variety. Nucleic Acids Res. 2011, 39, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Du, R.; Edwards, A.; Flemington, E.K.; Zhang, K. The Sequence Structures of Human MicroRNA Molecules and Their Implications. PLoS ONE 2013, 8, e54215. [Google Scholar] [CrossRef] [PubMed]
- Berezikov, E. Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 2011, 12, 846–860. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Hao, Q.; Wang, Y.; Zhou, P.; Zou, B.; Zhang, Y.-X. Regulation of p53 expression and apoptosis by vault RNA2-1-5p in cervical cancer cells. Oncotarget 2015, 6, 28371–28388. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-H.; Xiao, X.; Zhang, Y.-N.; Wang, Y.-M.; Feng, L.-M.; Wu, Y.-M.; Zhang, Y.-X. MicroRNA miR-886-5p inhibits apoptosis by down-regulating Bax expression in human cervical carcinoma cells. Gynecol. Oncol. 2011, 120, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-L.; Wu, J.; Liu, Q.; Zhang, Y.; Sun, Z.-L.; Jing, H. MiR-886-5p inhibition inhibits growth and induces apoptosis of MCF7 cells. Asian Pac. J. Cancer Prev. 2014, 15, 1511–1515. [Google Scholar] [CrossRef]
- Gao, Q.Q.; Putzbach, W.E.; Murmann, A.E.; Chen, S.; Sarshad, A.A.; Peter, J.M.; Bartom, E.T.; Hafner, M.; Peter, M.E. 6mer seed toxicity in tumor suppressive microRNAs. Nat. Commun. 2018, 9, 4504. [Google Scholar] [CrossRef]
- Aakula, A.; Kohonen, P.; Leivonen, S.K.; Mäkelä, R.; Hintsanen, P.; Mpindi, J.P.; Martens-Uzunova, E.; Aittokallio, T.; Jenster, G.; Perälä, M.; et al. Systematic Identification of MicroRNAs That Impact on Proliferation of Prostate Cancer Cells and Display Changed Expression in Tumor Tissue. Eur. Urol. 2015, 69, 1120–1128. [Google Scholar] [CrossRef]
- Lee, K.-S.; Park, J.-L.; Lee, K.; Richardson, L.E.; Johnson, B.H.; Lee, H.-S.; Lee, J.-S.; Kim, S.-B.; Kwon, O.-H.; Song, K.S.; et al. nc886, a non-coding RNA of anti-proliferative role, is suppressed by CpG DNA methylation in human gastric cancer. Oncotarget 2014, 5, 3944–3955. [Google Scholar] [CrossRef]
- Lee, H.-S.; Lee, K.; Jang, H.-J.; Lee, G.K.; Park, J.-L.; Kim, S.-Y.; Kim, S.-B.; Johnson, B.H.; Zo, J.I.; Lee, J.-S.; et al. Epigenetic silencing of the non-coding RNA nc886 provokes oncogenes during human esophageal tumorigenesis. Oncotarget 2014, 5, 3472–3481. [Google Scholar] [CrossRef]
- Kunkeaw, N.; Jeon, S.H.; Lee, K.; Johnson, B.H.; Tanasanvimon, S.; Javle, M.; Pairojkul, C.; Chamgramol, Y.; Wongfieng, W.; Gong, B.; et al. Cell death/proliferation roles for nc886, a non-coding RNA, in the protein kinase R pathway in cholangiocarcinoma. Oncogene 2012, 32, 3722–3731. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.-H.; Lee, H.-S.; Lee, J.-S.; Lee, Y.-S.; Park, J.-L.; Kim, S.-Y.; Hwang, J.-A.; Kunkeaw, N.; Jung, S.Y.; Kim, T.J.; et al. nc886 is induced by TGF-β and suppresses the microRNA pathway in ovarian cancer. Nat. Commun. 2018, 9, 1166. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.K.; Hong, S.-H.; Shin, S.; Lee, H.-S.; Lee, J.-S.; Park, E.J.; Choi, S.S.; Min, J.W.; Park, D.; Hwang, J.-A.; et al. nc886, a non-coding RNA and suppressor of PKR, exerts an oncogenic function in thyroid cancer. Oncotarget 2016, 7, 75000–75012. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hu, Z.; Zhang, H.; Tang, L.; Lou, M.; Geng, Y. Silencing nc886, a Non-Coding RNA, Induces Apoptosis of Human Endometrial Cancer Cells-1A In Vitro. Med. Sci. Monit. 2017, 23, 1317–1324. [Google Scholar] [CrossRef][Green Version]
- Xi, X.; Li, T.; Huang, Y.; Sun, J.; Zhu, Y.; Yang, Y.; Lu, Z.J. RNA Biomarkers: Frontier of Precision Medicine for Cancer. Non-Coding RNA 2017, 3, 9. [Google Scholar] [CrossRef]
- Xiang, P.; Liu, Y.; Liu, L.; Lin, Q.; Liu, X.; Zhang, H.; Xu, J.; Fang, B. The Biological Function and Clinical Significance of miR-886-5p in Multiple Myeloma. Acta Haematol. 2019, 142, 208–216. [Google Scholar] [CrossRef]
- Perfetti, A.; Greco, S.; Bugiardini, E.; Cardani, R.; Gaia, P.; Gaetano, C.; Meola, G.; Martelli, F. Plasma microRNAs as biomarkers for myotonic dystrophy type 1. Neuromuscul. Disord. 2014, 24, 509–515. [Google Scholar] [CrossRef][Green Version]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef]
- Tosar, J.P.; Gámbaro, F.; Sanguinetti, J.; Bonilla, B.; Witwer, K.W.; Cayota, A. Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucleic Acids Res. 2015, 43, 5601–5616. [Google Scholar] [CrossRef]
- Van Balkom, B.W.M.; Eisele, A.S.; Pegtel, D.M.; Bervoets, S.; Verhaar, M.C. Quantitative and qualitative analysis of small RNAs in human endothelial cells and exosomes provides insights into localized RNA processing, degradation and sorting. J. Extracell. Vesicles 2015, 4, 26760. [Google Scholar] [CrossRef]
- Nolte’T Hoen, E.N.M.; Buermans, H.P.J.; Waasdorp, M.; Stoorvogel, W.; Wauben, M.H.M.; Hoen, P.A.C. ’T Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012, 40, 9272–9285. [Google Scholar] [CrossRef] [PubMed]
- Lunavat, T.R.; Cheng, L.; Kim, D.-K.; Bhadury, J.; Jang, S.C.; Lässer, C.; Sharples, R.A.; López, M.D.; Nilsson, J.; Gho, Y.S.; et al. Small RNA deep sequencing discriminates subsets of extracellular vesicles released by melanoma cells—Evidence of unique microRNA cargos. RNA Boil. 2015, 12, 810–823. [Google Scholar] [CrossRef] [PubMed]
- Li, C.C.Y.; Eaton, S.A.; Young, P.E.; Lee, M.; Shuttleworth, R.; Humphreys, D.T.; Grau, G.E.; Combes, V.; Bebawy, M.; Gong, J.; et al. Glioma microvesicles carry selectively packaged coding and non-coding RNAs which alter gene expression in recipient cells. RNA Boil. 2013, 10, 1333–1344. [Google Scholar] [CrossRef]
- Dugué, P.-A.; Dowty, J.G.; Joo, J.E.; Wong, E.M.; Makalic, E.; Schmidt, D.F.; English, D.R.; Hopper, J.L.; Pedersen, J.; Severi, G.; et al. Heritable methylation marks associated with breast and prostate cancer risk. Prostate 2018, 78, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kallio, M.A.; Tuimala, J.T.; Hupponen, T.; Klemelä, P.; Gentile, M.; Scheinin, I.; Koski, M.; Käki, J.; Korpelainen, E.I. Chipster: User-friendly analysis software for microarray and other high-throughput data. BMC Genom. 2011, 12, 507. [Google Scholar] [CrossRef]
- Irizarry, R.A.; Hobbs, B.; Collin, F.; Beazer-Barclay, Y.D.; Antonellis, K.J.; Scherf, U.; Speed, T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003, 4, 249–264. [Google Scholar] [CrossRef]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]
- Mayr, C.; Bartel, D.P. Widespread Shortening of 3′UTRs by Alternative Cleavage and Polyadenylation Activates Oncogenes in Cancer Cells. Cell 2009, 138, 673–684. [Google Scholar] [CrossRef]
- Benhamed, M.; Herbig, U.; Ye, T.; Dejean, A.; Bischof, O. Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nature 2012, 14, 266–275. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Iorio, F.; Knijnenburg, T.A.; Vis, D.J.; Bignell, G.R.; Menden, M.P.; Schubert, M.; Aben, N.; Gonçalves, E.; Barthorpe, S.; Lightfoot, H.; et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 2016, 166, 740–754. [Google Scholar] [CrossRef] [PubMed]
| GEO ACC * | PUBMED ID | Platform | Total Samples | Benign | Cancer | Fold Change (N/B vs T) | p-Value | Analytical Tool |
|---|---|---|---|---|---|---|---|---|
| GSE45604 | 24518785 | Affymetrix | 60 | 10 Normal | 50 Cancer | −1.4 | 0.05 | GEO2R |
| GSE26964 | 21647377 | Capitalbio | 13 | 6 Primary PrCa | 7 Metastasis Bone | −2.7 | N.S. | GEO2R |
| GSE23022 | 21400514 | Affymetrix | 40 | 20 Normal | 20 Cancer | −1.3 | 0.03 | GEO2R |
| GSE55323 | 24967583 | Agilent | 40 | 20 Non-Recurrent | 20 Recurrent | −1.2 | N.S. | GEO2R |
| GSE62610 | 25416653 | Taqman qPCR | 36 | 14 Non-Recurrent | 22 Recurrent | −1.7 | N.S. | GEO2R |
| GSE21036 | 20579941 | Agilent | 140 | 28 Normal | 112 Cancer | −2.0 | 0.001 | GEO2R |
| GSE36802 | 23233736 | Affymetrix | 42 | 21 Benign | 21 Cancer | −1.8 | 0.0002 | GEO2R |
| TCGA data | 26544944 | Small-RNA-Seq | 24 a | 12 Normal | 12 Cancer | −3.2 | 0.03 | miRDeep2 |
| Gene Set Name | Genes in Gene Set (K) | Genes in Overlap (k) | p-Value | FDR q-Value |
|---|---|---|---|---|
| KEGG_NEUROTROPHIN_SIGNALING_PATHWAY | 126 | 21 | 1.26 × 10−9 | 2.34 × 10−7 |
| KEGG_INSULIN_SIGNALING_PATHWAY | 137 | 18 | 7.63 × 10−7 | 6.13 × 10−5 |
| KEGG_APOPTOSIS | 88 | 14 | 1.26 × 10−6 | 6.13 × 10−5 |
| KEGG_CELL_CYCLE | 128 | 17 | 1.32 × 10−6 | 6.13 × 10−5 |
| KEGG_PATHWAYS_IN_CANCER | 328 | 28 | 6.45 × 10−6 | 2.40 × 10−4 |
| KEGG_VALINE_LEUCINE_AND_ISOLEUCINE_DEGRADATION | 44 | 9 | 1.22 × 10−5 | 3.79 × 10−4 |
| KEGG_TGF_BETA_SIGNALING_PATHWAY | 86 | 12 | 2.81 × 10−5 | 6.73 × 10−4 |
| KEGG_CHRONIC_MYELOID_LEUKEMIA | 73 | 11 | 2.89 × 10−5 | 6.73 × 10−4 |
| KEGG_MAPK_SIGNALING_PATHWAY | 267 | 23 | 3.61 × 10−5 | 7.46 × 10−4 |
| KEGG_MTOR_SIGNALING_PATHWAY | 52 | 9 | 4.99 × 10−5 | 9.28 × 10−4 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fort, R.S.; Garat, B.; Sotelo-Silveira, J.R.; Duhagon, M.A. vtRNA2-1/nc886 Produces a Small RNA That Contributes to Its Tumor Suppression Action through the microRNA Pathway in Prostate Cancer. Non-Coding RNA 2020, 6, 7. https://doi.org/10.3390/ncrna6010007
Fort RS, Garat B, Sotelo-Silveira JR, Duhagon MA. vtRNA2-1/nc886 Produces a Small RNA That Contributes to Its Tumor Suppression Action through the microRNA Pathway in Prostate Cancer. Non-Coding RNA. 2020; 6(1):7. https://doi.org/10.3390/ncrna6010007
Chicago/Turabian StyleFort, Rafael Sebastián, Beatriz Garat, José Roberto Sotelo-Silveira, and María Ana Duhagon. 2020. "vtRNA2-1/nc886 Produces a Small RNA That Contributes to Its Tumor Suppression Action through the microRNA Pathway in Prostate Cancer" Non-Coding RNA 6, no. 1: 7. https://doi.org/10.3390/ncrna6010007
APA StyleFort, R. S., Garat, B., Sotelo-Silveira, J. R., & Duhagon, M. A. (2020). vtRNA2-1/nc886 Produces a Small RNA That Contributes to Its Tumor Suppression Action through the microRNA Pathway in Prostate Cancer. Non-Coding RNA, 6(1), 7. https://doi.org/10.3390/ncrna6010007




