DEAD-Box Helicase 3 Modulates the Non-Coding RNA Pool in Ribonucleoprotein Condensates During Stress Granule Formation
Abstract
1. Introduction
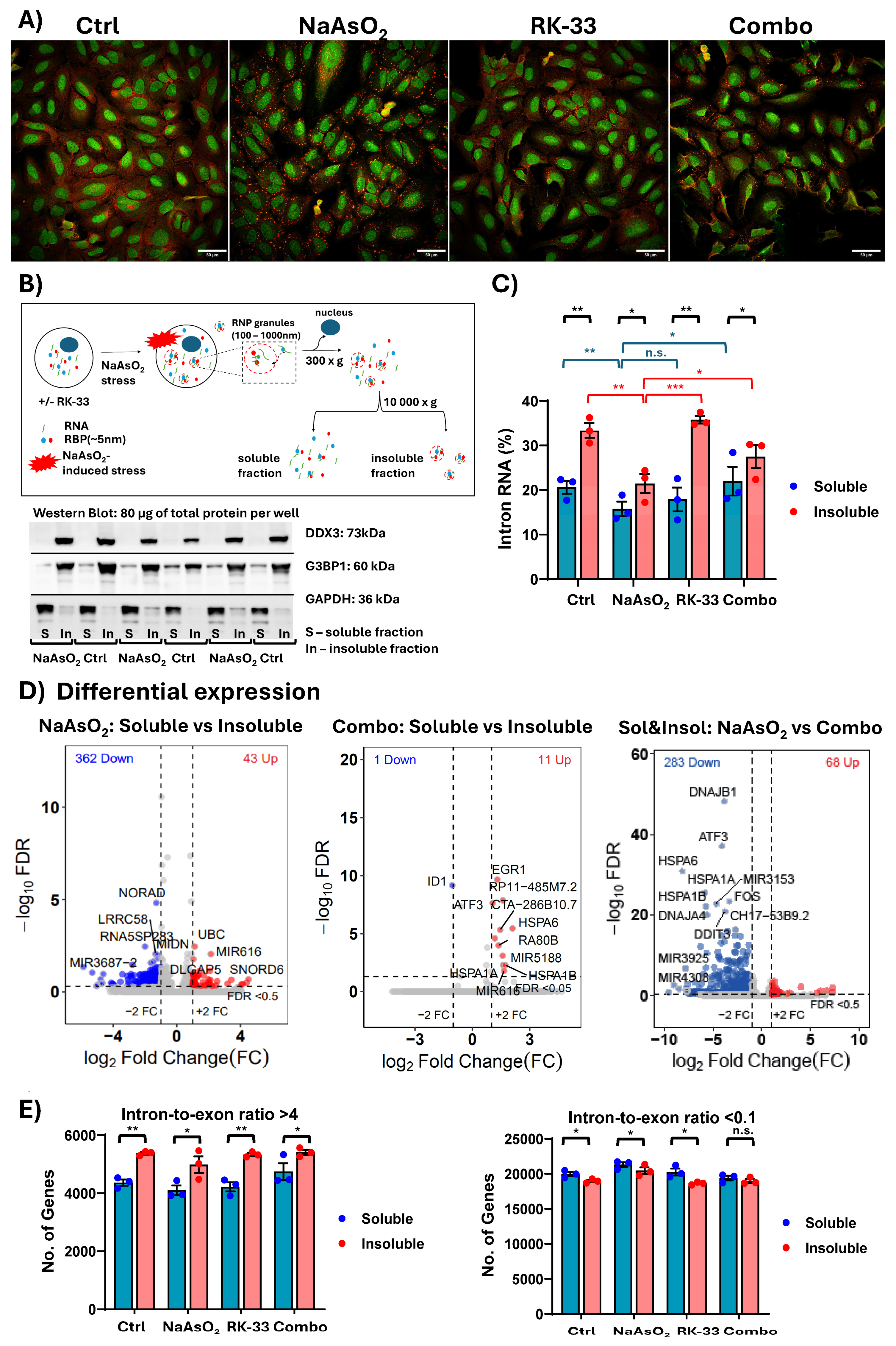
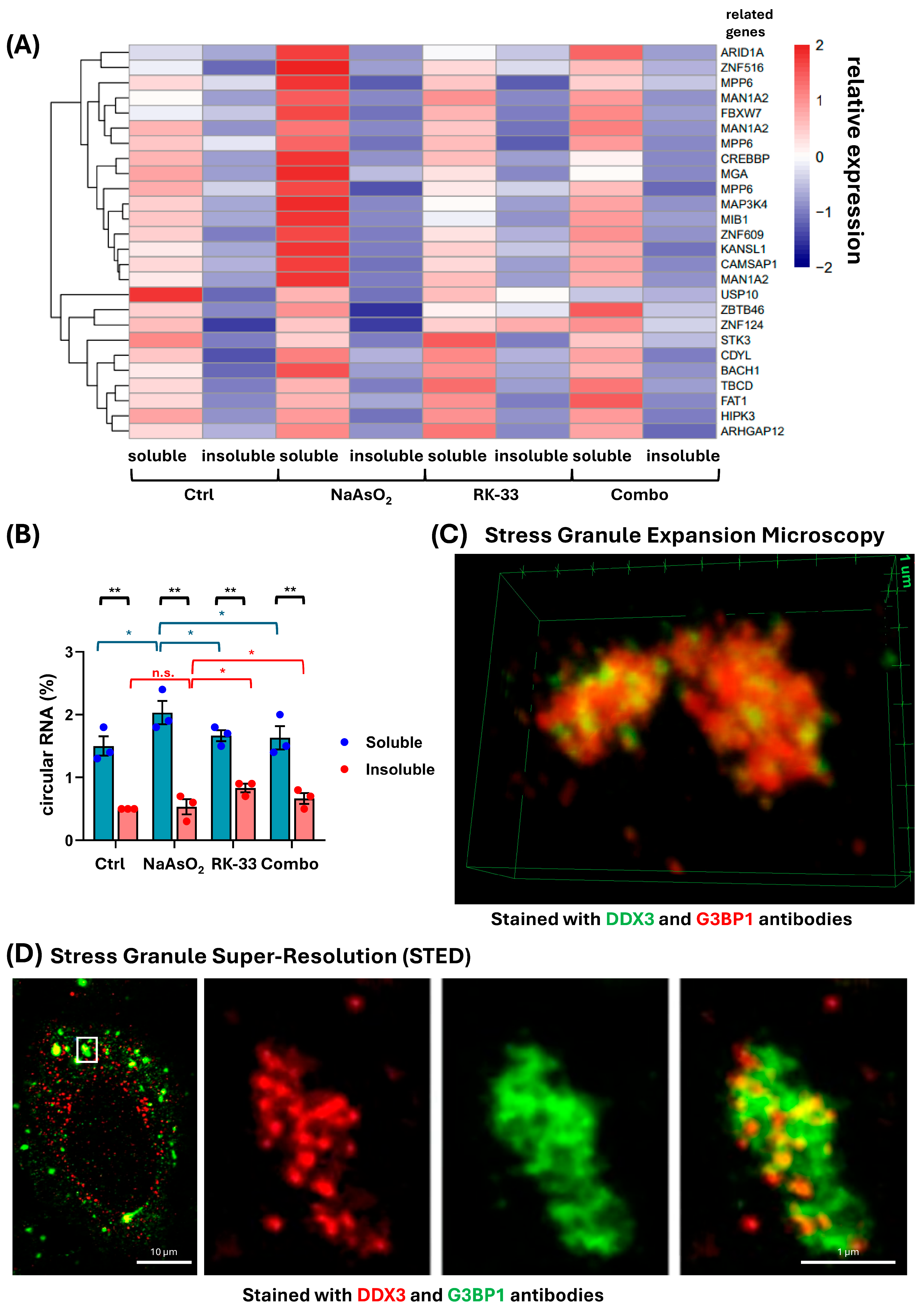
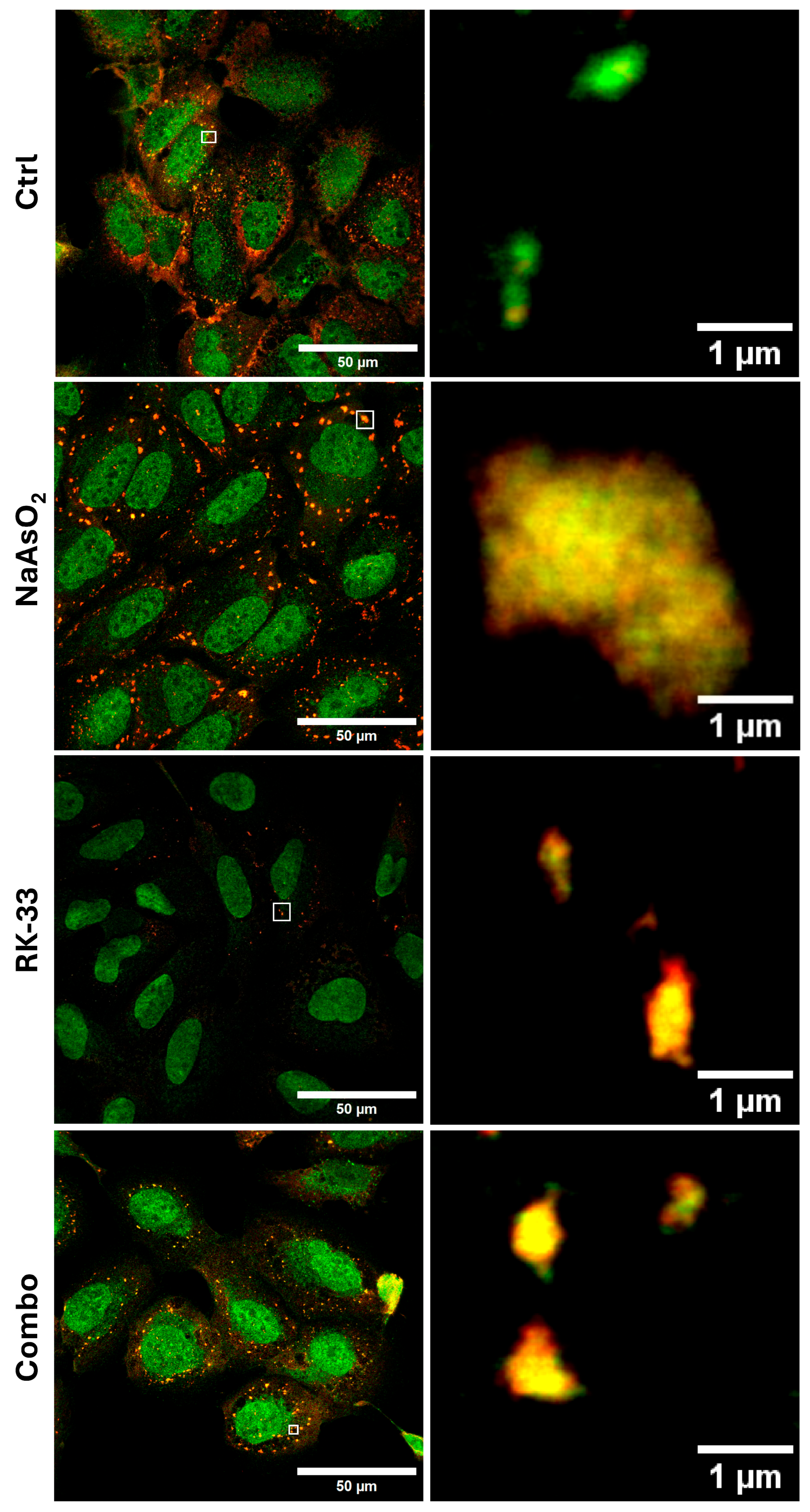
2. Results and Discussion
3. Methods
3.1. Cell Culture and Stress Granule Induction
3.2. Expansion Microscopy
3.3. STED and Confocal Microscopy
3.4. Stress Granule (SG) Isolation
3.5. Western Blot
3.6. Intron and Circular RNA Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kedersha, N.; Anderson, P. Regulation of Translation by Stress Granules and Processing Bodies. Prog. Mol. Biol. Transl. Sci. 2009, 90, 155–185. [Google Scholar] [CrossRef] [PubMed]
- Daniel, M.; Bastian, E.; Franka, V.; Jan, E.; Gregory, R.; Jeffrey, A.C. Single-Molecule Imaging Reveals Translation of Mrnas Localized to Stress Granules. Cell 2020, 183, 1801–1812. [Google Scholar] [CrossRef]
- Place, D.E.; Samir, P.; Malireddi, R.S.; Kanneganti, T.D. Integrated Stress Response Restricts Macrophage Necroptosis. Life Sci. Alliance 2022, 5, e202101260. [Google Scholar] [CrossRef] [PubMed]
- Samir, P.; Kesavardhana, S.; Patmore, D.M.; Gingras, S.; Malireddi, R.K.S.; Karki, R.; Guy, C.S.; Briard, B.; Place, D.E.; Bhattacharya, A.; et al. Ddx3x acts as a Live-or-Die Checkpoint in Stressed Cells by Regulating Nlrp3 Inflammasome. Nature 2019, 573, 590–594. [Google Scholar] [CrossRef] [PubMed]
- Arimoto, K.; Fukuda, H.; Imajoh-Ohmi, S.; Saito, H.; Takekawa, M. Formation of Stress Granules Inhibits Apoptosis by Suppressing Stress-Responsive Mapk Pathways. Nat. Cell Biol. 2008, 10, 1324–1332. [Google Scholar] [CrossRef] [PubMed]
- Omer, A.; Patel, D.; Lian, X.J.; Sadek, J.; Di Marco, S.; Pause, A.; Gorospe, M.; Gallouzi, I.E. Stress Granules Counteract Senescence by Sequestration of Pai-1. EMBO Rep. 2018, 19, e44722. [Google Scholar] [CrossRef] [PubMed]
- Amen, T.; Kaganovich, D. Stress Granules Inhibit Fatty Acid Oxidation by Modulating Mitochondrial Permeability. Cell Rep. 2021, 35, 109237. [Google Scholar] [CrossRef] [PubMed]
- Marcelo, A.; Koppenol, R.; de Almeida, L.P.; Matos, C.A.; Nóbrega, C. Stress Granules, Rna-Binding Proteins and Polyglutamine Diseases: Too Much Aggregation? Cell Death Dis. 2021, 12, 592. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.I.; Namkoong, S. Stress Granules Dynamics: Benefits in Cancer. BMB Rep. 2022, 55, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Farny, N.G. Connecting the Dots: Neuronal Senescence, Stress Granules, and Neurodegeneration. Gene 2023, 871, 147437. [Google Scholar] [CrossRef] [PubMed]
- White, J.P.; Lloyd, R.E. Regulation of Stress Granules in Virus Systems. Trends Microbiol. 2012, 20, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Wheeler, J.R.; Walters, R.W.; Agrawal, A.; Barsic, A.; Parker, R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 2016, 164, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Streit, L.; Kuhn, T.; Vomhof, T.; Bopp, V.; Ludolph, A.C.; Weishaupt, J.H.; Gebhardt, J.C.M.; Michaelis, J.; Danzer, K.M. Stress Induced Tdp-43 Mobility Loss Independent of Stress Granules. Nat. Commun. 2022, 13, 5480. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, C.; Olivia, L.; Juan-Pablo, C.; Fréderic, P.; Marie, T.; Emeline, S.; Carine, V.; Nathalie, S.-L.; Emeline, L.; Camille, L.; et al. The Proteome and Transcriptome of Stress Granules and P Bodies during Human T Lymphocyte Activation. Cell Rep. 2023, 42, 112211. [Google Scholar] [CrossRef] [PubMed]
- van Leeuwen, W.; VanInsberghe, M.; Battich, N.; Salmen, F.; van Oudenaarden, A.; Rabouille, C. Identification of the Stress Granule Transcriptome Via Rna-Editing in Single Cells and in Vivo. Cell Rep. Methods 2022, 2, 100235. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Liu, T.; Gresham, D.; Holt, L.J. Mrna Condensation Fluidizes the Cytoplasm. bioRxiv. 2023, 123, 219a–220a. [Google Scholar] [CrossRef]
- Law, J.O.; Jones, C.M.; Stevenson, T.; Williamson, T.A.; Turner, M.S.; Kusumaatmaja, H.; Grellscheid, S.N. A Bending Rigidity Parameter for Stress Granule Condensates. Sci. Adv. 2023, 9, eadg0432. [Google Scholar] [CrossRef] [PubMed]
- Aulas, A.; Fay, M.M.; Lyons, S.M.; Achorn, C.A.; Kedersha, N.; Anderson, P.; Ivanov, P. Stress-Specific Differences in Assembly and Composition of Stress Granules and Related Foci. J. Cell Sci. 2017, 130, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Linsenmeier, M.; Hondele, M.; Grigolato, F.; Secchi, E.; Weis, K.; Arosio, P. Dynamic Arrest and Aging of Biomolecular Condensates Are Modulated by Low-Complexity Domains, Rna and Biochemical Activity. Nat. Commun. 2022, 13, 3030. [Google Scholar] [CrossRef] [PubMed]
- Hondele, M.; Sachdev, R.; Heinrich, S.; Wang, J.; Vallotton, P.; Fontoura, B.M.A.; Weis, K. Dead-Box Atpases Are Global Regulators of Phase-Separated Organelles. Nature 2019, 573, 144–148. [Google Scholar] [CrossRef] [PubMed]
- Cui, B.C.; Sikirzhytski, V.; Aksenova, M.; Lucius, M.D.; Levon, G.H.; Mack, Z.T.; Pollack, C.; Odhiambo, D.; Broude, E.; Lizarraga, S.B.; et al. Pharmacological Inhibition of Dead-Box Rna Helicase 3 Attenuates Stress Granule Assembly. Biochem. Pharmacol. 2020, 182, 114280. [Google Scholar] [CrossRef] [PubMed]
- Martina, S. Human Dead-Box Protein 3 Has Multiple Functions in Gene Regulation and Cell Cycle Control and Is a Prime Target for Viral Manipulation. Biochem. Pharmacol. 2010, 79, 297–306. [Google Scholar] [CrossRef]
- Ivan, I.R.E.S.; Juliana Helena Costa, S.; Juliana, F. A Comprehensive Review on Ddx3x Liquid Phase Condensation in Health and Neurodevelopmental Disorders. Int. J. Biol. Macromol. 2024, 259, 129330. [Google Scholar] [CrossRef]
- Kedersha, N.; Stoecklin, G.; Ayodele, M.; Yacono, P.; Lykke-Andersen, J.; Fritzler, M.J.; Scheuner, D.; Kaufman, R.J.; Golan, D.E.; Anderson, P. Stress Granules and Processing Bodies Are Dynamically Linked Sites of Mrnp Remodeling. J. Cell Biol. 2005, 169, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Paul, T.W.; Farhad, V.; Guus, M.B.; Kathleen, L.G.; Georgia, C.-T.; Natalie, D.; Paul, J.; Venu, R. Targeting Rna Helicase Ddx3x with a Small Molecule Inhibitor for Breast Cancer Bone Metastasis Treatment. Cancer Lett. 2024, 604, 217260. [Google Scholar] [CrossRef] [PubMed]
- Bol, G.M.; Vesuna, F.; Xie, M.; Zeng, J.; Aziz, K.; Gandhi, N.; Levine, A.; Irving, A.; Korz, D.; Tantravedi, S.; et al. Targeting Ddx3 with a Small Molecule Inhibitor for Lung Cancer Therapy. EMBO Mol. Med. 2015, 7, 648–669. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.N.Y.; Atkinson, S.C.; Audsley, M.D.; Heaton, S.M.; Jans, D.A.; Borg, N.A. Rk-33 Is a Broad-Spectrum Antiviral Agent That Targets Dead-Box Rna Helicase Ddx3x. Cells 2020, 9, 170. [Google Scholar] [CrossRef] [PubMed]
- Namkoong, S.; Ho, A.; Woo, Y.M.; Kwak, H.; Lee, J.H. Systematic Characterization of Stress-Induced Rna Granulation. Mol. Cell 2018, 70, 175–187.e8. [Google Scholar] [CrossRef] [PubMed]
- Gwon, Y.; Maxwell, B.A.; Kolaitis, R.-M.; Zhang, P.; Kim, H.J.; Taylor, J.P. Ubiquitination of G3bp1 Mediates Stress Granule Disassembly in a Context-Specific Manner. Science 2021, 372, eabf6548. [Google Scholar] [CrossRef] [PubMed]
- Glauninger, H.; Hickernell, C.J.W.; Bard, J.A.; Drummond, D.A. Stressful Steps: Progress and Challenges in Understanding Stress-Induced Mrna Condensation and Accumulation in Stress Granules. Mol. Cell 2022, 82, 2544–2556. [Google Scholar] [CrossRef] [PubMed]
- Glauninger, H.; Bard, J.A.M.; Wong Hickernell, C.J.; Airoldi, E.M.; Li, W.; Singer, R.H.; Paul, S.; Fei, J.; Sosnick, T.R.; Wallace, E.W.J.; et al. Transcriptome-Wide Mrna Condensation Precedes Stress Granule Formation and Excludes Stress-Induced Transcripts. bioRxiv 2024. [Google Scholar] [CrossRef]
- Demeshkina, N.A.; Ferré-D’Amaré, A.R. Large-Scale Purifications Reveal Yeast and Human Stress Granule Cores Are Heterogeneous Particles With Complex Transcriptomes and Proteomes. Cell Rep. 2025, 44, 115738. [Google Scholar] [CrossRef] [PubMed]
- Talhouarne, G.J.S.; Gall, J.G. Lariat Intronic Rnas in the Cytoplasm of Vertebrate Cells. Proc. Natl. Acad. Sci. USA 2018, 115, E7970–E7977. [Google Scholar] [CrossRef] [PubMed]
- St Laurent, G.; Shtokalo, D.; Tackett, M.R.; Yang, Z.; Eremina, T.; Wahlestedt, C.; Urcuqui-Inchima, S.; Seilheimer, B.; McCaffrey, T.A.; Kapranov, P. Intronic Rnas Constitute the Major Fraction of the Non-Coding Rna in Mammalian Cells. BMC Genom. 2012, 13, 504. [Google Scholar] [CrossRef] [PubMed]
- Armakola, M.; Higgins, M.J.; Figley, M.D.; Barmada, S.J.; Scarborough, E.A.; Diaz, Z.; Fang, X.; Shorter, J.; Krogan, N.J.; Finkbeiner, S.; et al. Inhibition of Rna Lariat Debranching Enzyme Suppresses Tdp-43 Toxicity in Als Disease Models. Nat. Genet. 2012, 44, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Lungu, C.; Crespo, R.; Steijaert, T.H.; Gorska, A.; Palstra, R.-J.; Prins, H.A.B.; van Ijcken, W.; Mueller, Y.M.; van Kampen, J.J.A.; et al. Selective Cell Death in Hiv-1-Infected Cells by Ddx3 Inhibitors Leads to Depletion of the Inducible Reservoir. Nat. Commun. 2021, 12, 2475. [Google Scholar] [CrossRef] [PubMed]
- Vesuna, F.; Akhrymuk, I.; Smith, A.; Winnard, P.T.; Lin, S.-C.; Panny, L.; Scharpf, R.; Kehn-Hall, K.; Raman, V. RK-33, a Small Molecule Inhibitor of Host Rna Helicase Ddx3, Suppresses Multiple Variants of Sars-Cov-2. Front. Microbiol. 2022, 13, 959577. [Google Scholar] [CrossRef] [PubMed]
- Elguindy, M.M.; Mendell, J.T. Norad-Induced Pumilio Phase Separation is Required for Genome Stability. Nature 2021, 595, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Tatomer, D.C.; Luo, Z.; Wu, H.; Yang, L.; Chen, L.L.; Cherry, S.; Wilusz, J.E. The Output of Protein-Coding Genes Shifts to Circular Rnas When the Pre-Mrna Processing Machinery Is Limiting. Mol. Cell 2017, 68, 940–954.e3. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Yang, F.; Huang, B.; Pan, X.; Li, W.; Yu, T.; Wang, X.; Ran, L.; Qian, K.; Li, H.; et al. Circarid1a Binds to Igf2bp3 in Gastric Cancer and Promotes Cancer Proliferation by Forming a Circarid1a-Igf2bp3-Slc7a5 Rna-Protein Ternary Complex. J. Exp. Clin. Cancer Res. 2022, 41, 251. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Chen, J.; Wu, Y.; Luo, H.; Ke, Y. Decrease of Circarid1a Retards Glioblastoma Invasion by Modulating Mir-370-3p/Tgfbr2 Pathway. Int. J. Biol. Sci. 2022, 18, 5123–5135. [Google Scholar] [CrossRef] [PubMed]
- Her, P.H.; Li, T.; Huang, Z.; Xu, X.; Yang, W.; Chen, M.; Teng, M.; Chen, S.; Zeng, Y.; Liu, S.; et al. Functional Landscape of Circular Rnas in Human Cancer Cells. bioRxiv 2025. [Google Scholar] [CrossRef]
- Guo, R.; Cui, X.; Li, X.; Zang, W.; Chang, M.; Sun, Z.; Liu, Z.; Sun, Y.; Jia, J.; Li, W. Circman1a2 is Upregulated by Helicobacter Pylori and Promotes Development of Gastric Cancer. Cell Death Dis. 2022, 13, 409. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.M.; Wang, J.P.; Tang, Y.Y.; Zhao, J.; He, S.Y.; Xiong, F.; Guo, C.; Xiang, B.; Zhou, M.; Li, X.L.; et al. Circman1a2 Could Serve as a Novel Serum Biomarker for Malignant Tumors. Cancer Sci. 2019, 110, 2180–2188. [Google Scholar] [CrossRef] [PubMed]
- Ding, N.; Zhang, Y.; Huang, M.; Liu, J.; Wang, C.; Zhang, C.; Cao, J.; Zhang, Q.; Jiang, L. Circ-Crebbp Inhibits Sperm Apoptosis Via the Pi3k-Akt Signaling Pathway by Sponging Mir-10384 and Mir-143-3p. Commun. Biol. 2022, 5, 1339. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Liu, F.; Song, X.; Sun, R.; Zhang, M.; Huang, J.; Gu, W.; Shao, Y. M6a-Modified Circcrebbp Enhances Radiosensitivity of Esophageal Squamous cell Carcinoma by Reducing the Stability of Myc through Interaction with Igf2bp3. Int. J. Biol. Macromol. 2025, 286, 138534. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhang, H.; Wei, W.; Xiao, X.; Huang, C.; Wang, L.; Zhong, H.; Jiang, Y.; Zheng, F.; Yang, H.; et al. Regulation of Cd8+ T Cells Infiltration and Immunotherapy by Circmga/Hnrnpl Complex in Bladder Cancer. Oncogene 2023, 42, 1247–1262. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.-L.; Chen, W.; Xie, J.-J.; Zhang, M.-L.; Nie, R.-C.; Liang, H.; Mei, J.; Han, K.; Xiang, Z.-C.; Wang, F.-W.; et al. A Novel Peptide Encoded by N6-Methyladenosine Modified Circmap3k4 Prevents Apoptosis in Hepatocellular Carcinoma. Mol. Cancer 2022, 21, 93. [Google Scholar] [CrossRef] [PubMed]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-Znf609 Is a Circular Rna That Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e9. [Google Scholar] [CrossRef] [PubMed]
- Ayyildiz, D.; Bergonzoni, G.; Monziani, A.; Tripathi, T.; Döring, J.; Kerschbamer, E.; Di Leva, F.; Pennati, E.; Donini, L.; Kovalenko, M.; et al. Cag Repeat Expansion in the Huntington’s Disease Gene Shapes Linear and Circular Rnas Biogenesis. PLoS Genet. 2023, 19, e1010988. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.-X.; Xu, S.-D.; Deng, M.-H.; Hao, S.-H.; Chen, J.-W.; Ma, X.-D.; Zhuang, W.-T.; Cao, J.-H.; Lv, Y.-R.; Lin, J.-L.; et al. Mex-3 Rna Binding Family Member A (Mex3a)/Circmpp6 Complex Promotes Colorectal Cancer Progression by Inhibiting Autophagy. Signal Transduct. Target. Ther. 2024, 9, 80. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.; Danßmann, C.; Klironomos, F.; Winkler, A.; Fallmann, J.; Kruetzfeldt, L.-M.; Szymansky, A.; Naderi, J.; Bernhart, S.H.; Grunewald, L.; et al. Defining the Landscape of Circular Rnas in Neuroblastoma Unveils a Global Suppressive Function of Mycn. Nat. Commun. 2023, 14, 3936. [Google Scholar] [CrossRef] [PubMed]
- Taniuchi, K.; Ogasawara, M. Khsrp-Bound Small Nucleolar Rnas Associate with Promotion of Cell Invasiveness and Metastasis of Pancreatic Cancer. Oncotarget 2020, 11, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Irmela, R.E.A.T.; Andreas, H.; Christine, D.A.; Janani, N.; Charlotte, M.F.; Marta, A.; Hannes, A.; Tuomas, P.J.K.; Michael, S.; Titus, M.F.; et al. G3bp-Driven Rnp Granules Promote Inhibitory Rna-Rna Interactions Resolved by Ddx3x to Regulate Mrna Translatability. Mol. Cell 2025, 85, 585–601.e511. [Google Scholar] [CrossRef]
- Ania, M.O.; Arvind, H.P. Hepatitis C Virus Core Protein Interacts with a Human Dead Box Protein Ddx3. Virology 1999, 257, 330–340. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.C.; Lee, Y.H.; Tarn, W.Y. The Dead-Box Rna Helicase Ddx3 Associates with Export Messenger Ribonucleoproteins as Well as Tip-Associated Protein and Participates in Translational Control. Mol. Biol. Cell 2008, 19, 3847–3858. [Google Scholar] [CrossRef] [PubMed]
- Venkat, S.R.K.Y.; Christine, N.; Ya-hui, C.; Lawrence, K.; Kuan-Teh, J. Requirement of Ddx3 Dead Box Rna Helicase for Hiv-1 Rev-Rre Export Function. Cell 2004, 119, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Mo, J.; Liang, H.; Su, C.; Li, P.; Chen, J.; Zhang, B. Ddx3x: Structure, Physiologic Functions and Cancer. Mol. Cancer 2021, 20, 38. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Fang, E.; Mei, H.; Chen, Y.; Li, H.; Li, D.; Song, H.; Wang, J.; Hong, M.; Xiao, W.; et al. Cis-Acting Circ-Ctnnb1 Promotes β-Catenin Signaling and Cancer Progression Via Ddx3-Mediated Transactivation of Yy1. Cancer Res. 2019, 79, 557–571. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, J.; Zhao, F. Screening Linear and Circular Rna Transcripts from Stress Granules. Genom. Proteom. Bioinform. 2023, 21, 886–893. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, X.; Guo, M.; Yu, J.; Yan, C. The Common Stress Responsive Transcription Factor Atf3 Binds Genomic Sites Enriched with P300 and H3k27ac for Transcriptional Regulation. BMC Genom. 2016, 17, 335. [Google Scholar] [CrossRef] [PubMed]
- Batra, J.; Tripathi, S.; Kumar, A.; Katz, J.M.; Cox, N.J.; Lal, R.B.; Sambhara, S.; Lal, S.K. Human Heat Shock Protein 40 (Hsp40/Dnajb1) Promotes Influenza a Virus Replication by Assisting Nuclear Import of Viral Ribonucleoproteins. Sci. Rep. 2016, 6, 19063. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, V.P.; Stamatis, M.; Shmukler, A.; Aneskievich, B.J. Basal and Stress-Inducible Expression of Hspa6 in Human Keratinocytes is Regulated by Negative and Positive Promoter Regions. Cell Stress. Chaperones 2015, 20, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Solarz, A.; Majcher-Maślanka, I.; Kryst, J.; Chocyk, A. A Search for Biomarkers of Early-Life Stress-Related Psychopathology: Focus on 70-kDa Heat Shock Proteins. Neuroscience 2021, 463, 238–253. [Google Scholar] [CrossRef] [PubMed]
- McMahon, A.C.; Rahman, R.; Jin, H.; Shen, J.L.; Fieldsend, A.; Luo, W.; Rosbash, M. Tribe: Hijacking an Rna-Editing Enzyme to Identify Cell-Specific Targets of Rna-Binding Proteins. Cell 2016, 165, 742–753. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Yang, G.; Goodwin, D.R.; O’Flanagan, C.H.; Sinha, A.; Zhang, C.; Kitko, K.E.; Park, D.; Aparicio, S.; Consortium, I.; et al. A Multifunctional Anchor for Multimodal Expansion Microscopy. bioRxiv 2022. [Google Scholar] [CrossRef]
- Anthony, K.; Saumya, J.; Tyler, M.; Joshua, R.W.; Roy, P. Isolation of Mammalian Stress Granule Cores for Rna-Seq Analysis. Methods 2018, 137, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. Star: Ultrafast Universal Rna-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. The Subread Aligner: Fast, Accurate and Scalable Read Mapping by Seed-and-Vote. Nucleic Acids Res. 2013, 41, e108. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Metge, F.; Dieterich, C. Specific Identification and Quantification of Circular Rnas from Sequencing Data. Bioinformatics 2016, 32, 1094–1096. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential Expression Analysis of Multifactor Rna-Seq Experiments with Respect to Biological Variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Society. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Stevens, J.R.; Herrick, J.S.; Wolff, R.K.; Slattery, M.L. Power in Pairs: Assessing the Statistical Value of Paired Samples in Tests for Differential Expression. BMC Genom. 2018, 19, 953. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Korunova, E.; Cui, B.C.; Ji, H.; Sikirzhytskaya, A.; Samaddar, S.; Chen, M.; Sikirzhytski, V.; Shtutman, M. DEAD-Box Helicase 3 Modulates the Non-Coding RNA Pool in Ribonucleoprotein Condensates During Stress Granule Formation. Non-Coding RNA 2025, 11, 59. https://doi.org/10.3390/ncrna11040059
Korunova E, Cui BC, Ji H, Sikirzhytskaya A, Samaddar S, Chen M, Sikirzhytski V, Shtutman M. DEAD-Box Helicase 3 Modulates the Non-Coding RNA Pool in Ribonucleoprotein Condensates During Stress Granule Formation. Non-Coding RNA. 2025; 11(4):59. https://doi.org/10.3390/ncrna11040059
Chicago/Turabian StyleKorunova, Elizaveta, B. Celia Cui, Hao Ji, Aliaksandra Sikirzhytskaya, Srestha Samaddar, Mengqian Chen, Vitali Sikirzhytski, and Michael Shtutman. 2025. "DEAD-Box Helicase 3 Modulates the Non-Coding RNA Pool in Ribonucleoprotein Condensates During Stress Granule Formation" Non-Coding RNA 11, no. 4: 59. https://doi.org/10.3390/ncrna11040059
APA StyleKorunova, E., Cui, B. C., Ji, H., Sikirzhytskaya, A., Samaddar, S., Chen, M., Sikirzhytski, V., & Shtutman, M. (2025). DEAD-Box Helicase 3 Modulates the Non-Coding RNA Pool in Ribonucleoprotein Condensates During Stress Granule Formation. Non-Coding RNA, 11(4), 59. https://doi.org/10.3390/ncrna11040059





