Long Non-Coding RNA Levels Are Modulated in Schistosoma mansoni following In Vivo Praziquantel Exposure
Abstract
1. Introduction
2. Results
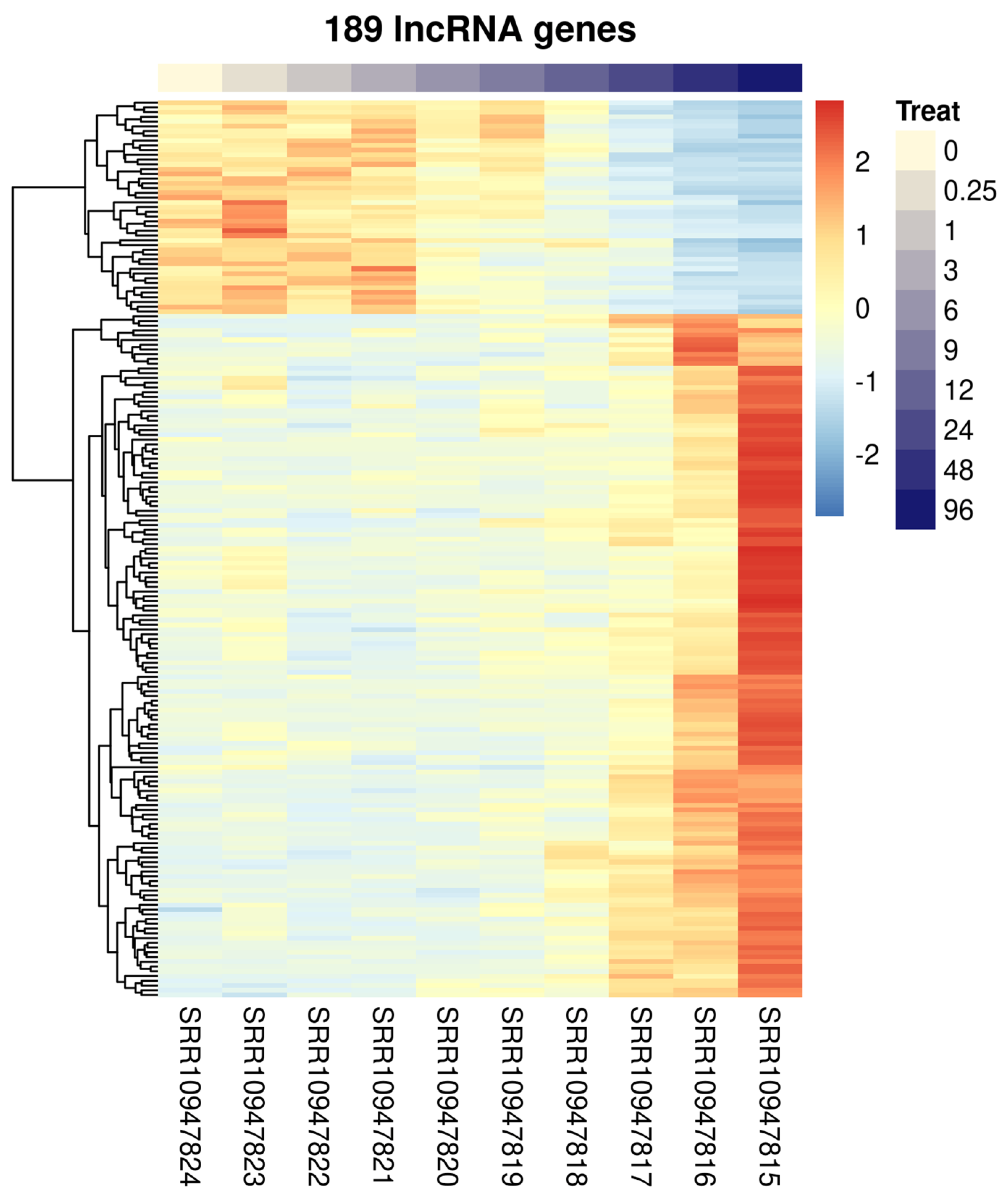
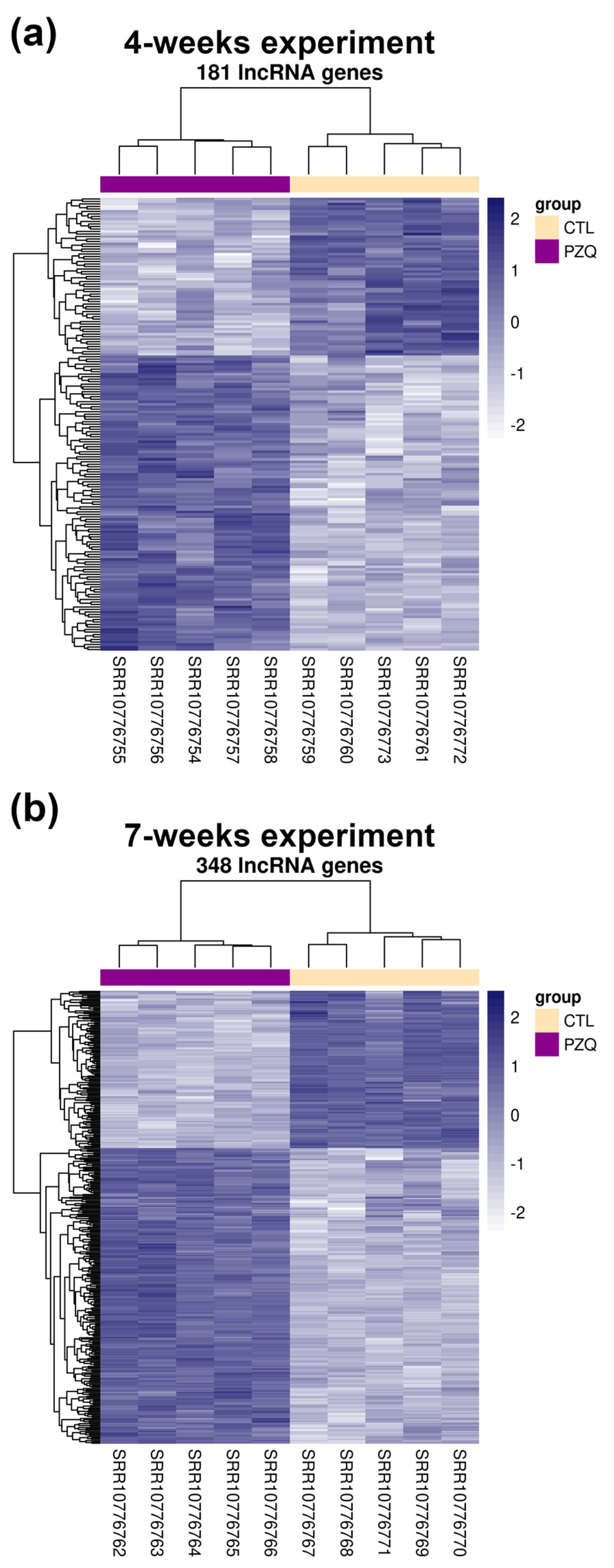
2.1. Sets of LncRNAs Are Differentially Expressed in S. mansoni following In Vivo Praziquantel Exposure
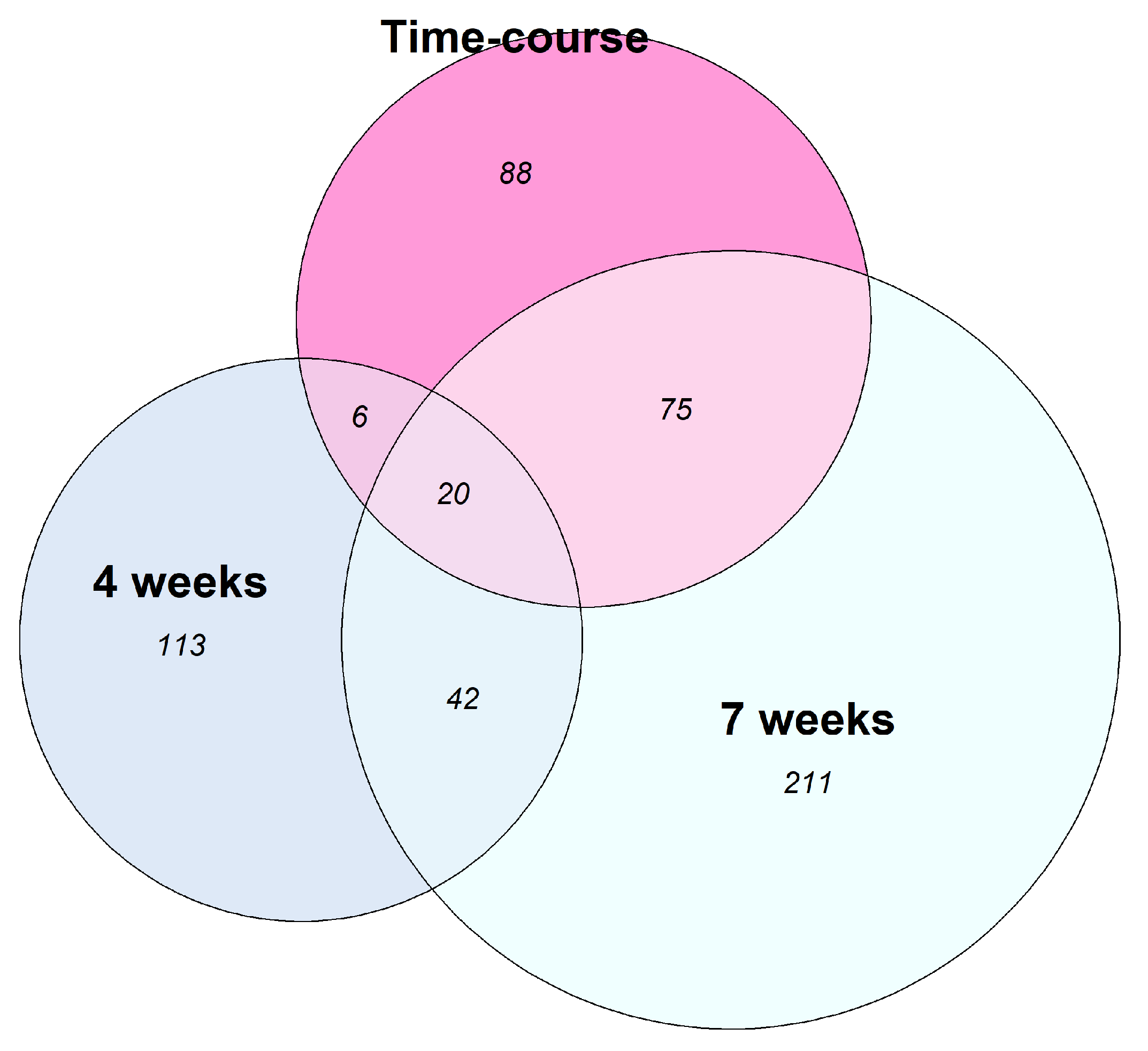
2.2. A Set of LncRNAs Is Concomitantly Differentially Expressed under Distinct PZQ Treatments
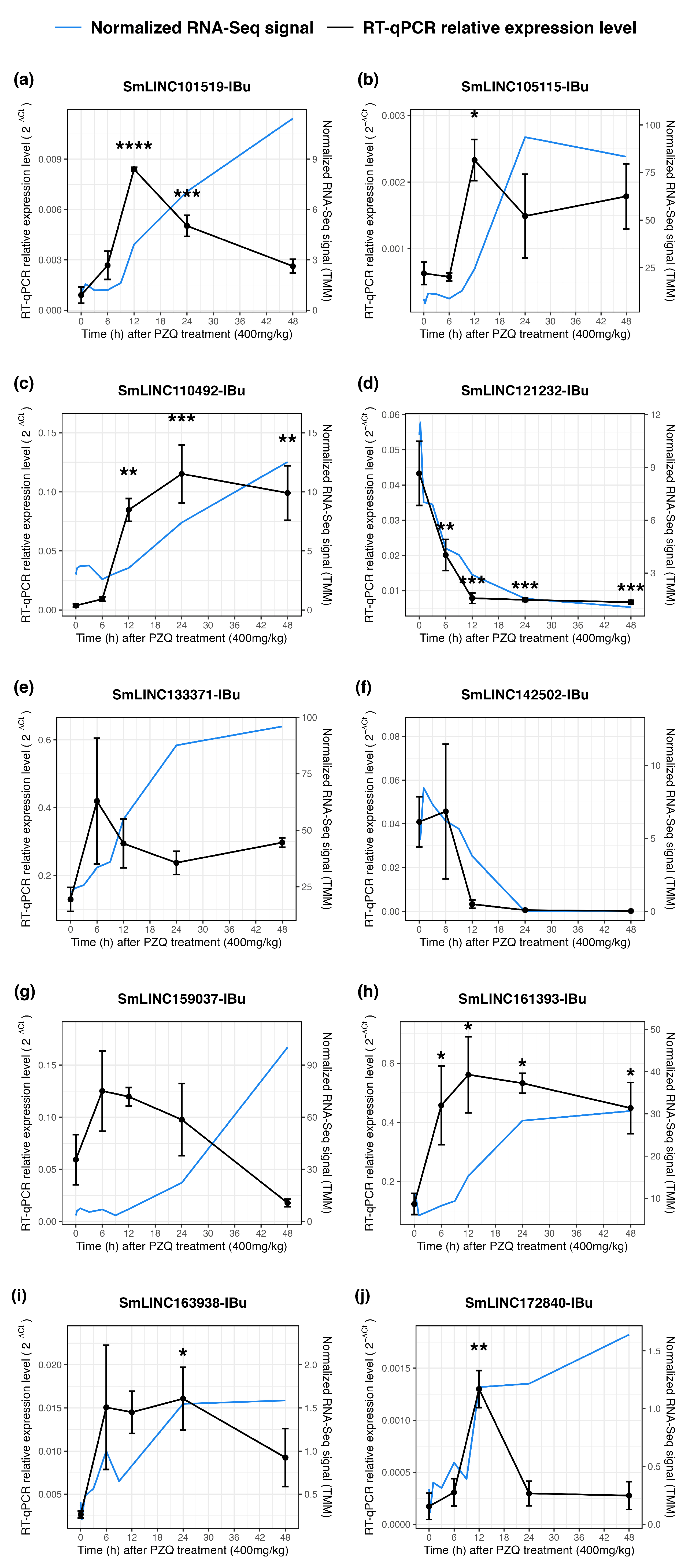
2.3. Validation by RT-qPCR of LncRNAs Differential Expression
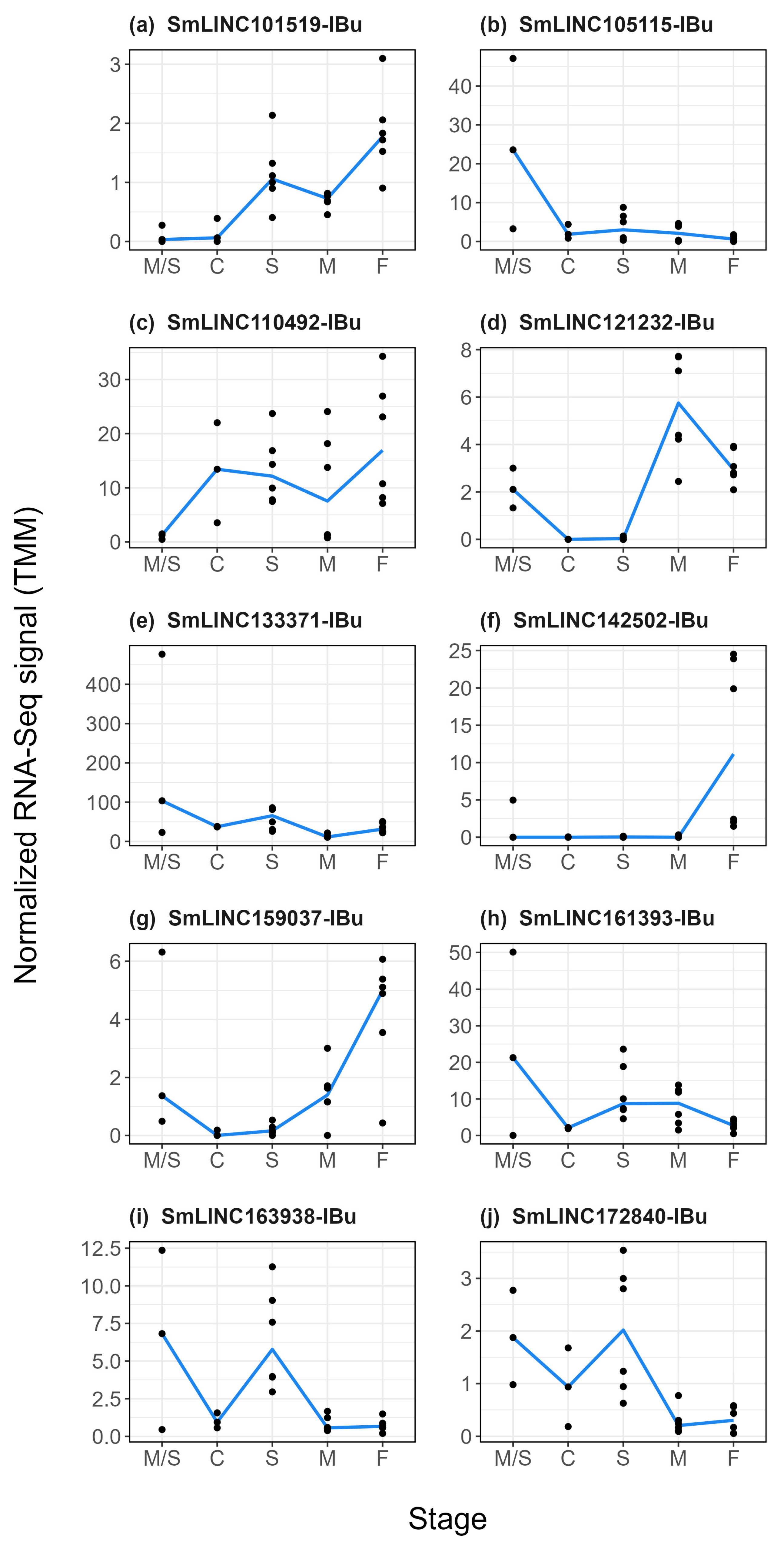
2.4. LncRNAs Modulated by Praziquantel Are Differentially Expressed along S. mansoni Life-Cycle Stages
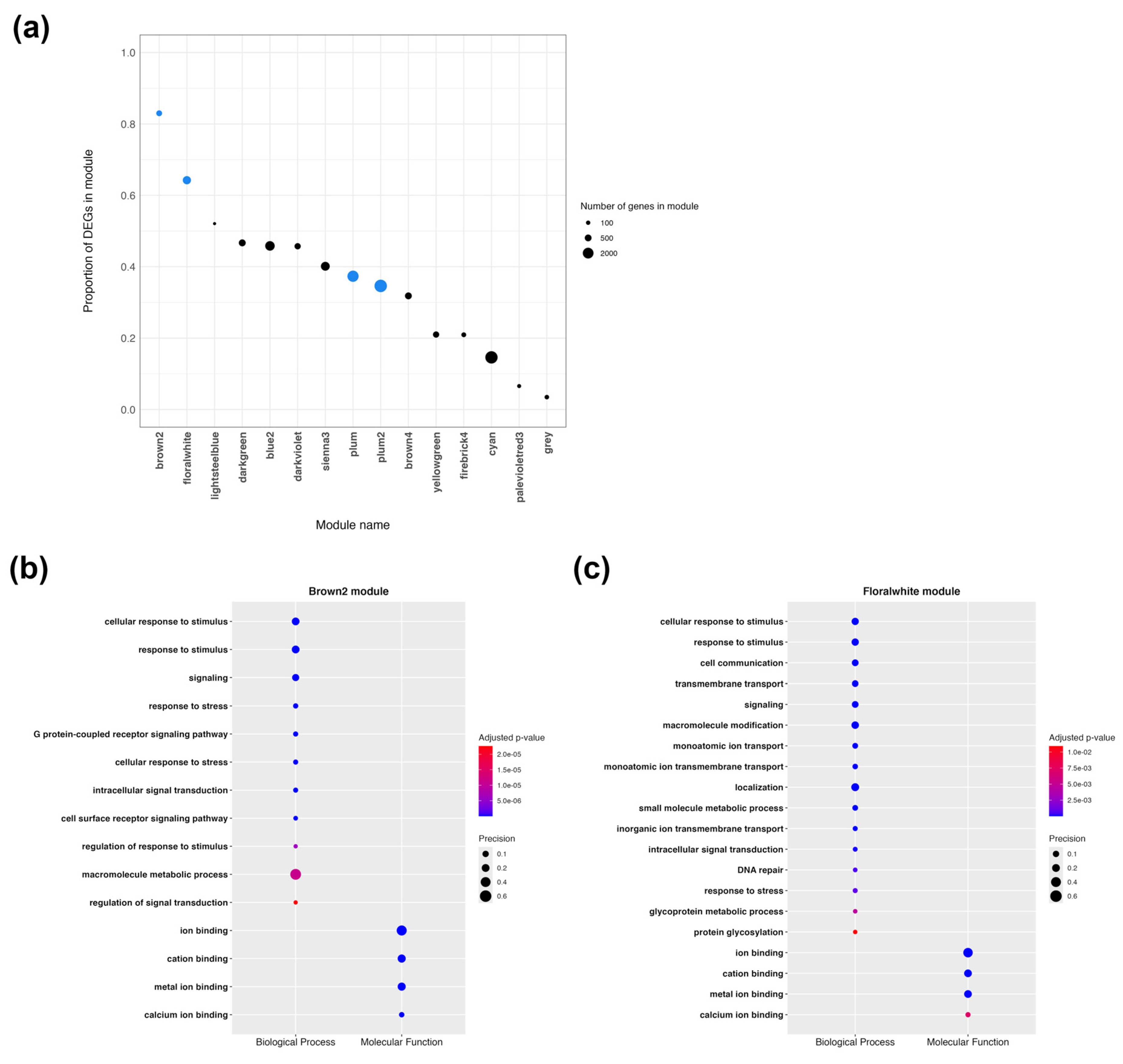
2.5. Weighted Gene Co-Expression Network Analysis Shows Terms Related to Drug Response Mechanisms
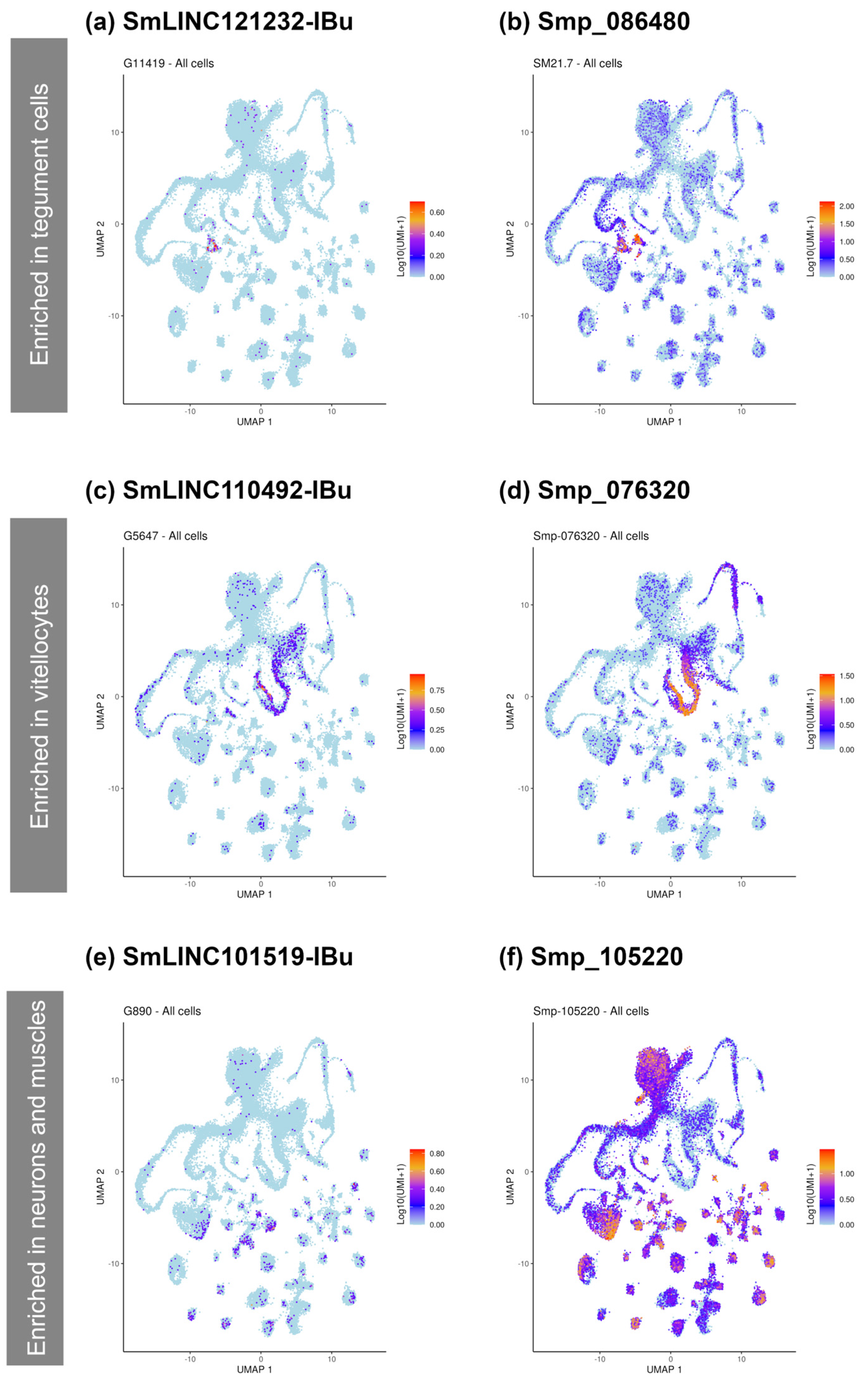
2.6. PZQ-Modulated lncRNAs Co-Localize with PZQ-Modulated Smps in Different Cell Types
3. Discussion
4. Materials and Methods
4.1. Re-Analyses of PZQ Treatment RNA-Seq Data
4.2. Selection of LncRNAs for RT-qPCR Validation
4.3. RNA Extraction, Quantification, and Quality Assessment
4.4. Primer Design, Reverse Transcription, and Quantitative PCR (RT-qPCR) Assays
4.5. Gene Expression Patterns of Selected Genes across S. mansoni Life-Cycle Stages
4.6. Weighted Gene Co-Expression Network Analysis (WGCNA)
4.7. Single-Cell Clusters Search of LncRNAs and Protein-Coding Genes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Colley, D.G.; Bustinduy, A.L.; Secor, W.E.; King, C.H. Human schistosomiasis. Lancet 2014, 383, 2253–2264. [Google Scholar] [CrossRef] [PubMed]
- WHO. Fact Sheets: Schistosomiasis; 2023. Available online: https://www.who.int/news-room/fact-sheets/detail/schistosomiasis (accessed on 9 January 2024).
- Zacharia, A.; Mushi, V.; Makene, T. A systematic review and meta-analysis on the rate of human schistosomiasis reinfection. PLoS ONE 2020, 15, e0243224. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Health Estimates 2020: Disease Burden by Cause, Age, Sex, by Country and by Region, 2000–2019. Geneva, World Health Organization. 2020. Available online: https://www.who.int/data/gho/data/themes/mortality-and-global-health-estimates/global-health-estimates-leading-causes-of-dalys (accessed on 9 January 2024).
- McManus, D.P.; Dunne, D.W.; Sacko, M.; Utzinger, J.; Vennervald, B.J.; Zhou, X.N. Schistosomiasis. Nat. Rev. Dis. Primers 2018, 4, 13. [Google Scholar] [CrossRef] [PubMed]
- Vale, N.; Gouveia, M.J.; Rinaldi, G.; Brindley, P.J.; Gärtner, F.; Correia da Costa, J.M. Praziquantel for Schistosomiasis: Single-Drug Metabolism Revisited, Mode of Action, and Resistance. Antimicrob. Agents Chemother. 2017, 61, e02582-16. [Google Scholar] [CrossRef]
- Lewis, F.A.; Tucker, M.S. Schistosomiasis. Adv. Exp. Med. Biol. 2014, 766, 47–75. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.A. Schistosomiasis then and now: What has changed in the last 100 years? Parasitology 2020, 147, 507–515. [Google Scholar] [CrossRef]
- Park, S.K.; Friedrich, L.; Yahya, N.A.; Rohr, C.M.; Chulkov, E.G.; Maillard, D.; Rippmann, F.; Spangenberg, T.; Marchant, J.S. Mechanism of praziquantel action at a parasitic flatworm ion channel. Sci. Transl. Med. 2021, 13, eabj5832. [Google Scholar] [CrossRef] [PubMed]
- Le Clec’h, W.; Chevalier, F.D.; Mattos, A.C.A.; Strickland, A.; Diaz, R.; McDew-White, M.; Rohr, C.M.; Kinung’hi, S.; Allan, F.; Webster, B.L.; et al. Genetic analysis of praziquantel response in schistosome parasites implicates a transient receptor potential channel. Sci. Transl. Med. 2021, 13, eabj9114. [Google Scholar] [CrossRef]
- McCusker, P.; Rohr, C.M.; Chan, J.D. Schistosoma mansoni alter transcription of immunomodulatory gene products following in vivo praziquantel exposure. PLoS Negl. Trop. Dis. 2021, 15, e0009200. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Gao, L.; Ma, X.; Huang, J.J.; Chen, J.; Zeng, L.; Ashby, C.R., Jr.; Zou, C.; Chen, Z.S. Long non-coding RNAs regulate drug resistance in cancer. Mol. Cancer 2020, 19, 54. [Google Scholar] [CrossRef] [PubMed]
- Smallegan, M.J.; Rinn, J.L. Linking long noncoding RNA to drug resistance. Proc. Natl. Acad. Sci. USA 2019, 116, 21963–21965. [Google Scholar] [CrossRef]
- Melé, M.; Mattioli, K.; Mallard, W.; Shechner, D.M.; Gerhardinger, C.; Rinn, J.L. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017, 27, 27–37. [Google Scholar] [CrossRef]
- Silveira, G.O.; Coelho, H.S.; Amaral, M.S.; Verjovski-Almeida, S. Long non-coding RNAs as possible therapeutic targets in protozoa, and in Schistosoma and other helminths. Parasitol Res. 2022, 121, 1091–1115. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics-challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, K.C.; Carvalho, M.L.; Maracaja-Coutinho, V.; Kitajima, J.P.; Verjovski-Almeida, S. Non-coding RNAs in schistosomes: An unexplored world. An. Acad. Bras. Cienc. 2011, 83, 673–694. [Google Scholar] [CrossRef]
- Kim, H.C.; Khalil, A.M.; Jolly, E.R. LncRNAs in molluscan and mammalian stages of parasitic schistosomes are developmentally-regulated and coordinately expressed with protein-coding genes. RNA Biol. 2020, 17, 805–815. [Google Scholar] [CrossRef]
- Amaral, M.S.; Maciel, L.F.; Silveira, G.O.; Olberg, G.G.O.; Leite, J.V.P.; Imamura, L.K.; Pereira, A.S.A.; Miyasato, P.A.; Nakano, E.; Verjovski-Almeida, S. Long non-coding RNA levels can be modulated by 5-azacytidine in Schistosoma mansoni. Sci. Rep. 2020, 10, 21565. [Google Scholar] [CrossRef]
- Silveira, G.O.; Coelho, H.S.; Pereira, A.S.A.; Miyasato, P.A.; Santos, D.W.; Maciel, L.F.; Olberg, G.G.G.; Tahira, A.C.; Nakano, E.; Oliveira, M.L.S.; et al. Long non-coding RNAs are essential for Schistosoma mansoni pairing-dependent adult worm homeostasis and fertility. PLoS Pathog. 2023, 19, e1011369. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Qu, Y.; Yang, Q.; Ma, X.; Meng, Q.; Xu, J.; Liu, X.; Wang, S. D-lnc: A comprehensive database and analytical platform to dissect the modification of drugs on lncRNA expression. RNA Biol. 2019, 16, 1586–1591. [Google Scholar] [CrossRef]
- Maciel, L.F.; Morales-Vicente, D.A.; Silveira, G.O.; Ribeiro, R.O.; Olberg, G.G.O.; Pires, D.S.; Amaral, M.S.; Verjovski-Almeida, S. Weighted Gene Co-Expression Analyses Point to Long Non-Coding RNA Hub Genes at Different Schistosoma mansoni Life-Cycle Stages. Front. Genet. 2019, 10, 823. [Google Scholar] [CrossRef]
- Geyer, K.K.; Rodriguez Lopez, C.M.; Chalmers, I.W.; Munshi, S.E.; Truscott, M.; Heald, J.; Wilkinson, M.J.; Hoffmann, K.F. Cytosine methylation regulates oviposition in the pathogenic blood fluke Schistosoma mansoni. Nat. Commun. 2011, 2, 424. [Google Scholar] [CrossRef]
- Lu, L.J.; Randerath, K. Mechanism of 5-azacytidine-induced transfer RNA cytosine-5-methyltransferase deficiency. Cancer Res. 1980, 40, 2701–2705. [Google Scholar] [PubMed]
- Taylor, S.M.; Jones, P.A. Mechanism of action of eukaryotic DNA methyltransferase. Use of 5-azacytosine-containing DNA. J. Mol. Biol. 1982, 162, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Geyer, K.K.; Munshi, S.E.; Vickers, M.; Squance, M.; Wilkinson, T.J.; Berrar, D.; Chaparro, C.; Swain, M.T.; Hoffmann, K.F. The anti-fecundity effect of 5-azacytidine (5-AzaC) on Schistosoma mansoni is linked to dis-regulated transcription, translation and stem cell activities. Int. J. Parasitol. Drugs Drug Resist. 2018, 8, 213–222. [Google Scholar] [CrossRef]
- Silveira, G.O.; Amaral, M.S.; Coelho, H.S.; Maciel, L.F.; Pereira, A.S.A.; Olberg, G.G.O.; Miyasato, P.A.; Nakano, E.; Verjovski-Almeida, S. Assessment of reference genes at six different developmental stages of Schistosoma mansoni for quantitative RT-PCR. Sci. Rep. 2021, 11, 16816. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Morales-Vicente, D.A.; Zhao, L.; Silveira, G.O.; Tahira, A.C.; Amaral, M.S.; Collins, J.J., 3rd; Verjovski-Almeida, S. Single-cell RNA-seq analyses show that long non-coding RNAs are conspicuously expressed in Schistosoma mansoni gamete and tegument progenitor cell populations. Front. Genet. 2022, 13, 924877. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.M.; Fitzsimmons, C.M.; Dunne, D.W.; Timson, D.J. Comparative biochemical analysis of three members of the Schistosoma mansoni TAL family: Differences in ion and drug binding properties. Biochimie 2015, 108, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Carson, J.; Thomas, C.M.; McGinty, A.; Takata, G.; Timson, D.J. The tegumental allergen-like proteins of Schistosoma mansoni: A biochemical study of SmTAL4-TAL13. Mol. Biochem. Parasitol. 2018, 221, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.M.; Timson, D.J. A mysterious family of calcium-binding proteins from parasitic worms. Biochem. Soc. Trans. 2016, 44, 1005–1010. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cao, X.; Zhou, X.; Hou, F.; Huang, Y.E.; Yuan, M.; Long, M.; Chen, S.; Lei, W.; Zhu, J.; Chen, J.; et al. ncRNADrug: A database for validated and predicted ncRNAs associated with drug resistance and targeted by drugs. Nucleic Acids Res. 2024, 52, D1393–D1399. [Google Scholar] [CrossRef] [PubMed]
- Ghiam, S.; Eslahchi, C.; Shahpasand, K.; Habibi-Rezaei, M.; Gharaghani, S. Identification of repurposed drugs targeting significant long non-coding RNAs in the cross-talk between diabetes mellitus and Alzheimer’s disease. Sci. Rep. 2022, 12, 18332. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, Z.; Zhang, J. In silico drug repositioning based on integrated drug targets and canonical correlation analysis. BMC Med. Genomics 2022, 15, 48. [Google Scholar] [CrossRef]
- Guo, H.; Liu, J.; Ben, Q.; Qu, Y.; Li, M.; Wang, Y.; Chen, W.; Zhang, J. The aspirin-induced long non-coding RNA OLA1P2 blocks phosphorylated STAT3 homodimer formation. Genome Biol. 2016, 17, 24. [Google Scholar] [CrossRef] [PubMed]
- Klempnauer, K.H.; Sippel, A.E. The highly conserved amino-terminal region of the protein encoded by the v-myb oncogene functions as a DNA-binding domain. EMBO J. 1987, 6, 2719–2725. [Google Scholar] [CrossRef]
- Biedenkapp, H.; Borgmeyer, U.; Sippel, A.E.; Klempnauer, K.H. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature 1988, 335, 835–837. [Google Scholar] [CrossRef]
- Boyer, L.A.; Latek, R.R.; Peterson, C.L. The SANT domain: A unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 2004, 5, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Beesley, N.J.; Cwiklinski, K.; Allen, K.; Hoyle, R.C.; Spithill, T.W.; La Course, E.J.; Williams, D.J.L.; Paterson, S.; Hodgkinson, J.E. A major locus confers triclabendazole resistance in Fasciola hepatica and shows dominant inheritance. PLoS Pathog. 2023, 19, e1011081. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, I.W.; Fitzsimmons, C.M.; Brown, M.; Pierrot, C.; Jones, F.M.; Wawrzyniak, J.M.; Fernandez-Fuentes, N.; Tukahebwa, E.M.; Dunne, D.W.; Khalife, J.; et al. Human IgG1 Responses to Surface Localised Schistosoma mansoni Ly6 Family Members Drop following Praziquantel Treatment. PLoS Negl. Trop. Dis. 2015, 9, e0003920. [Google Scholar] [CrossRef]
- Egesa, M.; Lubyayi, L.; Tukahebwa, E.M.; Bagaya, B.S.; Chalmers, I.W.; Wilson, S.; Hokke, C.H.; Hoffmann, K.F.; Dunne, D.W.; Yazdanbakhsh, M.; et al. Schistosoma mansoni schistosomula antigens induce Th1/Pro-inflammatory cytokine responses. Parasite Immunol. 2018, 40, e12592. [Google Scholar] [CrossRef]
- Farias, L.P.; Krautz-Peterson, G.; Tararam, C.A.; Araujo-Montoya, B.O.; Fraga, T.R.; Rofatto, H.K.; Silva, F.P., Jr.; Isaac, L.; Da’dara, A.A.; Wilson, R.A.; et al. On the three-finger protein domain fold and CD59-like proteins in Schistosoma mansoni. PLoS Negl. Trop. Dis. 2013, 7, e2482. [Google Scholar] [CrossRef]
- Kong, H.K.; Park, J.H. Characterization and function of human Ly-6/uPAR molecules. BMB Rep. 2012, 45, 595–603. [Google Scholar] [CrossRef]
- Loughner, C.L.; Bruford, E.A.; McAndrews, M.S.; Delp, E.E.; Swamynathan, S.; Swamynathan, S.K. Organization, evolution and functions of the human and mouse Ly6/uPAR family genes. Hum. Genomics 2016, 10, 10. [Google Scholar] [CrossRef]
- Fetterer, R.H.; Pax, R.A.; Strand, S.; Bennett, J.L. Schistosoma mansoni: Physical and chemical factors affecting the mechanical properties of the adult male musculature. Exp. Parasitol. 1978, 46, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.H.; Catto, B.A.; Webster, L.T., Jr. Effects of praziquantel on different developmental stages of Schistosoma mansoni in vitro and in vivo. J. Infect. Dis. 1985, 151, 1130–1137. [Google Scholar] [CrossRef]
- Olliaro, P.; Delgado-Romero, P.; Keiser, J. The little we know about the pharmacokinetics and pharmacodynamics of praziquantel (racemate and R-enantiomer). J. Antimicrob. Chemother. 2014, 69, 863–870. [Google Scholar] [CrossRef]
- Panic, G.; Ruf, M.T.; Keiser, J. Immunohistochemical Investigations of Treatment with Ro 13-3978, Praziquantel, Oxamniquine, and Mefloquine in Schistosoma mansoni-Infected Mice. Antimicrob. Agents. Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Reimers, N.; Homann, A.; Hoschler, B.; Langhans, K.; Wilson, R.A.; Pierrot, C.; Khalife, J.; Grevelding, C.G.; Chalmers, I.W.; Yazdanbakhsh, M.; et al. Drug-induced exposure of Schistosoma mansoni antigens SmCD59a and SmKK7. PLoS Negl. Trop. Dis. 2015, 9, e0003593. [Google Scholar] [CrossRef] [PubMed]
- Morais, S.B.; Figueiredo, B.C.; Assis, N.R.G.; Alvarenga, D.M.; de Magalhaes, M.T.Q.; Ferreira, R.S.; Vieira, A.T.; Menezes, G.B.; Oliveira, S.C. Schistosoma mansoni SmKI-1 serine protease inhibitor binds to elastase and impairs neutrophil function and inflammation. PLoS Pathog. 2018, 14, e1006870. [Google Scholar] [CrossRef]
- Wang, Y.; Qin, Z.; Cai, S.; Yu, L.; Hu, H.; Zeng, S. The role of non-coding RNAs in ABC transporters regulation and their clinical implications of multidrug resistance in cancer. Expert. Opin. Drug Metab. Toxicol. 2021, 17, 291–306. [Google Scholar] [CrossRef]
- He, J.; Zhu, S.; Liang, X.; Zhang, Q.; Luo, X.; Liu, C.; Song, L. LncRNA as a multifunctional regulator in cancer multi-drug resistance. Mol. Biol. Rep. 2021, 48, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Liu, X.; Liu, L.; Deng, H.; Zhang, J.; Xu, Q.; Cen, B.; Ji, A. Regulation of lncRNA expression. Cell Mol. Biol. Lett. 2014, 19, 561–575. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Kim, J.; Gorospe, M. Long noncoding RNA turnover. Biochimie 2015, 117, 15–21. [Google Scholar] [CrossRef]
- Zheng, Y.; Cai, X.; Bradley, J.E. microRNAs in parasites and parasite infection. RNA Biol. 2013, 10, 371–379. [Google Scholar] [CrossRef]
- Britton, C.; Winter, A.D.; Gillan, V.; Devaney, E. microRNAs of parasitic helminths-Identification, characterization and potential as drug targets. Int. J. Parasitol. Drugs Drug. Resist. 2014, 4, 85–94. [Google Scholar] [CrossRef]
- Tritten, L.; Burkman, E.; Moorhead, A.; Satti, M.; Geary, J.; Mackenzie, C.; Geary, T. Detection of circulating parasite-derived microRNAs in filarial infections. PLoS Negl. Trop. Dis. 2014, 8, e2971. [Google Scholar] [CrossRef]
- Marks, N.D.; Winter, A.D.; Gu, H.Y.; Maitland, K.; Gillan, V.; Ambroz, M.; Martinelli, A.; Laing, R.; MacLellan, R.; Towne, J.; et al. Profiling microRNAs through development of the parasitic nematode Haemonchus identifies nematode-specific miRNAs that suppress larval development. Sci. Rep. 2019, 9, 17594. [Google Scholar] [CrossRef]
- Meningher, T.; Barsheshet, Y.; Ofir-Birin, Y.; Gold, D.; Brant, B.; Dekel, E.; Sidi, Y.; Schwartz, E.; Regev-Rudzki, N.; Avni, O.; et al. Schistosomal extracellular vesicle-enclosed miRNAs modulate host T helper cell differentiation. EMBO Rep. 2020, 21, e47882. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, L.; Wang, J.; Qiu, L.; Chen, Y.; Davis, R.E.; Cheng, G. Schistosoma japonicum extracellular vesicle miRNA cargo regulates host macrophage functions facilitating parasitism. PLoS Pathog. 2019, 15, e1007817. [Google Scholar] [CrossRef] [PubMed]
- Leija-Montoya, A.G.; Gonzalez-Ramirez, J.; Martinez-Coronilla, G.; Mejia-Leon, M.E.; Isiordia-Espinoza, M.; Sanchez-Munoz, F.; Chavez-Cortez, E.G.; Pitones-Rubio, V.; Serafin-Higuera, N. Roles of microRNAs and Long Non-Coding RNAs Encoded by Parasitic Helminths in Human Carcinogenesis. Int J. Mol. Sci 2022, 23, 8173. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.C.; Xu, M.J.; Alasaad, S.; Song, H.Q.; Peng, L.; Tao, J.P.; Zhu, X.Q. Comparative analysis of microRNA profiles between adult Ascaris lumbricoides and Ascaris suum. BMC Vet. Res. 2014, 10, 99. [Google Scholar] [CrossRef]
- Fontenla, S.; Rinaldi, G.; Smircich, P.; Tort, J.F. Conservation and diversification of small RNA pathways within flatworms. BMC Evol. Biol. 2017, 17, 215. [Google Scholar] [CrossRef]
- Macchiaroli, N.; Cucher, M.; Kamenetzky, L.; Yones, C.; Bugnon, L.; Berriman, M.; Olson, P.D.; Rosenzvit, M.C. Identification and expression profiling of microRNAs in Hymenolepis. Int. J. Parasitol. 2019, 49, 211–223. [Google Scholar] [CrossRef]
- Holz, A.; Streit, A. Gain and Loss of Small RNA Classes-Characterization of Small RNAs in the Parasitic Nematode Family Strongyloididae. Genome Biol. Evol. 2017, 9, 2826–2843. [Google Scholar] [CrossRef]
- Santos, L.N.; Silva, E.S.; Santos, A.S.; De Sa, P.H.; Ramos, R.T.; Silva, A.; Cooper, P.J.; Barreto, M.L.; Loureiro, S.; Pinheiro, C.S.; et al. De novo assembly and characterization of the Trichuris trichiura adult worm transcriptome using Ion Torrent sequencing. Acta Tropica. 2016, 159, 132–141. [Google Scholar] [CrossRef]
- Azlan, A.; Halim, M.A.; Azzam, G. Genome-wide identification and characterization of long intergenic noncoding RNAs in the regenerative flatworm Macrostomum lignano. Genomics 2020, 112, 1273–1281. [Google Scholar] [CrossRef]
- Ross, E.; Blair, D.; Guerrero-Hernandez, C.; Sanchez Alvarado, A. Comparative and Transcriptome Analyses Uncover Key Aspects of Coding- and Long Noncoding RNAs in Flatworm Mitochondrial Genomes. G3 2016, 6, 1191–1200. [Google Scholar] [CrossRef] [PubMed]
- Rodelsperger, C.; Menden, K.; Serobyan, V.; Witte, H.; Baskaran, P. First insights into the nature and evolution of antisense transcription in nematodes. BMC Evol. Biol. 2016, 16, 165. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Chen, H.; Dzakah, E.E.; Yu, B.; Wang, X.; Fu, T.; Li, J.; Liu, L.; Fang, S.; Liu, W.; et al. Systematic evaluation of C. elegans lincRNAs with CRISPR knockout mutants. Genome Biol. 2019, 20, 7. [Google Scholar] [CrossRef] [PubMed]
- Amaral, M.S.; Santos, D.W.; Pereira, A.S.A.; Tahira, A.C.; Malvezzi, J.V.M.; Miyasato, P.A.; Freitas, R.P.; Kalil, J.; Tjon Kon Fat, E.M.; de Dood, C.J.; et al. Rhesus macaques self-curing from a schistosome infection can display complete immunity to challenge. Nat. Commun. 2021, 12, 6181. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Analysis Tool for High Throughput Sequencing Data, 0.11.9; 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. (accessed on 1 December 2023).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lun, A.T.; Smyth, G.K. From reads to genes to pathways: Differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res 2016, 5, 1438. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Liu, R.; Holik, A.Z.; Su, S.; Jansz, N.; Chen, K.; Leong, H.S.; Blewitt, M.E.; Asselin-Labat, M.L.; Smyth, G.K.; Ritchie, M.E. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 2015, 43, e97. [Google Scholar] [CrossRef] [PubMed]
- Leek, J.T. svaseq: Removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res. 2014, 42, e161. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, K.C.G.; Desgagné-Penix, I.; Germain, H. Custom selected reference genes outperform pre-defined reference genes in transcriptomic analysis. BMC Genom. 2020, 21, 35. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genomics 2023, 23, 125. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Pinheiro, J.; Bates, D. Mixed-Effects Models in S and S-PLUS; Springer: New York, NY, USA, 2000. [Google Scholar]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.; EISPACK_authors; Heisterkamp, S.; Van_Willigen, B.; Ranke, J.; R_Core_Team. nlme: Linear and Nonlinear Mixed Effects Models. 2023. Available online: https://CRAN.R-project.org/package=nlme (accessed on 1 December 2023).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014, 15, 550. [Google Scholar] [CrossRef]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Vilo, J.; Peterson, H. gprofiler2—An R package for gene list functional enrichment analysis and namespace conversion toolset g:Profiler. F1000Res 2020, 9, ELIXIR-709. [Google Scholar] [CrossRef]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jardim Poli, P.; Fischer-Carvalho, A.; Tahira, A.C.; Chan, J.D.; Verjovski-Almeida, S.; Sena Amaral, M. Long Non-Coding RNA Levels Are Modulated in Schistosoma mansoni following In Vivo Praziquantel Exposure. Non-Coding RNA 2024, 10, 27. https://doi.org/10.3390/ncrna10020027
Jardim Poli P, Fischer-Carvalho A, Tahira AC, Chan JD, Verjovski-Almeida S, Sena Amaral M. Long Non-Coding RNA Levels Are Modulated in Schistosoma mansoni following In Vivo Praziquantel Exposure. Non-Coding RNA. 2024; 10(2):27. https://doi.org/10.3390/ncrna10020027
Chicago/Turabian StyleJardim Poli, Pedro, Agatha Fischer-Carvalho, Ana Carolina Tahira, John D. Chan, Sergio Verjovski-Almeida, and Murilo Sena Amaral. 2024. "Long Non-Coding RNA Levels Are Modulated in Schistosoma mansoni following In Vivo Praziquantel Exposure" Non-Coding RNA 10, no. 2: 27. https://doi.org/10.3390/ncrna10020027
APA StyleJardim Poli, P., Fischer-Carvalho, A., Tahira, A. C., Chan, J. D., Verjovski-Almeida, S., & Sena Amaral, M. (2024). Long Non-Coding RNA Levels Are Modulated in Schistosoma mansoni following In Vivo Praziquantel Exposure. Non-Coding RNA, 10(2), 27. https://doi.org/10.3390/ncrna10020027





