Incorporation of Natural and Recombinant Collagen Proteins within Fmoc-Based Self-Assembling Peptide Hydrogels
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterization of the Proteins Used in Hydrogel Modification
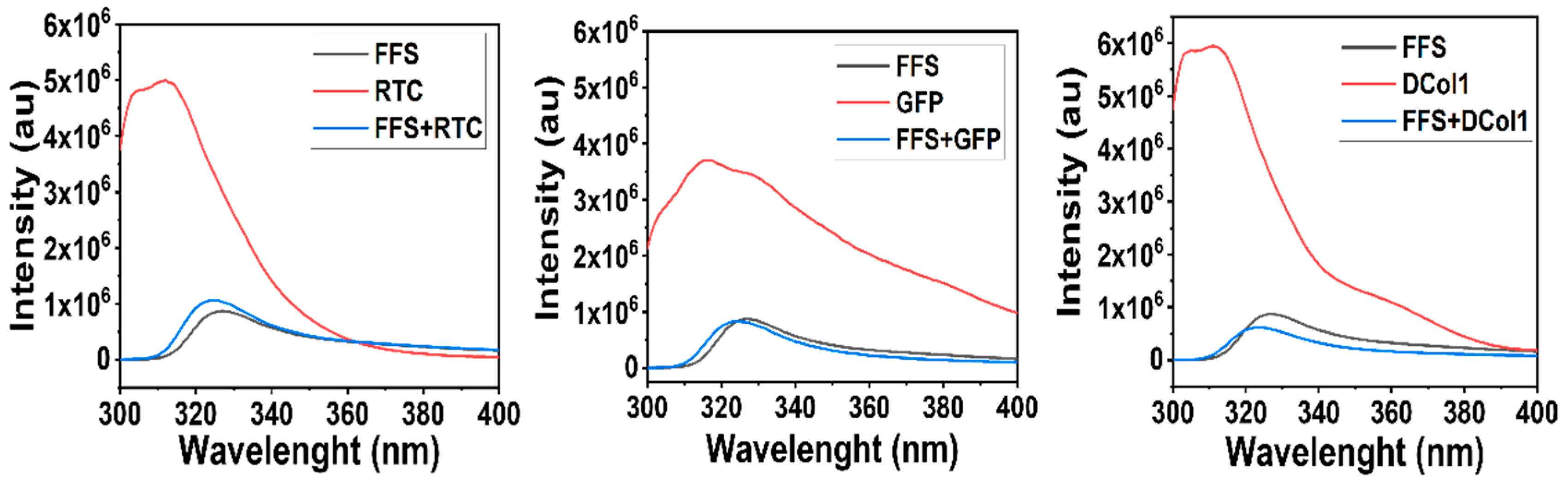
2.2. Protein Incorporation into Self-Assembly Peptide Hydrogels
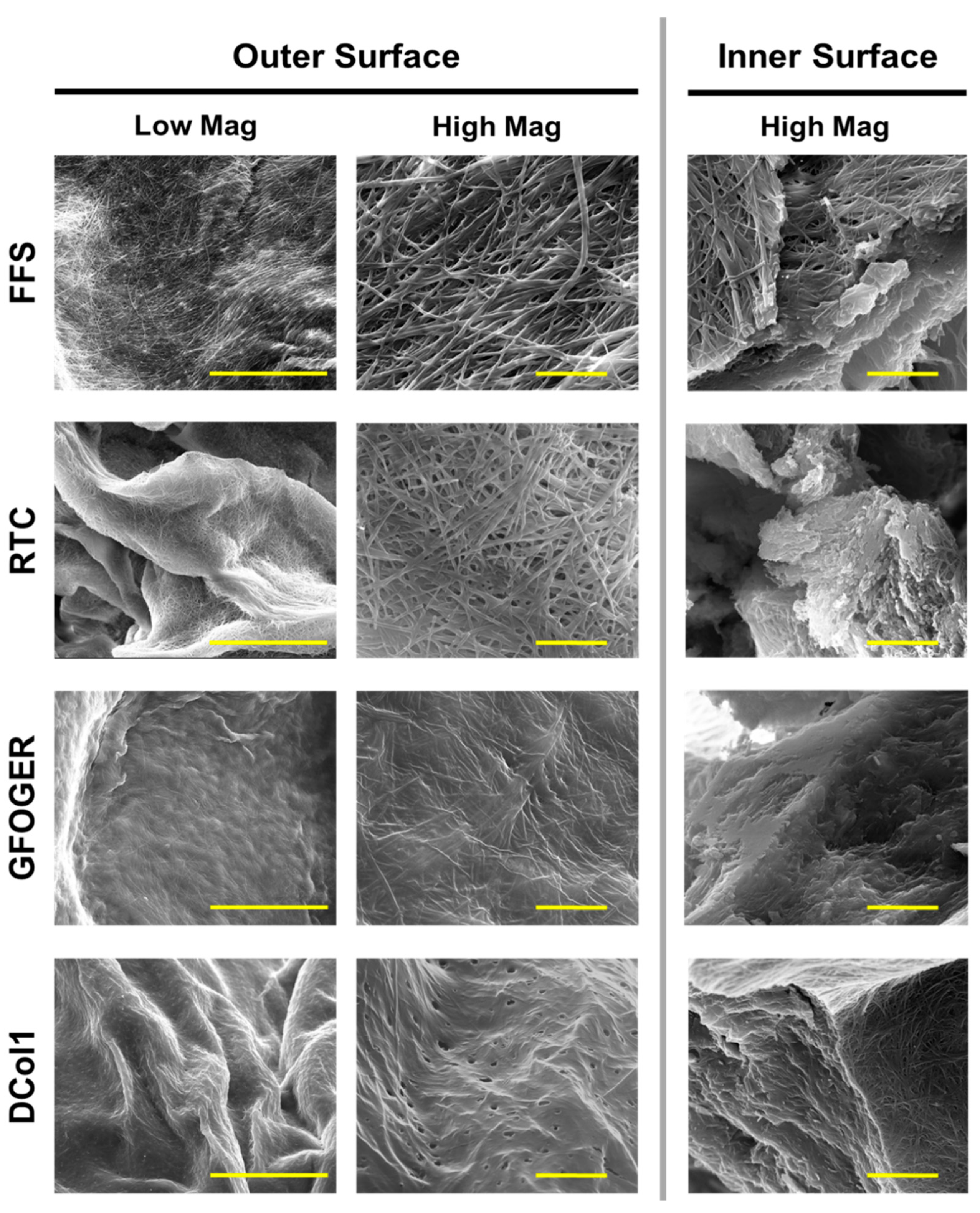
2.3. Hydrogels Microstrusture
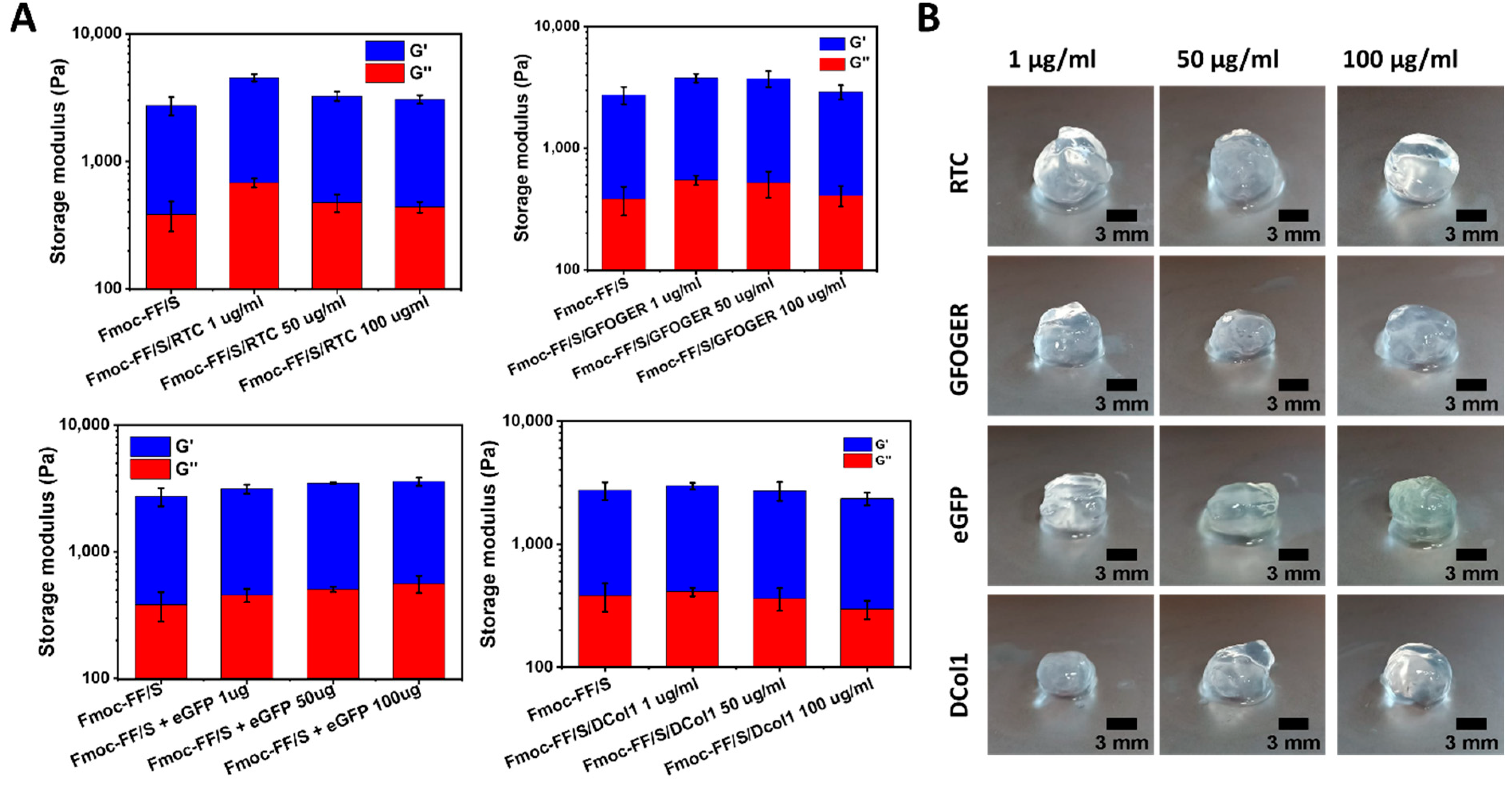
2.4. Mechanical Characterization of the Hydrogels
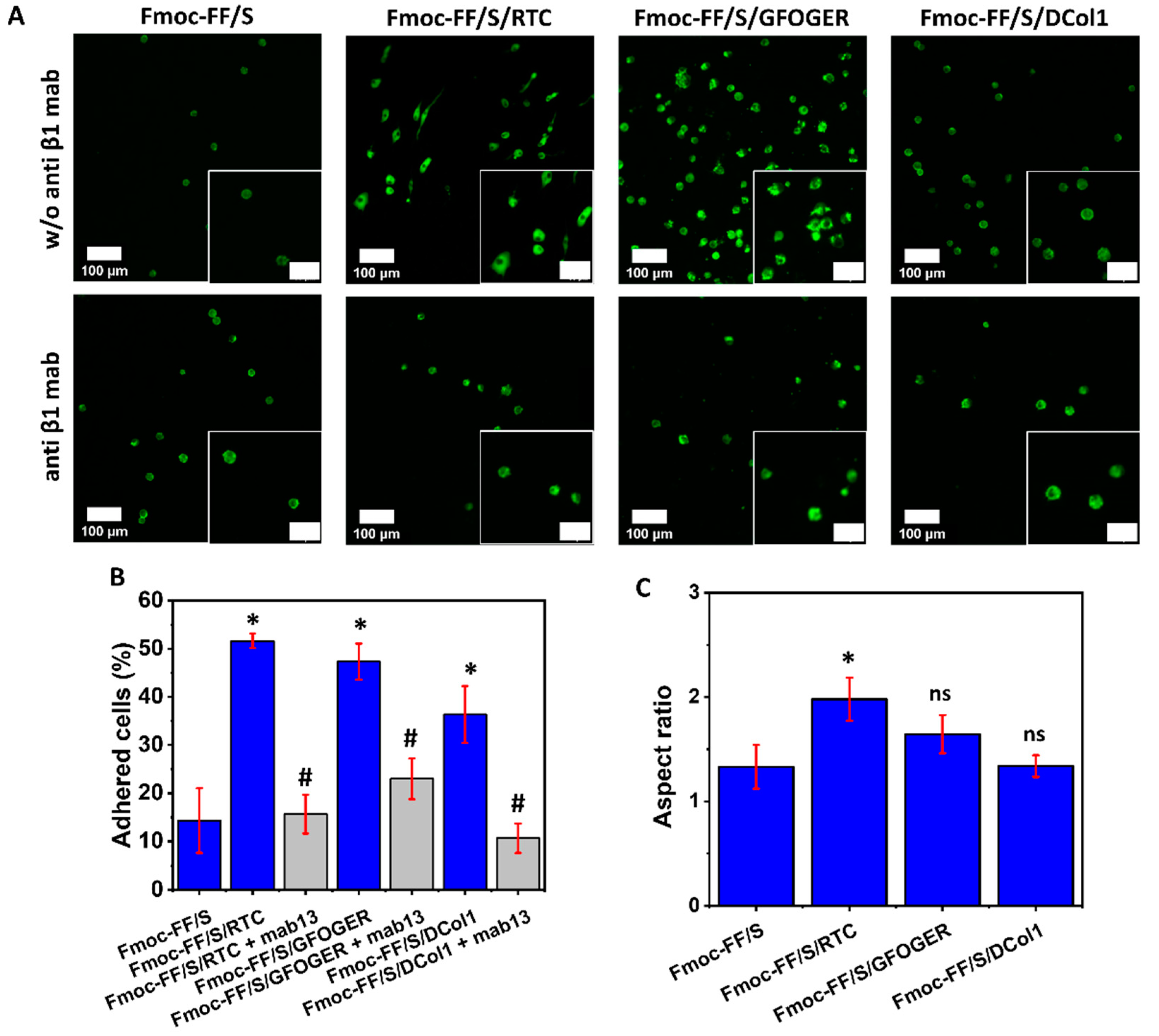
2.5. Collagen-Modified Hydrogels as Platform for Cell Culture
3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Recombinant Collagen Design and Purification
4.3. SDS-PAGE
4.4. Circular Dichroism Spectroscopy
4.5. Hydrogel Modification Protocol
4.6. Fluorescence Spectroscopy
4.7. Scanning Electron Microscopy (SEM)
4.8. Mechanical Properties of the Hydrogels
4.9. Hydrogel Cell Adhesion and Spreading
4.10. Integrin-Dependent Cell Adhesion Assay
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Ahmed, E.M. Hydrogel: Preparation, characterization, and applications: A review. J. Adv. Res. 2015, 6, 105–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mann, S.; Webb, J.; Williams, R.J.P.; Falini, G.; Albeck, S.; Weiner, S.; Addadi, L.; Aizenberg, J.; Hanson, J.; Koetzle, T.F.; et al. Porphyrin Amphiphiles as Templates for the Nucleation of Calcium Carbonate. 1987. Available online: https://pubs.acs.org/sharingguidelines (accessed on 7 February 2022).
- Zhang, S. Fabrication of novel biomaterials through molecular self-assembly. Nat. Biotechnol. 2003, 21, 1171–1178. [Google Scholar] [CrossRef] [PubMed]
- Guler, M.O.; Hsu, L.; Soukasene, S.; Harrington, D.A.; Hulvat, J.F.; Stupp, S.I. Presentation of RGDS Epitopes on Self-Assembled Nanofibers of Branched Peptide Amphiphiles. Biomacromolecules 2006, 7, 1855–1863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Storrie, H.; Guler, M.O.; Abu-Amara, S.N.; Volberg, T.; Rao, M.; Geiger, B.; Stupp, S.I. Supramolecular crafting of cell adhesion. Biomaterials 2007, 28, 4608–4618. [Google Scholar] [CrossRef] [PubMed]
- Benoit, D.; Schwartz, M.; Durney, A.R.; Anseth, K.S. Small functional groups for controlled differentiation of hydrogel-encapsulated human mesenchymal stem cells. Nat. Mater. 2008, 7, 816–823. [Google Scholar] [CrossRef] [Green Version]
- Lutolf, M.P.; Lauer-Fields, J.L.; Schmoekel, H.G.; Metters, A.T.; Weber, F.E.; Fields, G.B.; Hubbell, J.A. Synthetic matrix metalloproteinase-sensitive hydrogels for the conduction of tissue regeneration: Engineering cell-invasion characteristics. Proc. Natl. Acad. Sci. USA 2003, 100, 5413–5418. [Google Scholar] [CrossRef] [Green Version]
- Salinas, C.N.; Anseth, K.S. The influence of the RGD peptide motif and its contextual presentation in PEG gels on human mesenchymal stem cell viability. J. Tissue Eng. Regen. Med. 2008, 2, 296–304. [Google Scholar] [CrossRef]
- Ligorio, C.; Zhou, M.; Wychowaniec, J.K.; Zhu, X.; Bartlam, C.; Miller, A.F.; Vijayaraghavan, A.; Hoyland, J.A.; Saiani, A. Graphene oxide containing self-assembling peptide hybrid hydrogels as a potential 3D injectable cell delivery platform for intervertebral disc repair applications. Acta Biomater. 2019, 92, 92–103. [Google Scholar] [CrossRef]
- Bai, X.; Gao, M.; Syed, S.; Zhuang, J.; Xu, X.; Zhang, X.-Q. Bioactive hydrogels for bone regeneration. Bioact. Mater. 2018, 3, 401–417. [Google Scholar] [CrossRef]
- Xavier, J.R.; Thakur, T.; Desai, P.; Jaiswal, M.K.; Sears, N.; Cosgriff-Hernandez, E.; Kaunas, R.; Gaharwar, A.K. Bioactive nanoengineered hydrogels for bone tissue engineering: A growth-factor-free approach. ACS Nano 2015, 9, 3109–3118. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, J.; Fan, T.; Zhang, Y.; Zhao, Y.; Shi, X.; Zhang, Q. Biomimetic mineralized hierarchical hybrid scaffolds based on in situ synthesis of nano-hydroxyapatite/chitosan/chondroitin sulfate/hyaluronic acid for bone tissue engineering. Colloids Surf. B Biointerfaces 2017, 157, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Vitale, M.; Ligorio, C.; McAvan, B.; Hodson, N.W.; Allan, C.; Richardson, S.M.; Hoyland, J.A.; Bella, J. Hydroxyapatite-decorated Fmoc-hydrogel as a bone-mimicking substrate for osteoclast differentiation and culture. Acta Biomater. 2021, 138, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Ligorio, C.; O’Brien, M.; Hodson, N.W.; Mironov, A.; Iliut, M.; Miller, A.F.; Vijayaraghavan, A.; Hoyland, J.A.; Saiani, A. TGF-β3-loaded graphene oxide—Self-assembling peptide hybrid hydrogels as functional 3D scaffolds for the regeneration of the nucleus pulposus. Acta Biomater. 2021, 127, 116–130. [Google Scholar] [CrossRef] [PubMed]
- Ligorio, C.; Vijayaraghavan, A.; Hoyland, J.A.; Saiani, A. Acidic and basic self-assembling peptide and peptide-graphene oxide hydrogels: Characterisation and effect on encapsulated nucleus pulposus cells. Acta Biomater. 2022, 92, 92–103. [Google Scholar] [CrossRef]
- Gordon, M.K.; Hahn, R.A. Collagens. Cell Tissue Res. 2010, 339, 247–257. [Google Scholar] [CrossRef]
- Kadler, K.E.; Baldock, C.; Bella, J.; Boot-Handford, R.P. Collagens at a glance. Cell Sci. Glance 2005, 247, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Wojtowicz, A.M.; Shekaran, A.; Oest, M.; Dupont, K.M.; Templeman, K.L.; Hutmacher, D.W.; Guldberg, R.E.; García, A.J. Coating of biomaterial scaffolds with the collagen-mimetic peptide GFOGER for bone defect repair. Biomaterials 2010, 31, 2574–2582. [Google Scholar] [CrossRef] [Green Version]
- Chiu, L.-H.; Chen, S.-C.; Wu, K.-C.; Yang, C.-B.; Fang, C.-L.; Lai, W.-F.T.; Tsai, Y.-H. Differential effect of ECM molcules on re-expression of cartilaginous markers in near quiescent human chondrocytes. J. Cell. Physiol. 2010, 226, 1981–1988. [Google Scholar] [CrossRef]
- Drury, J.L.; Mooney, D.J. Hydrogels for tissue engineering: Scaffold design variables and applications. Biomaterials 2003, 24, 4337–4351. [Google Scholar] [CrossRef]
- Peng, Y.Y.; Glattauer, V.; Ramshaw, J.A.M.; Werkmeister, J.A. Evaluation of the immunogenicity and cell compatibility of avian collagen for biomedical applications. J. Biomed. Mater. Res. Part A 2009, 93, 1235–1244. [Google Scholar] [CrossRef]
- Terzi, A.; Gallo, N.; Bettini, S.; Sibillano, T.; Altamura, D.; Madaghiele, M.; De Caro, L.; Valli, L.; Salvatore, L.; Sannino, A.; et al. Sub and supramolecular X-ray characterization of engineered tissues from equine tendons, bovine dermis and fish skin type-I collagen. Macromol. Biosci. 2020, 20, 2000017. [Google Scholar] [CrossRef] [PubMed]
- Toman, P.D.; Chisholm, G.; McMullin, H.; Giere, L.M.; Olsen, D.R.; Kovach, R.J.; Leigh, S.D.; Fong, B.E.; Chang, R.; Daniels, G.A.; et al. Production of Recombinant Human Type I Procollagen Trimers Using a Four-gene Expression System in the Yeast Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 23303–23309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brodsky, B.; Kaplan, D.L. Shining Light on Collagen: Expressing Collagen in Plants. Tissue Eng. Part A 2013, 19, 1499–1501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruggiero, F.; Koch, M. Making recombinant extracellular matrix proteins. Methods 2008, 45, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Reches, M.; Gazit, E. Casting Metal Nanowires Within Discrete Self-Assembled Peptide Nanotubes. Science 2003, 300, 625–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayawarna, V.; Ali, M.; Jowitt, T.; Miller, A.F.; Saiani, A.; Gough, J.E.; Ulijn, R.V. Nanostructured Hydrogels for Three-Dimensional Cell Culture Through Self-Assembly of Fluorenylmethoxycarbonyl-Dipeptides. Adv. Mater. 2006, 18, 611–614. [Google Scholar] [CrossRef]
- Diaferia, C.; Morelli, G.; Accardo, A. Fmoc-diphenylalanine as a suitable building block for the preparation of hybrid materials and their potential applications. J. Mater. Chem. B 2019, 7, 5142–5155. [Google Scholar] [CrossRef]
- Remington, S.J. Green fluorescent protein: A perspective. Protein Sci. 2011, 20, 1509–1519. [Google Scholar] [CrossRef] [Green Version]
- Elsdale, T.; Bard, J. Collagen substrata for studies on cell behavior. J. Cell Biol. 1972, 54, 626–637. [Google Scholar] [CrossRef]
- Rajan, N.; Habermehl, J.; Coté, M.-F.; Doillon, C.J.; Mantovani, D. Preparation of ready-to-use, storable and reconstituted type I collagen from rat tail tendon for tissue engineering applications. Nat. Protoc. 2006, 1, 2753–2758. [Google Scholar] [CrossRef]
- Knight, C.G.; Morton, L.F.; Onley, D.J.; Peachey, A.R.; Messent, A.J.; Smethurst, P.A.; Tuckwell, D.S.; Farndale, R.W.; Barnes, M.J. Identification in Collagen Type I of an Integrin α2β1-binding Site Containing an Essential GER Sequence. J. Biol. Chem. 1998, 273, 33287–33294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenfield, N.J. Analysis of the kinetics of folding of proteins and peptides using circular dichroism. Nat. Protoc. 2006, 1, 2891–2899. [Google Scholar] [CrossRef] [Green Version]
- Drzewiecki, K.E.; Grisham, D.R.; Parmar, A.S.; Nanda, V.; Shreiber, D.I. Circular Dichroism Spectroscopy of Collagen Fibrillogenesis: A New Use for an Old Technique. Biophys. J. 2016, 111, 2377–2386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelly, S.; Price, N. The Use of Circular Dichroism in the Investigation of Protein Structure and Function. Curr. Protein Pept. Sci. 2000, 1, 349–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sreerama, N.; Woody, R.W. Poly (Pro) II helixes in globular proteins: Identification and circular dichroic analysis. Biochemistry 1994, 33, 10022–10025. [Google Scholar] [CrossRef]
- Yoon, H.H.; Bhang, S.H.; Kim, T.; Yu, T.; Hyeon, T.; Kim, B.-S. Dual Roles of Graphene Oxide in Chondrogenic Differentiation of Adult Stem Cells: Cell-Adhesion Substrate and Growth Factor-Delivery Carrier. Adv. Funct. Mater. 2014, 24, 6455–6464. [Google Scholar] [CrossRef]
- Capito, R.M.; Azevedo, H.S.; Velichko, Y.S.; Mata, A.; Stupp, S.I. Self-Assembly of Large and Small Molecules into Hierarchically Ordered Sacs and Membranes. Science 2008, 319, 1812–1816. [Google Scholar] [CrossRef]
- Teale, F.W.J.; Weber, G. Ultraviolet fluorescence of the aromatic amino acids. Biochem. J. 1957, 65, 476–482. [Google Scholar] [CrossRef]
- Sassi, A.P.; Blanch, H.W.; Prausnitz, J.M. Phase equilibria for aqueous protein/polyelectrolyte gel systems. AIChE J. 1996, 42, 2335–2353. [Google Scholar] [CrossRef] [Green Version]
- Inostroza-Brito, K.E.; Collin, E.; Siton-Mendelson, O.; Smith, K.; Monge-Marcet, A.; Ferreira, D.S.; Rodriguez, R.P.; Alonso, M.; Rodriguez-Cabello, J.C.; Reis, R.L.; et al. Co-assembly, spatiotemporal control and morphogenesis of a hybrid protein–peptide system. Nat. Chem. 2015, 7, 897–904. [Google Scholar] [CrossRef]
- Tuckwell, D.S.; Reid, K.B.M.; Barnes, M.J.; Humphries, M.J. The A-Domain of Integrin alpha2 Binds Specifically to a Range of Collagens but is not a General Receptor for the Collagenous Motif. J. Biol. Inorg. Chem. 1996, 241, 732–739. [Google Scholar] [CrossRef] [PubMed]
- Tuckwell, D.; Calderwood, D.A.; Green, L.J.; Humphries, M. Integrin alpha 2 I-domain is a binding site for collagens. J. Cell Sci. 1995, 108, 1629–1637. [Google Scholar] [CrossRef] [PubMed]
- Shimomura, O.; Johnson, F.H.; Saiga, Y. Extraction, Purification and Properties of Aequorin, a Bioluminescent Protein from the Luminous Hydromedusan, Aequorea. J. Cell. Comp. Physiol. 1962, 59, 223–239. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, K.; Pan, Z.; Guan, E.; Ge, S.; Liu, Y.; Nakamura, T.; Ren, X.-D.; Rafailovich, M.; Clark, R.A. Cell adaptation to a physiologically relevant ECM mimic with different viscoelastic properties. Biomaterials 2007, 28, 671–679. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, N.; McKillop, T.J.; Jowitt, T.; Howard, M.; Davies, H.; Holmes, D.F.; Roberts, I.S.; Bella, J. Collagen-like Proteins in Pathogenic, E. coli Strains. PLoS ONE 2012, 7, e37872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuckwell, D.S.; Smith, L.; Korda, M.; Askari, J.A.; Santoso, S.; Barnes, M.J.; Farndale, R.W.; Humphries, M.J. Monoclonal antibodies identify residues 199–216 of the integrin alpha2 vWFA domain as a functionally important region within alpha2beta1. Biochem. J. 2000, 350, 485–493. [Google Scholar] [CrossRef]
- Gullberg, D. I Domain Integrins. In Advances in Experimental Medicine and Biology; Springer: Dordrecht, The Netherland, 2014; Volume 819, pp. 41–60. [Google Scholar] [CrossRef]
- Ingber, D.E. Cellular mechanotransduction: Putting all the pieces together again. FASEB J. 2006, 20, 811–827. [Google Scholar] [CrossRef]
- Caliari, S.R.; Burdick, J.A. A practical guide to hydrogels for cell culture. Nat. Methods 2016, 13, 405–414. [Google Scholar] [CrossRef] [Green Version]
- Slaughter, B.V.; Fisher, O.Z. Hydrogels in regenerative medicine. Adv Mater. 2009, 21, 3307–3329. [Google Scholar] [CrossRef] [Green Version]
- Knight, C.G.; Morton, L.F.; Peachey, A.R.; Tuckwell, D.S.; Farndale, R.W.; Barnes, M.J. The collagen-binding a-domains of integrins α1/β1 and α2/β1 recognize the same specific amino acid sequence, GFOGER, in native (triple-helical) collagens. J. Biol. Chem. 2000, 275, 35–40. [Google Scholar] [CrossRef] [Green Version]
- John, T. Eyes. Ph.D. Thesis, University of Manchester, Manchester, UK, 2014. [Google Scholar]
- ProteoGenix. Available online: https://www.proteogenix.science/ (accessed on 18 March 2022).
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Humphries, M.J. Cell-substrate adhesion assays. Curr. Protoc. Cell Biol. 1998, 9, 9.1.1–9.1.11. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- OriginLab—Origin and OriginPro—Data Analysis and Graphing Software. Available online: https://www.originlab.com/ (accessed on 9 September 2021).


| Molecule | Sequence/Access IDs (Integrin Binding Sites in Bold Type) | Amino Acids (aa) | Mw (kDa) | Isoelectric Point (pI) |
|---|---|---|---|---|
| Fmoc-FF/S hydrogel Fmoc-FF peptide Fmoc-S peptide | Fmoc-FF Fmoc-S | 2 1 | 0.53 0.33 | 7.81 7.81 |
| Rat tail collagen 1 α1 (I) chain α2 (I) chain | P02454, NP_445756 P02466, NP_445808 | 1056 3 1040 3 | 300 2 | 9.52 |
| GFOGER peptide | Ac-GPCGPPGPPGPPGPPGPPGFOGERGPPGPPGPPGPPGPPGPC-NH2 | 42 | 11.2 2 | 6.96 |
| DCol1 recombinant protein | MGSHHHHHHSGLVPRGSGPPGPPGPQGPAGPRGEPGPAGPKGEPGPAGPPGFPGERGPPGPQGPAGPIGPKGEPGPIGPQGPKGDPGETQIRFRLGPASIIETNSHGWFPGTDGALITGLTFLAPKDATRVQVFFQHLQVRFGDGPWQDVKGLDEVGSDTGRTGE | 165 | 50.0 2 | 6.97 |
| eGFP recombinant protein | MGSSHHHHHHSSGLVPRGSHMVSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFICTTGKLPVPWPTLVTTLTYGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTIFFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIKVNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAAGITLGMDELYK | 259 | 29.1 | 6.61 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vitale, M.; Ligorio, C.; Smith, I.P.; Richardson, S.M.; Hoyland, J.A.; Bella, J. Incorporation of Natural and Recombinant Collagen Proteins within Fmoc-Based Self-Assembling Peptide Hydrogels. Gels 2022, 8, 254. https://doi.org/10.3390/gels8050254
Vitale M, Ligorio C, Smith IP, Richardson SM, Hoyland JA, Bella J. Incorporation of Natural and Recombinant Collagen Proteins within Fmoc-Based Self-Assembling Peptide Hydrogels. Gels. 2022; 8(5):254. https://doi.org/10.3390/gels8050254
Chicago/Turabian StyleVitale, Mattia, Cosimo Ligorio, Ian P. Smith, Stephen M. Richardson, Judith A. Hoyland, and Jordi Bella. 2022. "Incorporation of Natural and Recombinant Collagen Proteins within Fmoc-Based Self-Assembling Peptide Hydrogels" Gels 8, no. 5: 254. https://doi.org/10.3390/gels8050254
APA StyleVitale, M., Ligorio, C., Smith, I. P., Richardson, S. M., Hoyland, J. A., & Bella, J. (2022). Incorporation of Natural and Recombinant Collagen Proteins within Fmoc-Based Self-Assembling Peptide Hydrogels. Gels, 8(5), 254. https://doi.org/10.3390/gels8050254






