Protein Kinase PoxMKK1 Regulates Plant-Polysaccharide-Degrading Enzyme Biosynthesis, Mycelial Growth and Conidiation in Penicillium oxalicum
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Media and Growth Conditions
2.2. Yeast-Two Hybrid Assay
2.3. Construction of Gene Deletion Mutant and Its Complementary Strain
2.4. Molecular Manipulation
2.5. RNA Sequencing and RT-qPCR Analysis
2.6. Determination of Enzyme Activity and Protein Concentration
2.7. Phenotypic and Growth Analyses
2.8. Quantification of Conidia Production
2.9. Protein Sequence Analysis
2.10. Accession Numbers
3. Results
3.1. POX07948 Interacting with PoxMK1 Is Required for FPase Production in P. oxalicum
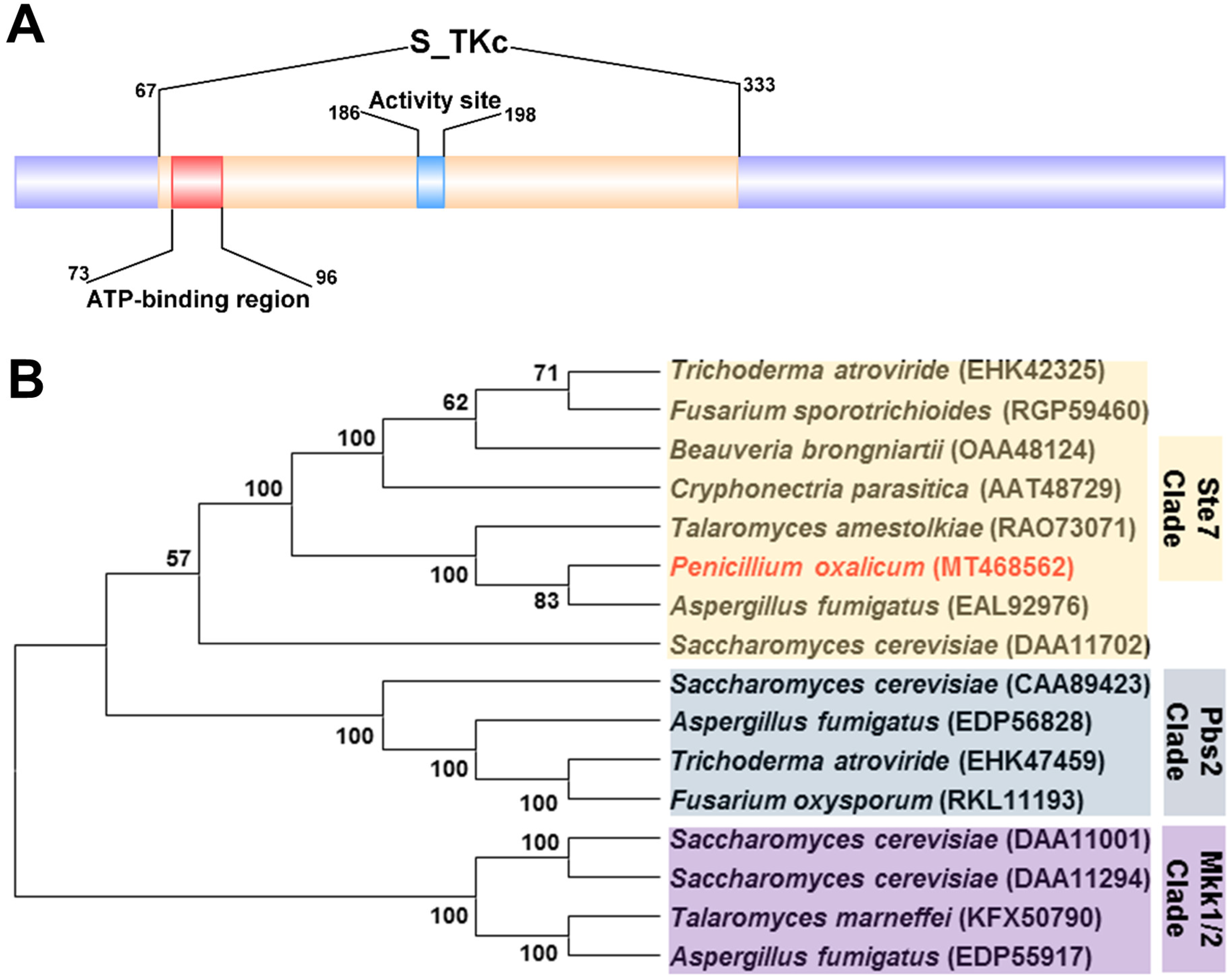
3.2. Characterization and Phylogenetic Analyses of PoxMKK1 in P. oxalicum
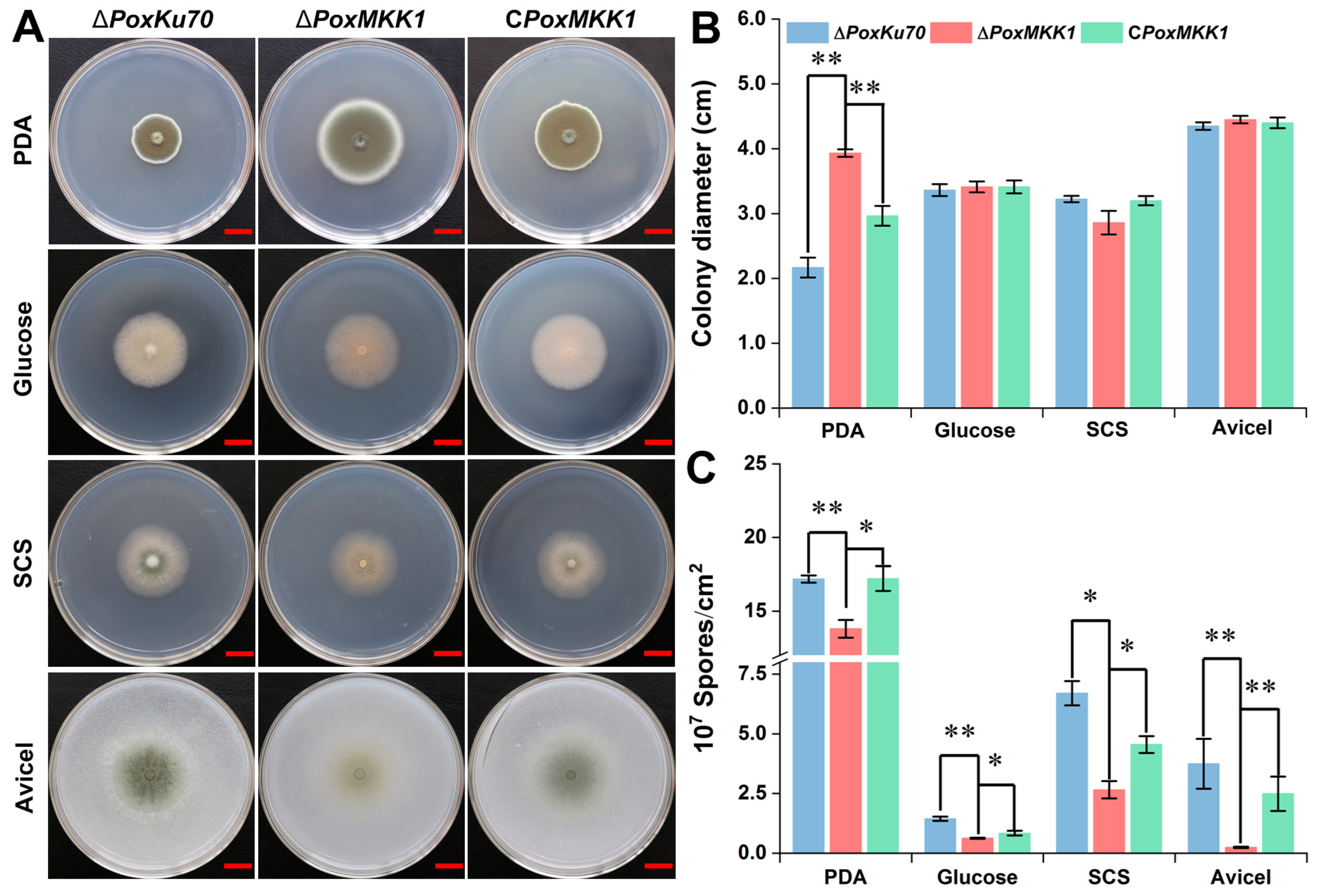
3.3. PoxMKK1 Is Involved in Mycelial Growth and Conidiation in P. oxalicum
3.4. Loss of PoxMKK1 Alters PPDE Production of P. oxalicum under SmF and SSF
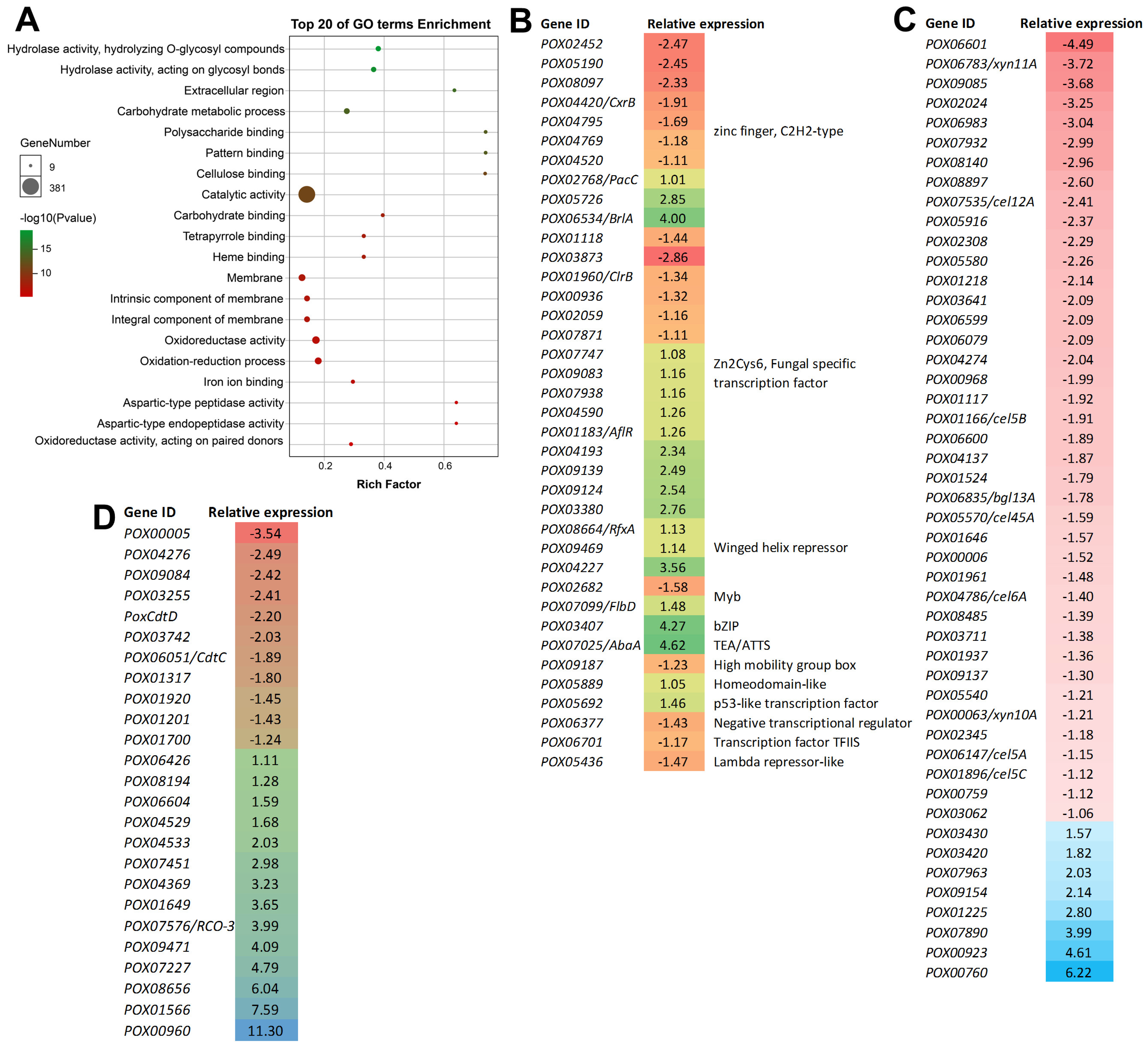
3.5. RNA-seq Analyses Revealed the Global Regulation of PoxMKK1 in P. oxalicum
3.6. Regulation Kinetics of PoxMKK1 on the Expression of Major Genes Encoding PPDE and TFs in P. oxalicum under SmF and SSF
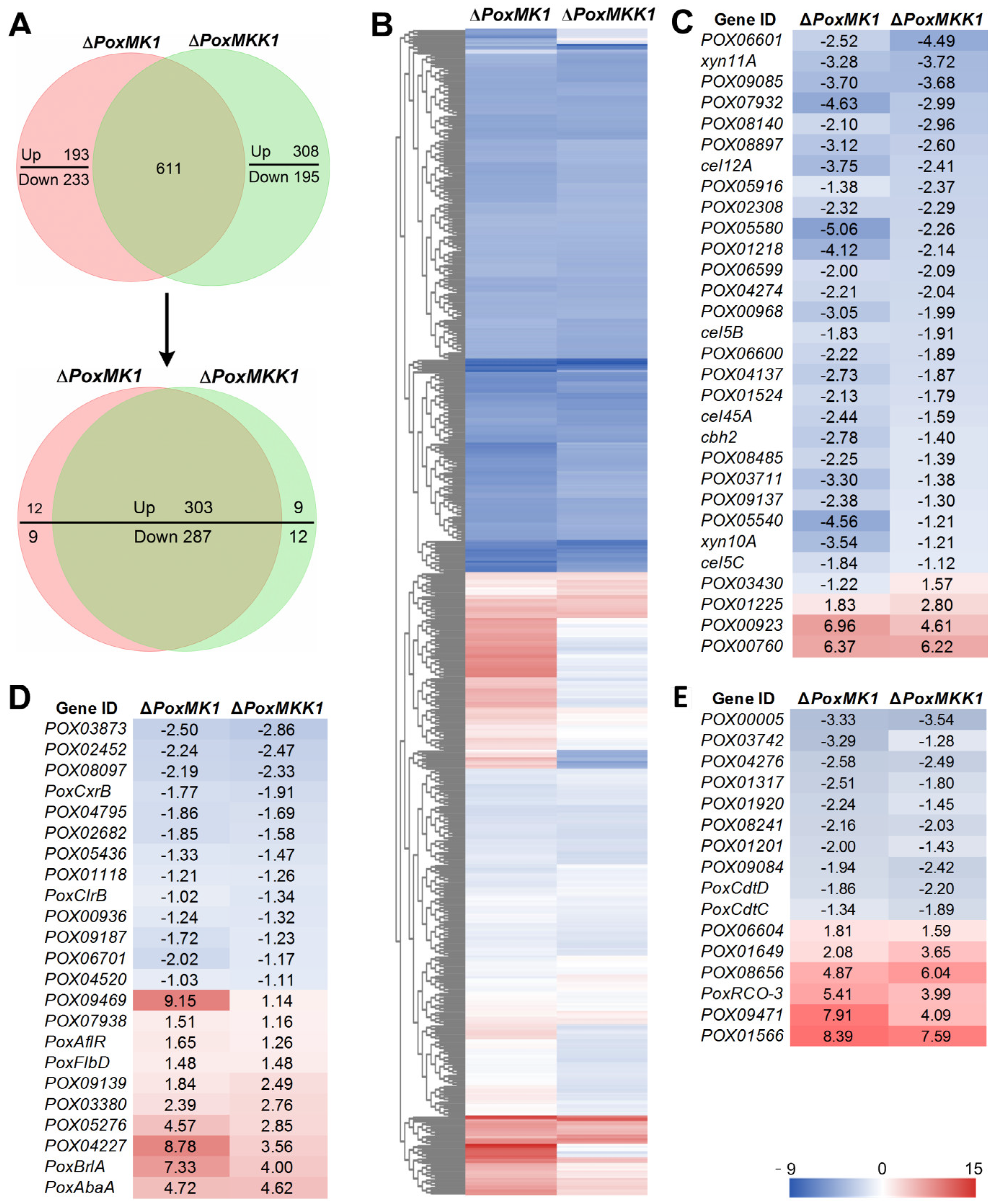
3.7. Comparative Analysis of PoxMK1 and PoxMKK1 Regulons in P. oxalicum
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Vries, R.P.; Mäkelä, M.R. Genomic and Postgenomic Diversity of Fungal Plant Biomass Degradation Approaches. Trends Microbiol. 2020, 6, 487–499. [Google Scholar] [CrossRef]
- Francois, J.M.; Alkim, C.; Morin, N. Engineering Microbial Pathways for Production of Bio-Based Chemicals from Lignocellulosic Sugars: Current Status and Perspectives. Biotechnol. Biofuels 2020, 13, 118. [Google Scholar] [CrossRef]
- Bryant, N.D.; Pu, Y.Q.; Tschaplinski, T.J.; Tuskan, G.A.; Muchero, W.; Kalluri, U.C.; Yoo, C.G.; Ragauskas, A.J. Transgenic Poplar Designed for Biofuels. Trends Plant Sci. 2020, 9, 881–896. [Google Scholar] [CrossRef] [PubMed]
- Benocci, T.; Aguilar-Pontes, M.V.; Zhou, M.M.; Seiboth, B.; de Vries, R.P. Regulators of Plant Biomass Degradation in Ascomycetous Fungi. Biotechnol. Biofuels 2017, 10, 152. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.C.; Wang, S.S.; Li, X.L.; He, Q.; Benz, J.P.; Tian, C.G. STK-12 Acts as a Transcriptional Brake to Control the Expression of Cellulase-Encoding Genes in Neurospora crassa. PLoS Genet. 2019, 11, e1008510. [Google Scholar] [CrossRef]
- Rodriguez-Iglesias, A.; Schmoll, M. Protein Phosphatases Regulate Growth, Development, Cellulases and Secondary Metabolism in Trichoderma reesei. Sci. Rep. 2019, 1, 10995. [Google Scholar] [CrossRef]
- Martínez-Soto, D.; Ruiz-Herrera, J. Functional Analysis of the MAPK Pathways in Fungi. Rev. Iberoam. Micol. 2017, 4, 192–202. [Google Scholar] [CrossRef] [PubMed]
- González-Rubio, G.; Fernández-Acero, T.; Martín, H.; Molina, M. Mitogen-Activated Protein Kinase Phosphatases (Mkps) in Fungal Signaling: Conservation, Function, and Regulation. Int. J. Mol. Sci. 2019, 7, 1709. [Google Scholar] [CrossRef]
- Tong, S.M.; Feng, M.G. Insights into Regulatory Roles of MAPK-Cascaded Pathways in Multiple Stress Responses and Life Cycles of Insect and Nematode Mycopathogens. Appl. Microbiol. Biotechnol. 2019, 2, 577–587. [Google Scholar] [CrossRef]
- Frawley, D.; Bayram, Ö. The Pheromone Response Module, a Mitogen-Activated Protein Kinase Pathway Implicated in the Regulation of Fungal Development, Secondary Metabolism and Pathogenicity. Fungal. Genet. Biol. 2020, 144, 103469. [Google Scholar] [CrossRef]
- Izumitsu, K.; Yoshimi, A.; Kubo, D.; Morita, A.; Saitoh, Y.; Tanaka, C. The MAPKK Kinase Chste11 Regulates Sexual/Asexual Development, Melanization, Pathogenicity, and Adaptation to Oxidative Stress in Cochliobolus heterostrophus. Curr. Genet. 2009, 4, 439–448. [Google Scholar] [CrossRef]
- Wasserstrom, L.; Lengeler, K.B.; Walther, A.; Wendland, J. Molecular Determinants of Sporulation in Ashbya gossypii. Genetics 2013, 1, 87–99. [Google Scholar] [CrossRef]
- Xu, H.J.; Zhang, Q.Q.; Cui, W.J.; Zhang, X.F.; Liu, W.Y.; Zhang, L.; Islam, M.N.; Baek, K.H.; Wang, Y.J. AbSte7, a MAPKK Gene of Alternaria brassicicola, is Involved in Conidiation, Salt/Oxidative Stress, and Pathogenicity. J. Microbiol. Biotechnol. 2016, 7, 1311–1319. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sun, H.H.; Ying, S.H.; Feng, M.G. Characterization of Three Mitogen-Activated Protein Kinase Kinase-Like Proteins in Beauveria bassiana. Fungal. Genet. Biol. 2018, 113, 24–31. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Ge, Q.W.; Cao, Q.C.; Cui, H.F.; Hu, P.; Yu, X.P.; Ye, Z.H. Cloning and Characterization of Two MAPK Genes UeKpp2 and UeKpp6 in Ustilago esculenta. Curr. Microbiol. 2018, 8, 1016–1024. [Google Scholar] [CrossRef]
- Yu, L.; Xiong, D.G.; Han, Z.; Liang, Y.M.; Tian, C.G. The Mitogen-Activated Protein Kinase Gene CcPmk1 is Required for Fungal Growth, Cell Wall Integrity and Pathogenicity in Cytospora chrysosperma. Fungal. Genet. Biol. 2019, 128, 1–13. [Google Scholar] [CrossRef]
- Wang, X.L.; Lu, D.X.; Tian, C.M. Mitogen-Activated Protein Kinase Cascade Cgste50-Ste11-Ste7-Mk1 Regulates Infection-Related Morphogenesis in the Poplar Anthracnose Fungus Colletotrichum gloeosporioides. Microbiol. Res. 2021, 248, 126748. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.F.; Qin, X.F.; Zhu, F.; Zhang, Y.F.; Zhang, X.C.; Hartley, W.; Xue, S.G. Halving Gypsum Dose by Penicillium oxalicum on Alkaline Neutralization and Microbial Community Reconstruction in Bauxite Residue. Chem. Eng. J. 2023, 451, 139008. [Google Scholar] [CrossRef]
- Hao, S.F.; Wang, P.Y.; Ge, F.; Li, F.; Deng, S.Q.; Zhang, D.Y.; Tian, J. Enhanced Lead (Pb) Immobilization in Red Soil by Phosphate Solubilizing Fungi Associated with Tricalcium Phosphate Influencing Microbial Community Composition and Pb Translocation in Lactuca sativa L. J. Hazard Mater. 2022, 424, 127720. [Google Scholar] [CrossRef]
- Li, C.X.; Liu, L.; Zhang, T.; Luo, X.M.; Feng, J.X.; Zhao, S. Three-Dimensional Genome Map of the Filamentous Fungus Penicillium oxalicum. Microbiol. Spectr. 2022, 3, e0212121. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.B.; Liu, G.D.; Li, Z.H.; Qin, Y.Q.; Qu, Y.B.; Song, X. G Protein-cAMP Signaling Pathway Mediated by PGA3 Plays Different Roles in Regulating the Expressions of Amylases and Cellulases in Penicillium decumbens. Fungal Genet. Biol. 2013, 58–59, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.M.; Tian, D.; Zhang, T.; Liao, L.S.; Li, C.X.; Luo, X.M.; Feng, J.X.; Zhao, S. G Protein γ Subunit Modulates Expression of Plant-Biomass-Degrading Enzyme Genes and Mycelial-Development-Related Genes in Penicillium oxalicum. Appl. Microbiol. Biotechnol. 2021, 11, 4675–4691. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Ning, Y.N.; Li, C.X.; Tian, D.; Guo, H.; Pang, X.M.; Luo, X.M.; Zhao, S.; Feng, J.X. A Mitogen-Activated Protein Kinase PoxMK1 Mediates Regulation of the Production of Plant-Biomass-Degrading Enzymes, Vegetative growth, and Pigment Biosynthesis in Penicillium oxalicum. Appl. Microbiol. Biotechnol. 2021, 2, 661–678. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Yan, Y.S.; He, Q.P.; Yang, L.; Yin, X.; Li, C.X.; Mao, L.C.; Liao, L.S.; Huang, J.Q.; Xie, S.B.; et al. Comparative Genomic, Transcriptomic and Secretomic Profiling of Penicillium oxalicum HP7-1 and its Cellulase and Xylanase Hyper-Producing Mutant EU2106, and Identification of Two Novel Regulatory Genes of Cellulase and Xylanase Gene Expression. Biotechnol. Biofuels 2016, 9, 203. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Mai, R.M.; Fang, Q.Q.; Ou, J.F.; Mo, L.X.; Tian, D.; Li, C.X.; Gu, L.S.; Luo, X.M.; Feng, J.X.; et al. Regulatory function of the Novel Transcription Factor CxrC in Penicillium oxalicum. Mol. Microbiol. 2021, 6, 1512–1532. [Google Scholar] [CrossRef]
- Yan, Y.S.; Zhao, S.; Liao, L.S.; He, Q.P.; Xiong, Y.R.; Wang, L.; Li, C.X.; Feng, J.X. Transcriptomic Profiling and Genetic Analyses Reveal Novel Key Regulators of Cellulase and Xylanase Gene Expression in Penicillium oxalicum. Biotechnol. Biofuels 2017, 1, 279. [Google Scholar] [CrossRef]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-Throughput Functional Annotation and Data Mining with the Blast2GO suite. Nucleic Acids Res. 2008, 10, 3420–3435. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 4, 402–408. [Google Scholar] [CrossRef]
- Su, L.H.; Zhao, S.; Jiang, S.X.; Liao, X.Z.; Duan, C.J.; Feng, J.X. Cellulase with High β-Glucosidase Activity by Penicillium oxalicum Under Solid State Fermentation and Its Use in Hydrolysis of Cassava Residue. World J. Microbiol. Biotechnol. 2017, 2, 37. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 21, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol. Biol. Evol. 2018, 6, 1547–1549. [Google Scholar] [CrossRef]
- Li, Z.H.; Yao, G.S.; Wu, R.M.; Gao, L.W.; Kan, Q.B.; Liu, M.; Yang, P.; Liu, G.D.; Qin, Y.Q.; Song, X.; et al. Synergistic and Dose-Controlled Regulation of Cellulase Gene Expression in Penicillium oxalicum. PLoS Genet. 2015, 9, e1005509. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.Y.; Zhao, S.; Zhang, T.; Li, C.X.; Liao, L.S.; Zhang, F.F.; Luo, X.M.; Feng, J.X. The Transcription Factor TpRfx1 is an Essential Regulator of Amylase and Cellulase Gene Expression in Talaromyces pinophilus. Biotechnol. Biofuels 2018, 11, 276. [Google Scholar] [CrossRef] [PubMed]
- Kunitake, E.; Hagiwara, D.; Miyamoto, K.; Kanamaru, K.; Kimura, M.; Kobayashi, T. Regulation of Genes Encoding Cellulolytic Enzymes by Pal-PacC Signaling in Aspergillus nidulans. Appl. Microbiol. Biotechnol. 2016, 8, 3621–3635. [Google Scholar] [CrossRef] [PubMed]
- Li, C.X.; Zhao, S.; Luo, X.M.; Feng, J.X. Weighted Gene Co-Expression Network Analysis Identifies Critical Genes for the Production of Cellulase and Xylanase in Penicillium oxalicum. Front. Microbiol. 2020, 11, 520. [Google Scholar] [CrossRef]
- Zhao, S.; Liu, Q.; Wang, J.X.; Liao, X.Z.; Guo, H.; Li, C.X.; Zhang, F.F.; Liao, L.S.; Luo, X.M.; Feng, J.X. Differential Transcriptomic Profiling of Filamentous Fungus During Solid-State and Submerged Fermentation and Identification of an Essential Regulatory Gene PoxMBF1 that Directly Regulated Cellulase and Xylanase Gene Expression. Biotechnol. Biofuels 2019, 12, 103. [Google Scholar] [CrossRef]
- Qin, Y.Q.; Bao, L.F.; Gao, M.R.; Chen, M.; Lei, Y.F.; Liu, G.D.; Qu, Y.B. Penicillium decumbens BrlA Extensively Regulates Secondary Metabolism and Functionally Associates with the Expression of Cellulase Genes. Appl. Microbiol. Biotechnol. 2013, 24, 10453–10467. [Google Scholar] [CrossRef]
- Park, H.S.; Yu, J.H. Genetic Control of Asexual Sporulation in Filamentous Fungi. Curr. Opin. Microbiol. 2012, 6, 669–677. [Google Scholar] [CrossRef]
- Wang, M.Y.; Zhang, M.L.; Li, L.; Dong, Y.M.; Jiang, Y.; Liu, K.M.; Zhang, R.Q.; Jiang, B.J.; Niu, K.L.; Fang, X. Role of Trichoderma reesei Mitogen-Activated Protein Kinases (MAPKs) in Cellulase Formation. Biotechnol. Biofuels 2017, 10, 99. [Google Scholar] [CrossRef]
- Wang, M.Y.; Dong, Y.M.; Zhao, Q.S.; Wang, F.Z.; Liu, K.M.; Jiang, B.J.; Fang, X. Identification of the Role of a MAP kinase Tmk2 in Hypocrea jecorina (Trichoderma reesei). Sci. Rep. 2014, 4, 6732. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.Y.; Zhao, Q.S.; Yang, J.; Jiang, B.J.; Wang, F.Z.; Liu, K.M.; Fang, X. A Mitogen-Activated Protein Kinase Tmk3 Participates in High Osmolarity Resistance, Cell Wall Integrity Maintenance and Cellulase Production Regulation in Trichoderma reesei. PLoS ONE 2013, 8, e72189. [Google Scholar] [CrossRef]
- de Paula, R.G.; Antoniêto, A.C.C.; Carraro, C.B.; Lopes, D.C.B.; Persinoti, G.F.; Peres, N.T.A.; Martinez-Rossi, N.M.; Silva-Rocha, R.; Silva, R.N. The Duality of the MAPK Signaling Pathway in the Control of Metabolic Processes and Cellulase Production in Trichoderma reesei. Sci. Rep. 2018, 1, 14931. [Google Scholar] [CrossRef]
- Wang, Z.X.; An, N.; Xu, W.; Zhang, W.Q.; Meng, X.F.; Chen, G.J.; Liu, W.F. Functional Characterization of the Upstream Components of the Hog1-Like Kinase Cascade in Hyperosmotic and Carbon Sensing in Trichoderma reesei. Biotechnol. Biofuels 2018, 1, 97. [Google Scholar] [CrossRef] [PubMed]
- de Assis, L.J.; Silva, L.P.; Liu, L.; Schmitt, K.; Valerius, O.; Braus, G.H.; Ries, L.N.A.; Goldman, G.H. The High Osmolarity Glycerol Mitogen-Activated Protein Kinase Regulates Glucose Catabolite Repression in Filamentous Fungi. PLoS Genet. 2020, 8, e1008996. [Google Scholar] [CrossRef]
- Gu, Q.; Chen, Y.; Liu, Y.; Zhang, C.Q.; Ma, Z.H. The Transmembrane Protein FgSho1 Regulates Fungal Development and Pathogenicity via the MAPK Module Ste50-Ste11-Ste7 in Fusarium graminearum. New Phytol. 2015, 1, 315–328. [Google Scholar] [CrossRef]
- Lu, K.; Zhang, M.; Yang, R.; Zhang, M.; Guo, Q.; Baek, K.H.; Xu, H.J. The MAP Kinase Kinase Gene AbSte7 Regulates Multiple Aspects of Alternaria brassicicola Pathogenesis. Plant Pathol. J. 2019, 2, 91–99. [Google Scholar] [CrossRef]
- Cho, Y.; Cramer, R.A., Jr.; Kim, K.H.; Davis, J.; Mitchell, T.K.; Figuli, P.; Pryor, B.M.; Lemasters, E.; Lawrence, C.B. The Fus3/Kss1 MAP Kinase Homolog Amk1 Regulates the Expression of Genes Encoding Hydrolytic Enzymes in Alternaria brassicicola. Fungal Genet. Biol. 2007, 6, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.X.; Xu, L.S.; Liu, J.; Yin, Z.Y.; Gao, X.N.; Feng, H.; Huang, L.L. A Mitogen-Activated Protein Kinase Gene (VmPmk1) Regulates Virulence and Cell Wall Degrading Enzyme Expression in Valsa mali. Microb. Pathog. 2017, 111, 298–306. [Google Scholar] [CrossRef]
- Diaz, A.B.; Blandino, A.; Webb, C.; Caro, I. Modelling of Different Enzyme Productions by Solid-State Fermentation on Several Agro-Industrial Residues. Appl. Microbiol. Biotechnol. 2016, 22, 9555–9566. [Google Scholar] [CrossRef]
- Salgado-Bautista, D.; Volke-Sepúlveda, T.; Figueroa-Martínez, F.; Carrasco-Navarro, U.; Chagolla-López, A.; Favela-Torres, E. Solid-State Fermentation Increases Secretome Complexity in Aspergillus brasiliensis. Fungal Biol. 2020, 8, 723–734. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.T.; Hu, S.; Yu, X.Y.; Jin, L.; Zhu, Y.J.; Jin, F.J. Studies of Cellulose and Starch Utilization and the Regulatory Mechanisms of Related Enzymes in Fungi. Polymers 2020, 3, 530. [Google Scholar] [CrossRef]
- Kunitake, E.; Kobayashi, T. Conservation and Diversity of the Regulators of Cellulolytic Enzyme Genes in Ascomycete Fungi. Curr. Genet. 2017, 6, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, G.; Chen, M.; Li, Z.; Qin, Y.; Qu, Y. Cellodextrin Transporters Play Important Roles in Cellulase Induction in the Cellulolytic Fungus Penicillium oxalicum. Appl. Microbiol. Biotechnol. 2013, 24, 10479–10488. [Google Scholar] [CrossRef]
- Li, J.Y.; Liu, Q.; Li, J.G.; Lin, L.C.; Li, X.L.; Zhang, Y.L.; Tian, C.G. RCO-3 and COL-26 Form an External-to-Internal Module That Regulates the Dual-Affinity Glucose Transport System in Neurospora crassa. Biotechnol. Biofuels 2021, 1, 33. [Google Scholar] [CrossRef] [PubMed]
- Maerz, S.; Ziv, C.; Vogt, N.; Helmstaedt, K.; Cohen, N.; Gorovits, R.; Yarden, O.; Seiler, S. The Nuclear Dbf2-Related Kinase COT1 and the Mitogen-Activated Protein Kinases MAK1 and MAK2 Genetically Interact to Regulate Filamentous Growth, Hyphal Fusion and Sexual Development in Neurospora crassa. Genetics 2008, 3, 1313–1325. [Google Scholar] [CrossRef]
- Frawley, D.; Greco, C.; Oakley, B.; Alhussain, M.M.; Fleming, A.B.; Keller, N.P.; Bayram, Ö. The Tetrameric Pheromone Module SteC-MkkB-MpkB-SteD Regulates Asexual Sporulation, Sclerotia Formation and Aflatoxin Production in Aspergillus flavus. Cell. Microbiol. 2020, 6, e13192. [Google Scholar] [CrossRef]
- Frawley, D.; Stroe, M.C.; Oakley, B.R.; Heinekamp, T.; Strassburger, M.; Fleming, A.B.; Brakhage, A.A.; Bayram, O. The Pheromone Module SteC-MkkB-MpkB-SteD-HamE Regulates Development, Stress Responses and Secondary Metabolism in Aspergillus fumigatus. Front. Microbiol. 2020, 11, 811. [Google Scholar] [CrossRef] [PubMed]
- Priegnitz, B.E.; Brandt, U.; Pahirulzaman, K.A.; Dickschat, J.S.; Fleissner, A. The AngFus3 Mitogen-Activated Protein Kinase Controls Hyphal Differentiation and Secondary Metabolism in Aspergillus niger. Eukaryot. Cell 2015, 6, 602–615. [Google Scholar] [CrossRef]
- Schamber, A.; Leroch, M.; Diwo, J.; Mendgen, K.; Hahn, M. The Role of Mitogen-Activated Protein (MAP) Kinase Signalling Components and the Ste12 Transcription Factor in Germination and Pathogenicity of Botrytis cinerea. Mol. Plant Pathol. 2010, 1, 105–119. [Google Scholar] [CrossRef]
- Moretti, M.; Rossi, M.; Ciuffo, M.; Turina, M. Functional Characterization of the Three Mitogen-Activated Protein Kinase Kinases (MAP2Ks) Present in the Cryphonectria parasitica genome Reveals the Necessity of Cpkk1 and Cpkk2, but not Cpkk3, for Pathogenesis on Chestnut (Castanea spp.). Mol. Plant Pathol. 2014, 5, 500–512. [Google Scholar] [CrossRef]
- Yuan, Q.F.; Chen, M.J.; Yan, Y.Q.; Gu, Q.N.; Huang, J.B.; Zheng, L. ChSte7 is Required for Vegetative Growth and Various Plant Infection Processes in Colletotrichum higginsianum. Biomed. Res. Int. 2016, 2016, 7496569. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, B.; Luo, X.-M.; Zhao, S.; Feng, J.-X. Protein Kinase PoxMKK1 Regulates Plant-Polysaccharide-Degrading Enzyme Biosynthesis, Mycelial Growth and Conidiation in Penicillium oxalicum. J. Fungi 2023, 9, 397. https://doi.org/10.3390/jof9040397
Ma B, Luo X-M, Zhao S, Feng J-X. Protein Kinase PoxMKK1 Regulates Plant-Polysaccharide-Degrading Enzyme Biosynthesis, Mycelial Growth and Conidiation in Penicillium oxalicum. Journal of Fungi. 2023; 9(4):397. https://doi.org/10.3390/jof9040397
Chicago/Turabian StyleMa, Bo, Xue-Mei Luo, Shuai Zhao, and Jia-Xun Feng. 2023. "Protein Kinase PoxMKK1 Regulates Plant-Polysaccharide-Degrading Enzyme Biosynthesis, Mycelial Growth and Conidiation in Penicillium oxalicum" Journal of Fungi 9, no. 4: 397. https://doi.org/10.3390/jof9040397
APA StyleMa, B., Luo, X.-M., Zhao, S., & Feng, J.-X. (2023). Protein Kinase PoxMKK1 Regulates Plant-Polysaccharide-Degrading Enzyme Biosynthesis, Mycelial Growth and Conidiation in Penicillium oxalicum. Journal of Fungi, 9(4), 397. https://doi.org/10.3390/jof9040397






