Small GTPase FoSec4-Mediated Protein Secretion Is Important for Polarized Growth, Reproduction and Pathogenicity in the Banana Fusarium Wilt Fungus Fusarium odoratissimum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Growth and Conidiation Assays
2.3. Construction of Gene Deletion Mutants and Complementation Strains
2.4. Yeast Two-Hybrid (Y2H) Assay
2.5. Virulence Assays
2.6. Extraction and Collection of Secreted Protein
2.7. Fusaric Acid Production Assay
2.8. Extracellular Hydrolase Activity Detection
2.9. Light and Epifluorescence Microscopy
3. Results
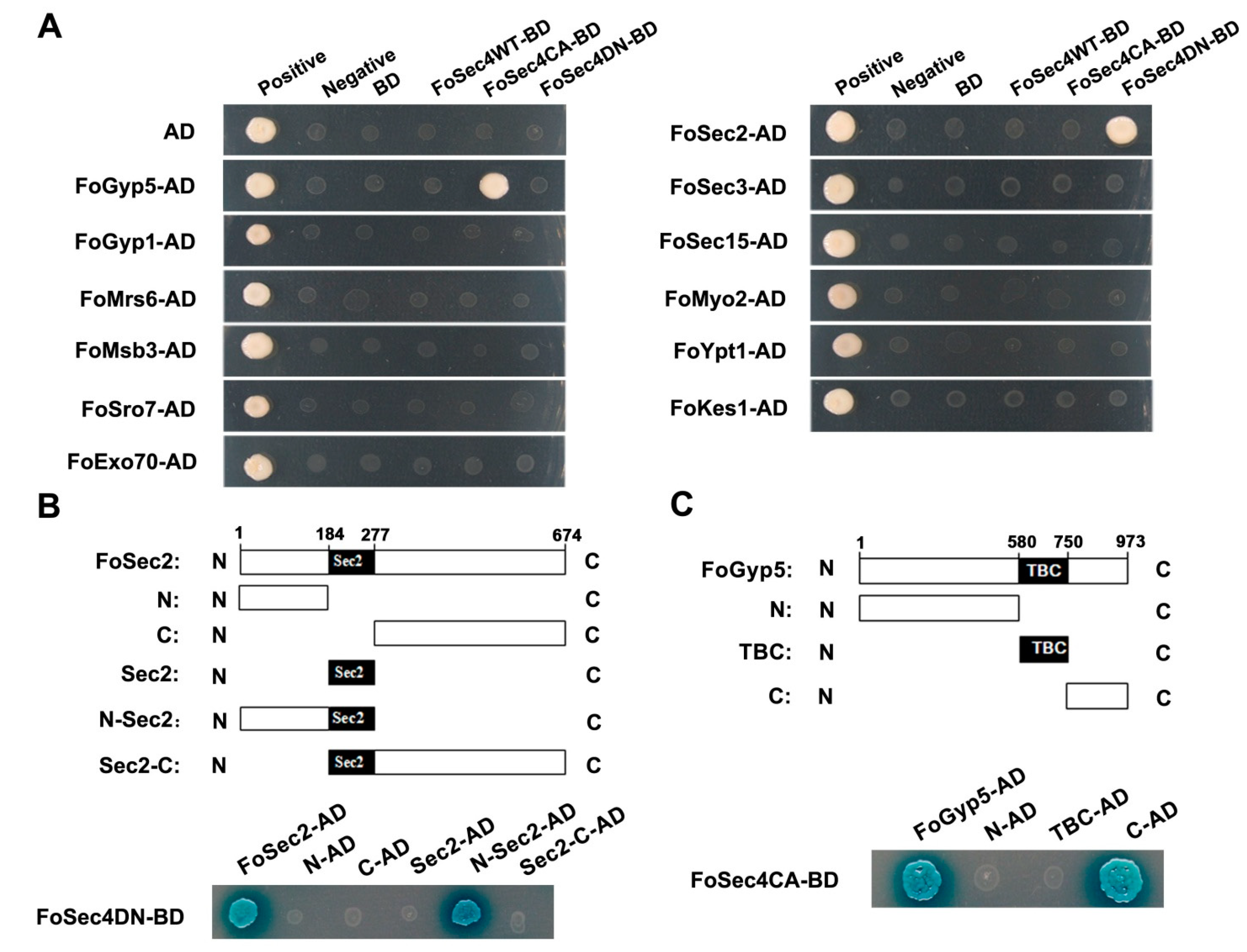
3.1. FoSec2 and FoGyp5 Interact with FoSec4DN and FoSec4CA, Respectively
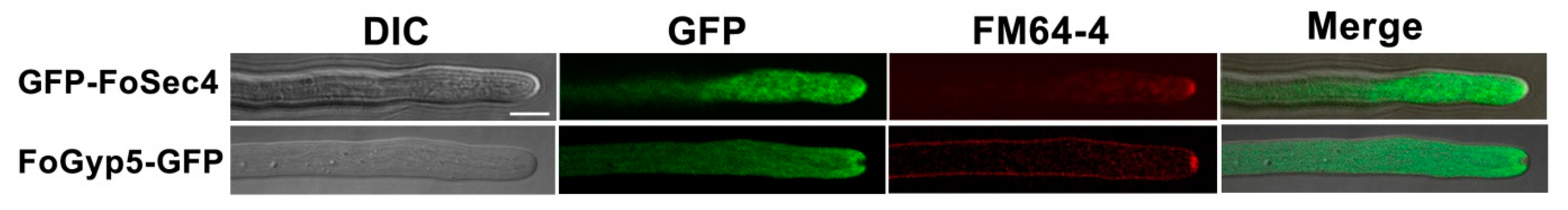
3.2. FoSec4 and FoGyp5 Are Well Distributed in the Cytoplasm but Exhibit Different Patterns in the Hyphal Tips of F. odoratissimum
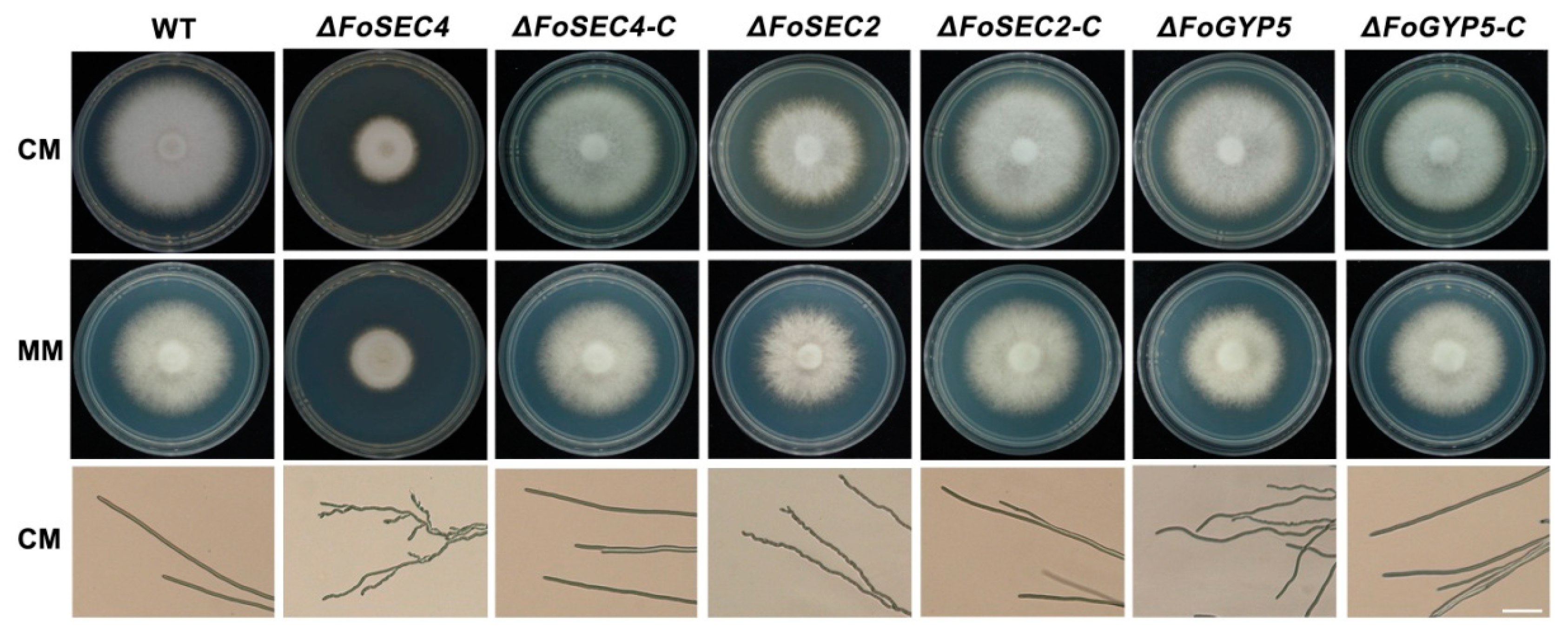
3.3. FoSec4 and FoSec2 Are Important for Vegetative Growth and Polarized Growth in F. odoratissimum
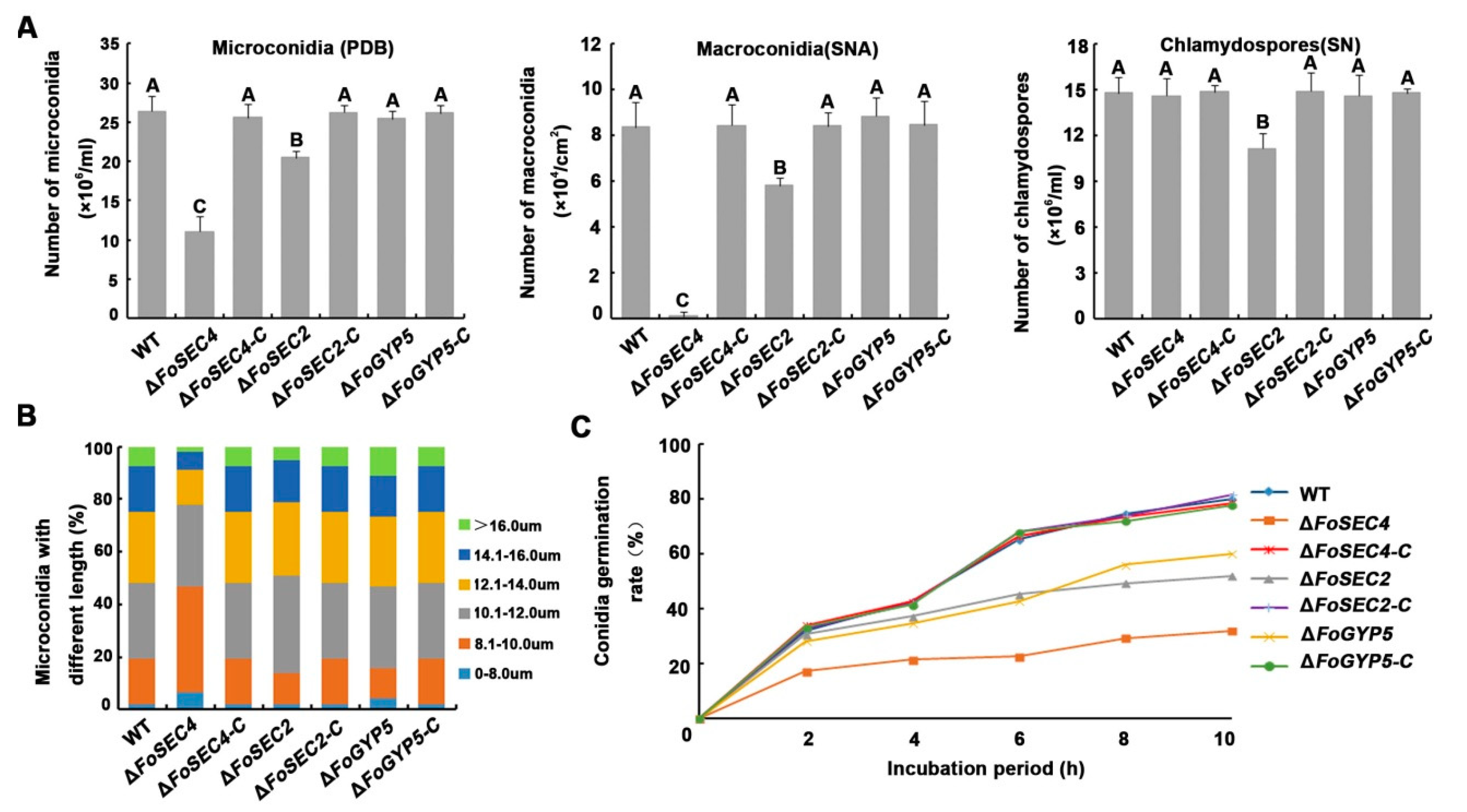
3.4. FoSec2 and FoSec4 Participate in the Regulation of the Reproduction Process of F. odoratissimum
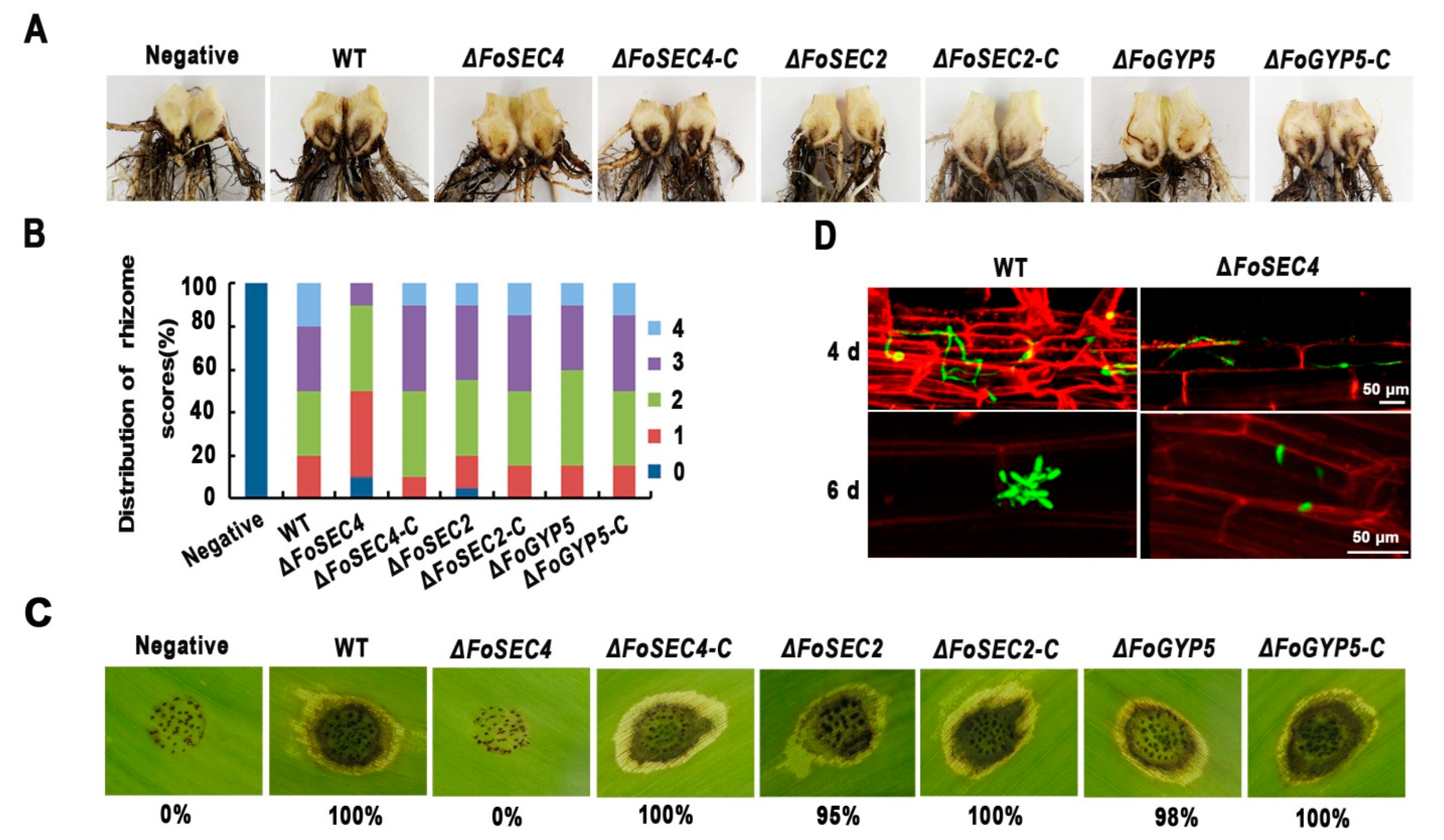
3.5. Deletion of FoSEC4, but Not FoSEC2 or FoGYP5, Leads to Notably Decreased Virulence
3.6. Deletion of FoSEC4 Results in Attenuated Activities of Extracellular Cell Wall-Degrading Enzymes and Reduced Protein Secretion
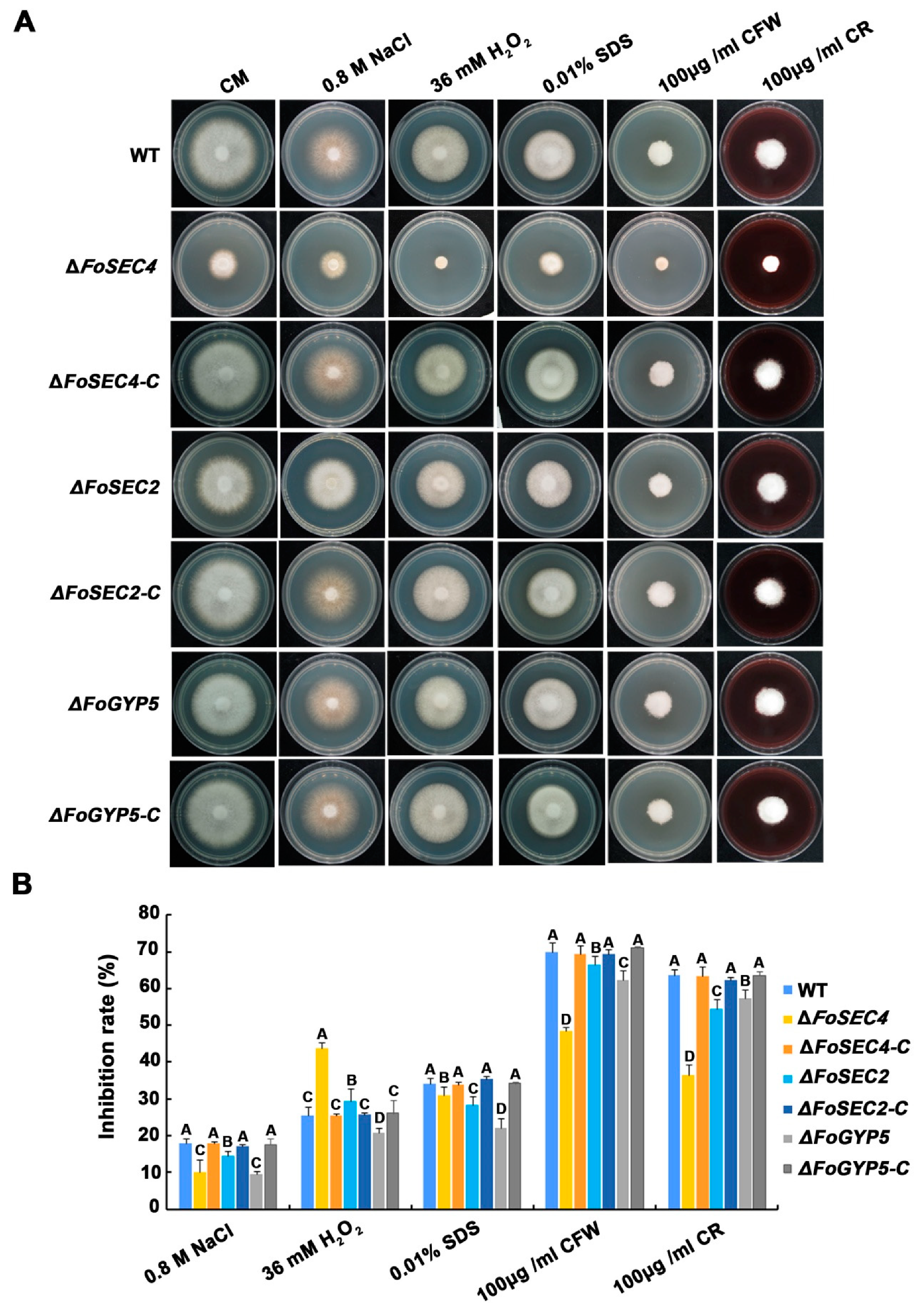
3.7. FoSec4, FoSec2 and FoGyp5 Participate in the Response to Environmental Stressors
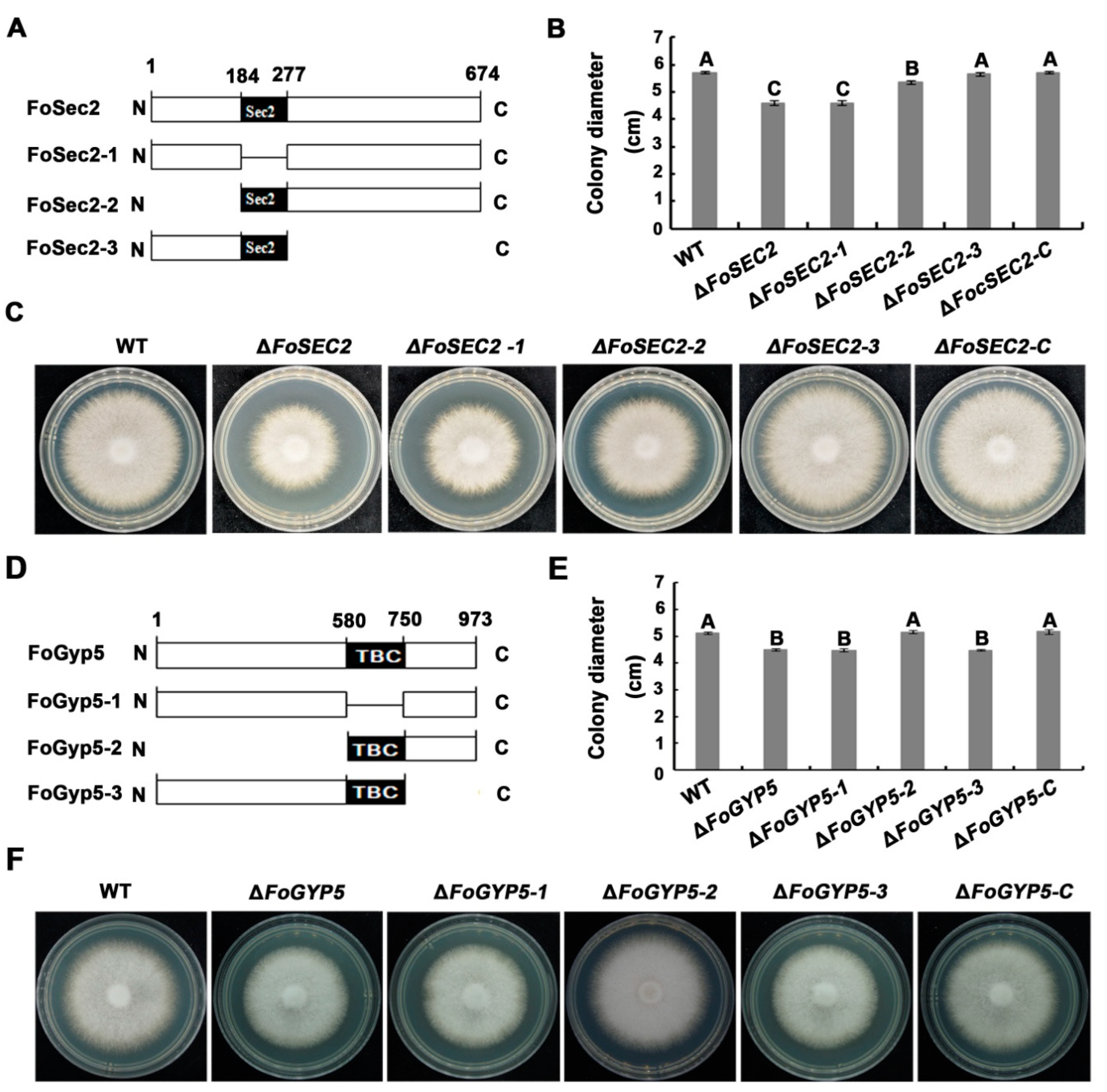
3.8. The Amino-Terminal Region and Sec2 Domain Are Essential for the Biological Functions of FoSec2, While the Carboxyl-Terminal Region and TBC Domain Are Essential for the Biological Functions of FoGyp5
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stenmark, H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009, 10, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Grosshans, B.L.; Ortiz, D.; Novick, P. Rabs and their effectors: Achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. USA 2006, 103, 11821–11827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, S.L.; Cao, C.; Pylypenko, O.; Rak, A.; Wandinger-Ness, A. Rab GTPases at a glance. J. Cell Sci. 2007, 120, 3905–3910. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, M. Regulation of secretory vesicle traffic by Rab small GTPases. Cell. Mol. Life Sci. CMLS 2008, 65, 2801–2813. [Google Scholar] [CrossRef] [PubMed]
- Novick, P.; Field, C.; Schekman, R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell 1980, 21, 205–215. [Google Scholar] [CrossRef]
- Gallwitz, D.; Donath, C.; Sander, C. A yeast gene encoding a protein homologous to the human c-has/bas proto-oncogene product. Nature 1983, 306, 704–707. [Google Scholar] [CrossRef]
- TerBush, D.R.; Maurice, T.; Roth, D.; Novick, P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996, 15, 6483–6494. [Google Scholar] [CrossRef]
- Hsu, S.C.; Ting, A.E.; Hazuka, C.D.; Davanger, S.; Kenny, J.W.; Kee, Y.; Scheller, R.H. The mammalian brain rsec6/8 complex. Neuron 1996, 17, 1209–1219. [Google Scholar] [CrossRef] [Green Version]
- Punt, P.J.; Seiboth, B.; Weenink, X.O.; van Zeijl, C.; Lenders, M.; Konetschny, C.; Ram, A.F.; Montijn, R.; Kubicek, C.P.; van den Hondel, C.A. Identification and characterization of a family of secretion-related small GTPase-encoding genes from the filamentous fungus Aspergillus niger: A putative SEC4 homologue is not essential for growth. Mol. Microbiol. 2001, 41, 513–525. [Google Scholar] [CrossRef]
- Powers-Fletcher, M.V.; Feng, X.; Krishnan, K.; Askew, D.S. Deletion of the sec4 homolog srgA from Aspergillus fumigatus is associated with an impaired stress response, attenuated virulence and phenotypic heterogeneity. PLoS ONE 2013, 8, e66741. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Qin, G.; Li, B.; Tian, S. Knocking out Bcsas1 in Botrytis cinerea impacts growth, development, and secretion of extracellular proteins, which decreases virulence. Mol. Plant-Microbe Interact. MPMI 2014, 27, 590–600. [Google Scholar] [PubMed] [Green Version]
- Siriputthaiwan, P.; Jauneau, A.; Herbert, C.; Garcin, D.; Dumas, B. Functional analysis of CLPT1, a Rab/GTPase required for protein secretion and pathogenesis in the plant fungal pathogen Colletotrichum lindemuthianum. J. Cell Sci. 2005, 118, 323–329. [Google Scholar] [PubMed] [Green Version]
- Dumas, B.; Borel, C.; Herbert, C.; Maury, J.; Jacquet, C.; Balsse, R.; Esquerré-Tugayé, M.T. Molecular characterization of CLPT1, a SEC4-like Rab/GTPase of the phytopathogenic fungus Colletotrichum lindemuthianum which is regulated by the carbon source. Gene 2001, 272, 219–225. [Google Scholar] [PubMed]
- Zheng, H.; Chen, S.; Chen, X.; Liu, S.; Dang, X.; Yang, C.; Giraldo, M.C.; Oliveira-Garcia, E.; Zhou, J.; Wang, Z.; et al. The small GTPase MoSec4 is involved in vegetative development and pathogenicity by regulating the extracellular protein secretion in Magnaporthe oryzae. Front. Plant Sci. 2016, 7, 1458. [Google Scholar]
- Soldati, T.; Shapiro, A.D.; Svejstrup, A.B.; Pfeffer, S.R. Membrane targeting of the small GTPase Rab9 is accompanied by nucleotide exchange. Nature 1994, 369, 76–78. [Google Scholar] [PubMed] [Green Version]
- Ullrich, O.; Horiuchi, H.; Bucci, C.; Zerial, M. Membrane association of Rab5 mediated by GDP-dissociation inhibitor and accompanied by GDP/GTP exchange. Nature 1994, 369, 157–160. [Google Scholar]
- Segev, N. Ypt/rab gtpases: Regulators of protein trafficking. Sci. STKE Signal Transduct. Knowl. Environ. 2001, 2001, re11. [Google Scholar]
- Cherfils, J.; Zeghouf, M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 2013, 93, 269–309. [Google Scholar]
- Barr, F.; Lambright, D.G. Rab GEFs and GAPs. Curr. Opin. Cell Biol. 2010, 22, 461–470. [Google Scholar]
- Walch-Solimena, C.; Collins, R.N.; Novick, P.J. Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J. Cell Biol. 1997, 137, 1495–1509. [Google Scholar]
- Ortiz, D.; Medkova, M.; Walch-Solimena, C.; Novick, P. Ypt32 recruits the Sec4p guanine nucleotide exchange factor, Sec2p, to secretory vesicles; evidence for a Rab cascade in yeast. J. Cell Biol. 2002, 157, 1005–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, Y.; Shirakawa, R.; Horiuchi, H.; Dohmae, N.; Fukai, S.; Nureki, O. Asymmetric coiled-coil structure with Guanine nucleotide exchange activity. Structure 2007, 15, 245–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, Y.; Fukai, S.; Ishitani, R.; Nureki, O. Crystal structure of the Sec4p. Sec2p complex in the nucleotide exchanging intermediate state. Proc. Natl. Acad. Sci. USA 2007, 104, 8305–8310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, L.L.; Collins, R.N.; Novick, P.J. Identification of a Sec4p GTPase-activating protein (GAP) as a novel member of a Rab GAP family. J. Biol. Chem. 1998, 273, 3253–3256. [Google Scholar] [CrossRef] [Green Version]
- Chesneau, L.; Dupré, S.; Burdina, A.; Roger, J.; Le Panse, S.; Jacquet, M.; Cuif, M.H. Gyp5p and Gyl1p are involved in the control of polarized exocytosis in budding yeast. J. Cell Sci. 2004, 117, 4757–4767. [Google Scholar]
- Gao, X.D.; Albert, S.; Tcheperegine, S.E.; Burd, C.G.; Gallwitz, D.; Bi, E. The GAP activity of Msb3p and Msb4p for the Rab GTPase Sec4p is required for efficient exocytosis and actin organization. J. Cell Biol. 2003, 162, 635–646. [Google Scholar]
- Richardson, P.M.; Zon, L.I. Molecular cloning of a cDNA with a novel domain present in the tre-2 oncogene and the yeast cell cycle regulators BUB2 and cdc16. Oncogene 1995, 11, 1139–1148. [Google Scholar]
- Yan, H.; Huang, J.; Zhang, H.; Shim, W.B. A Rab GTPase protein FvSec4 is necessary for fumonisin B1 biosynthesis and virulence in Fusarium verticillioides. Curr. Genet. 2020, 66, 205–216. [Google Scholar]
- Zheng, H.; Li, L.; Miao, P.; Wu, C.; Chen, X.; Yuan, M.; Fang, T.; Norvienyeku, J.; Li, G.; Zheng, W.; et al. FgSec2A, a guanine nucleotide exchange factor of FgRab8, is important for polarized growth, pathogenicity and deoxynivalenol production in Fusarium graminearum. Environ. Microbiol. 2018, 20, 3378–3392. [Google Scholar] [CrossRef]
- Zheng, H.; Li, L.; Yu, Z.; Yuan, Y.; Zheng, Q.; Xie, Q.; Li, G.; Abubakar, Y.S.; Zhou, J.; Wang, Z.; et al. FgSpa2 recruits FgMsb3, a Rab8 GAP, to the polarisome to regulate polarized trafficking, growth and pathogenicity in Fusarium graminearum. New Phytol. 2021, 229, 1665–1683. [Google Scholar] [CrossRef]
- Kwon, M.J.; Arentshorst, M.; Fiedler, M.; de Groen, F.L.M.; Punt, P.J.; Meyer, V.; Ram, A.F.J. Molecular genetic analysis of vesicular transport in Aspergillus niger reveals partial conservation of the molecular mechanism of exocytosis in fungi. Microbiology 2014, 160, 316–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callejas-Negrete, O.A.; Castro-Longoria, E. The role of GYP-3 in cellular morphogenesis of Neurospora crassa: Analyzing its relationship with the polarisome. Fungal Genet. Biol. FGB 2019, 128, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Lozowicka, B.; Iwaniuk, P.; Konecki, R.; Kaczynski, P.; Kuldybayev, N.; Dutbayev, Y. Impact of diversified chemical and biostimulator protection on yield, health status, mycotoxin level, and economic profitability in spring wheat (Triticum aestivum L.) cultivation. Agronomy 2022, 12, 258. [Google Scholar] [CrossRef]
- Ordonez, N.; Seidl, M.F.; Waalwijk, C.; Drenth, A.; Kilian, A.; Thomma, B.P.; Ploetz, R.C.; Kema, G.H. Worse comes to worst: Bananas and Panama disease--when plant and pathogen clones meet. PLoS Pathog. 2015, 11, e1005197. [Google Scholar] [CrossRef] [Green Version]
- Hwang, S.C.; Ko, W.H. Cavendish banana cultivars resistant to Fusarium wilt acquired through somaclonal variation in Taiwan. Plant Dis. 2004, 88, 580–588. [Google Scholar] [CrossRef] [Green Version]
- Maryani, N.; Lombard, L.; Poerba, Y.S.; Subandiyah, S.; Crous, P.W.; Kema, G.H.J. Phylogeny and genetic diversity of the banana Fusarium wilt pathogen Fusarium oxysporum f. sp. cubense in the Indonesian centre of origin. Stud. Mycol. 2019, 92, 155–194. [Google Scholar]
- Widinugraheni, S.; Niño-Sánchez, J.; van der Does, H.C.; van Dam, P.; García-Bastidas, F.A.; Subandiyah, S.; Meijer, H.J.G.; Kistler, H.C.; Kema, G.H.J. A SIX1 homolog in Fusarium oxysporum f. sp. cubense tropical race 4 contributes to virulence towards Cavendish banana. PLoS ONE 2018, 13, e0205896. [Google Scholar]
- An, B.; Hou, X.; Guo, Y.; Zhao, S.; Luo, H.; He, C.; Wang, Q. The effector SIX8 is required for virulence of Fusarium oxysporum f.sp. cubense tropical race 4 to Cavendish banana. Fungal Biol. 2019, 123, 423–430. [Google Scholar]
- Yang, S.; Zhou, X.; Guo, P.; Lin, Y.; Fan, Q.; Zuriegat, Q.; Lu, S.; Yang, J.; Yu, W.; Liu, H.; et al. The exocyst regulates hydrolytic enzyme secretion at hyphal tips and septa in the banana Fusarium Wilt Fungus Fusarium odoratissimum. Appl. Environ. Microbiol. 2021, 87, e0308820. [Google Scholar] [CrossRef]
- Cheng, C.; Liu, F.; Sun, X.; Tian, N.; Mensah, R.A.; Li, D.; Lai, Z. Identification of Fusarium oxysporum f. sp. cubense tropical race 4 (Foc TR4) responsive miRNAs in banana root. Sci. Rep. 2019, 9, 13682. [Google Scholar]
- Yun, Y.; Liu, Z.; Yin, Y.; Jiang, J.; Chen, Y.; Xu, J.R.; Ma, Z. Functional analysis of the Fusarium graminearum phosphatome. New Phytol. 2015, 207, 119–134. [Google Scholar] [CrossRef] [PubMed]
- Leslie, J.F.; Summerell, B.A. Fusarium laboratory workshops-A recent history. Mycotoxin Res. 2006, 22, 73–74. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Cao, Z.; Huang, L.; Liu, S.; Shen, Z.; Wang, Y.; Wang, H.; Zhang, H.; Li, D.; Song, F. CCR4-Not complex subunit Not2 plays critical roles in vegetative growth, conidiation and virulence in watermelon Fusarium wilt pathogen Fusarium oxysporum f. sp. niveum. Front. Microbiol. 2016, 7, 1449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.H.; Hamari, Z.; Han, K.H.; Seo, J.A.; Reyes-Domínguez, Y.; Scazzocchio, C. Double-joint PCR: A PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. FGB 2004, 41, 973–981. [Google Scholar] [CrossRef]
- Zheng, W.; Zheng, H.; Zhao, X.; Zhang, Y.; Xie, Q.; Lin, X.; Chen, A.; Yu, W.; Lu, G.; Shim, W.B.; et al. Retrograde trafficking from the endosome to the trans-Golgi network mediated by the retromer is required for fungal development and pathogenicity in Fusarium graminearum. New Phytol. 2016, 210, 1327–1343. [Google Scholar] [CrossRef]
- Zhao, S.; An, B.; Guo, Y.; Hou, X.; Luo, H.; He, C.; Wang, Q. Label free proteomics and systematic analysis of secretome reveals effector candidates regulated by SGE1 and FTF1 in the plant pathogen Fusarium oxysporum f. sp. cubense tropical race 4. BMC Genom. 2020, 21, 275. [Google Scholar] [CrossRef]
- Yun, Y.; Zhou, X.; Yang, S.; Wen, Y.; You, H.; Zheng, Y.; Norvienyeku, J.; Shim, W.B.; Wang, Z. Fusarium oxysporum f. sp. lycopersici C(2)H(2) transcription factor FolCzf1 is required for conidiation, fusaric acid production, and early host infection. Curr. Genet. 2019, 65, 773–783. [Google Scholar]
- Eveleigh, D.E.; Mandels, M.; Andreotti, R.; Roche, C. Measurement of saccharifying cellulase. Biotechnol. Biofuels 2009, 2, 21. [Google Scholar] [CrossRef] [Green Version]
- Chi, M.H.; Park, S.Y.; Kim, S.; Lee, Y.H. A novel pathogenicity gene is required in the rice blast fungus to suppress the basal defenses of the host. PLoS Pathog. 2009, 5, e1000401. [Google Scholar] [CrossRef] [Green Version]
- Fischer-Parton, S.; Parton, R.M.; Hickey, P.C.; Dijksterhuis, J.; Atkinson, H.A.; Read, N.D. Confocal microscopy of FM4-64 as a tool for analysing endocytosis and vesicle trafficking in living fungal hyphae. J. Microsc. 2000, 198, 246–259. [Google Scholar] [CrossRef] [Green Version]
- Riquelme, M.; Sánchez-León, E. The Spitzenkörper: A choreographer of fungal growth and morphogenesis. Curr. Opin. Microbiol. 2014, 20, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Ling, N.; Wang, M.; Shen, Q.; Guo, S. Fusaric acid is a crucial factor in the disturbance of leaf water imbalance in Fusarium-infected banana plants. Plant Physiol. Biochem. PPB 2012, 60, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, M. Tip growth in filamentous fungi: A road trip to the apex. Annu. Rev. Microbiol. 2013, 67, 587–609. [Google Scholar] [CrossRef] [Green Version]
- Song, W.; Dou, X.; Qi, Z.; Wang, Q.; Zhang, X.; Zhang, H.; Guo, M.; Dong, S.; Zhang, Z.; Wang, P.; et al. R-SNARE homolog MoSec22 is required for conidiogenesis, cell wall integrity, and pathogenesis of Magnaporthe oryzae. PLoS ONE 2010, 5, e13193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bickle, M.; Delley, P.A.; Schmidt, A.; Hall, M.N. Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. EMBO J. 1998, 17, 2235–2245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lussier, M.; White, A.M.; Sheraton, J.; di Paolo, T.; Treadwell, J.; Southard, S.B.; Horenstein, C.I.; Chen-Weiner, J.; Ram, A.F.; Kapteyn, J.C.; et al. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 1997, 147, 435–450. [Google Scholar] [CrossRef]
- Ram, A.F.; Wolters, A.; Ten Hoopen, R.; Klis, F.M. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to calcofluor white. Yeast 1994, 10, 1019–1030. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Zheng, W.; Wu, C.; Yang, J.; Xi, Y.; Xie, Q.; Zhao, X.; Deng, X.; Lu, G.; Li, G.; et al. Rab GTPases are essential for membrane trafficking-dependent growth and pathogenicity in Fusarium graminearum. Environ. Microbiol. 2015, 17, 4580–4599. [Google Scholar] [CrossRef]
- Li, C.; Yang, J.; Li, W.; Sun, J.; Peng, M. Direct root penetration and rhizome vascular colonization by Fusarium oxysporum f. sp. cubense are the key steps in the successful infection of Brazil Cavendish. Plant Dis. 2017, 101, 2073–2078. [Google Scholar]
- Nair, J.; Müller, H.; Peterson, M.; Novick, P. Sec2 protein contains a coiled-coil domain essential for vesicular transport and a dispensable carboxy terminal domain. J. Cell Biol. 1990, 110, 1897–1909. [Google Scholar] [CrossRef]
- De Antoni, A.; Schmitzová, J.; Trepte, H.H.; Gallwitz, D.; Albert, S. Significance of GTP hydrolysis in Ypt1p-regulated endoplasmic reticulum to Golgi transport revealed by the analysis of two novel Ypt1-GAPs. J. Biol. Chem. 2002, 277, 41023–41031. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Protein in Saccharomyces cerevisiae | Homologs in Fusarium odoratissimum | Protein Identity |
|---|---|---|
| Gyp5 | FoGyp5 (FOIG_01503) | 40.67% |
| Gyp1 | FoGyp1 (FOIG_07346) | 39.32% |
| Mrs6 | FoMrs6 (FOIG_15902) | 26.32% |
| Msb3 | FoMsb3 (FOIG_04024) | 41.76% |
| Sro7 | FoSro7 (FOIG_01252) | 28.13% |
| Exo70 | FoExo70 (FOIG_06688) | 23.43% |
| Sec2 | FoSec2 (FOIG_11948) | 27.92% |
| Sec3 | FoSec3 (FOIG_06126) | 36.14% |
| Sec15 | FoSec15 (FOIG_08908) | 24.20% |
| Myo2 | FoMyo2 (FOIG_02660) | 46.22% |
| Ypt1 | FoYpt1 (FOIG_07693) | 79.01% |
| Osh4 | FoKes1 (FOIG_10637) | 45.56% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Y.; Guo, P.; Deng, H.; Lin, Y.; Huang, G.; Wu, J.; Lu, S.; Yang, S.; Zhou, J.; Zheng, W.; et al. Small GTPase FoSec4-Mediated Protein Secretion Is Important for Polarized Growth, Reproduction and Pathogenicity in the Banana Fusarium Wilt Fungus Fusarium odoratissimum. J. Fungi 2022, 8, 880. https://doi.org/10.3390/jof8080880
Zheng Y, Guo P, Deng H, Lin Y, Huang G, Wu J, Lu S, Yang S, Zhou J, Zheng W, et al. Small GTPase FoSec4-Mediated Protein Secretion Is Important for Polarized Growth, Reproduction and Pathogenicity in the Banana Fusarium Wilt Fungus Fusarium odoratissimum. Journal of Fungi. 2022; 8(8):880. https://doi.org/10.3390/jof8080880
Chicago/Turabian StyleZheng, Yuru, Pingting Guo, Huobing Deng, Yaqi Lin, Guilan Huang, Jie Wu, Songmao Lu, Shuai Yang, Jie Zhou, Wenhui Zheng, and et al. 2022. "Small GTPase FoSec4-Mediated Protein Secretion Is Important for Polarized Growth, Reproduction and Pathogenicity in the Banana Fusarium Wilt Fungus Fusarium odoratissimum" Journal of Fungi 8, no. 8: 880. https://doi.org/10.3390/jof8080880
APA StyleZheng, Y., Guo, P., Deng, H., Lin, Y., Huang, G., Wu, J., Lu, S., Yang, S., Zhou, J., Zheng, W., Wang, Z., & Yun, Y. (2022). Small GTPase FoSec4-Mediated Protein Secretion Is Important for Polarized Growth, Reproduction and Pathogenicity in the Banana Fusarium Wilt Fungus Fusarium odoratissimum. Journal of Fungi, 8(8), 880. https://doi.org/10.3390/jof8080880







