The Elongator Subunit Elp3 Regulates Development, Stress Tolerance, Cell Cycle, and Virulence in the Entomopathogenic Fungus Beauveria bassiana
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bioinformatic Analysis of Elp3 Homolog in B. bassiana
2.2. BbElp3 Subcellular Localization Examination
2.3. Generation of B. bassiana Elp3 Mutants
2.4. Western Blotting for Histone H3 Acetylation Detection
2.5. Investigation of Vegetative Growth and Conidial Properties
2.6. Bioassays Using Gellaria Mellonella and Investigation of Virulence-Related Events
2.7. Examination of Cell Cycle, Hyphal Septation Pattern, and Autophagosome Formation
2.8. Transcriptomic Analysis
3. Results
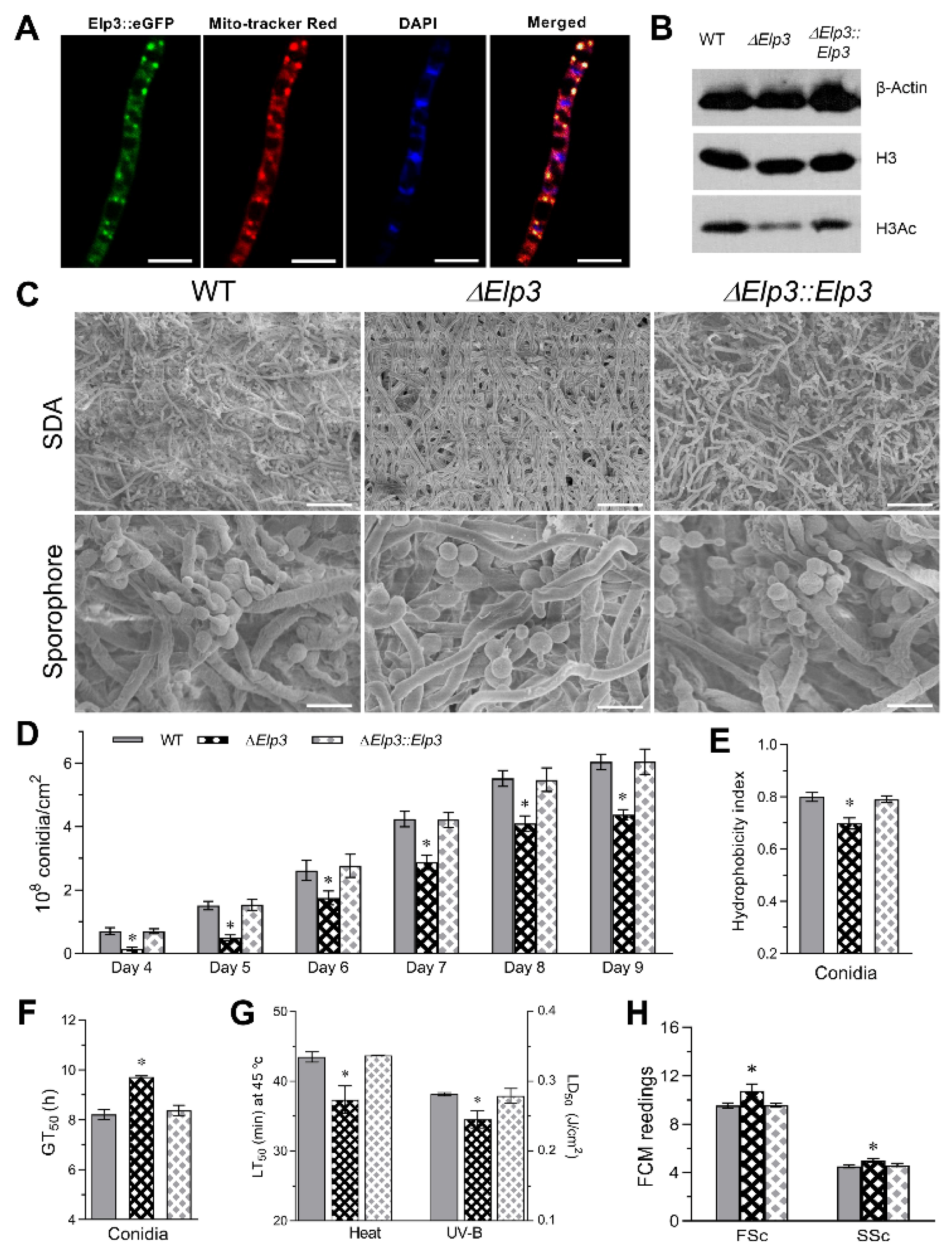
3.1. Bioinformatic Features and Subcellular Localization of B. bassiana Elp3 and Construction of Deletion and Complemented Strains
3.2. Role of BbElp3 in Histone H3 Acetylation, Sporulation, and Conidial Property
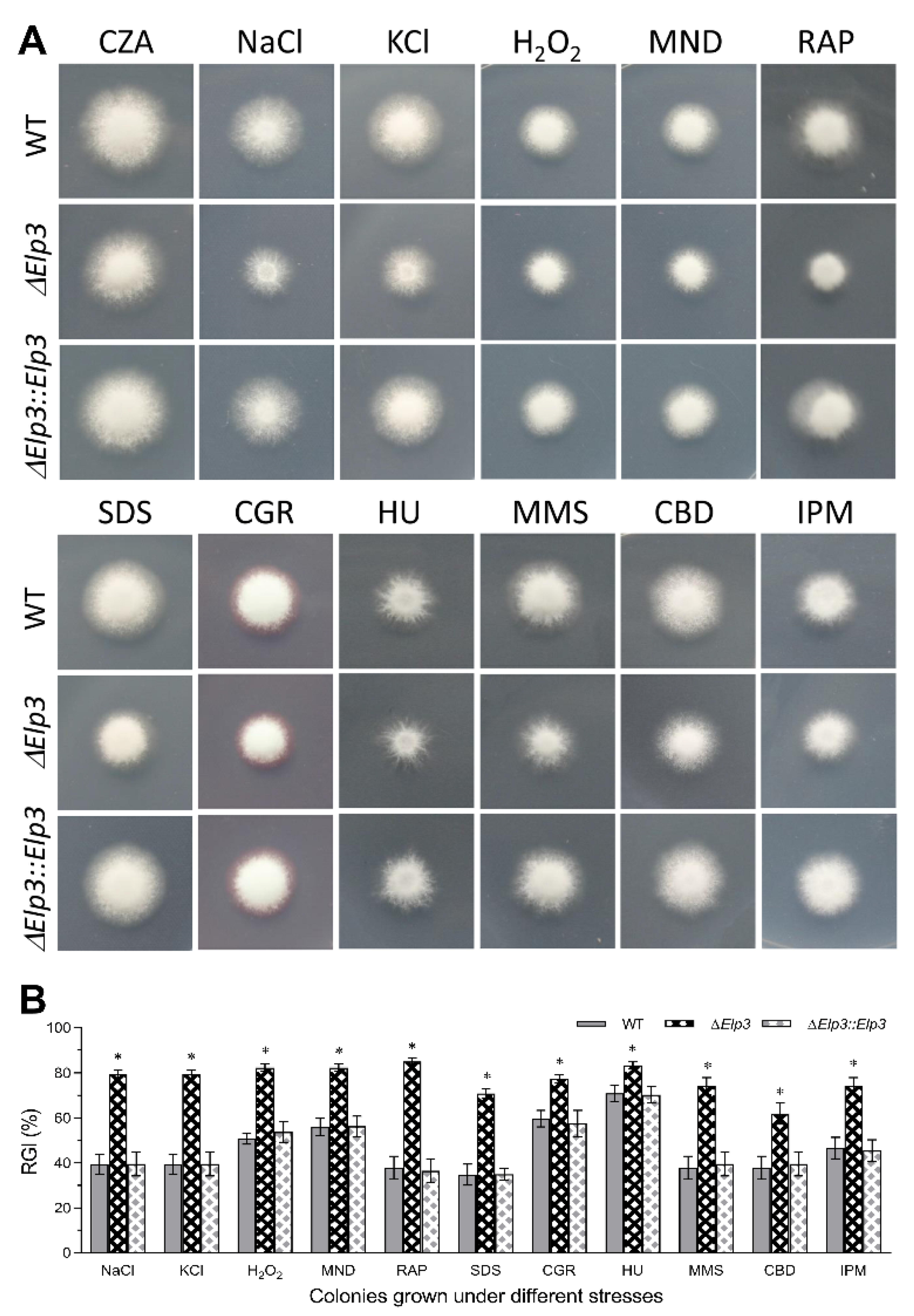
3.3. Role of BbElp3 in Carbon/Nitrogen Metabolism and Multi-Stress Tolerance
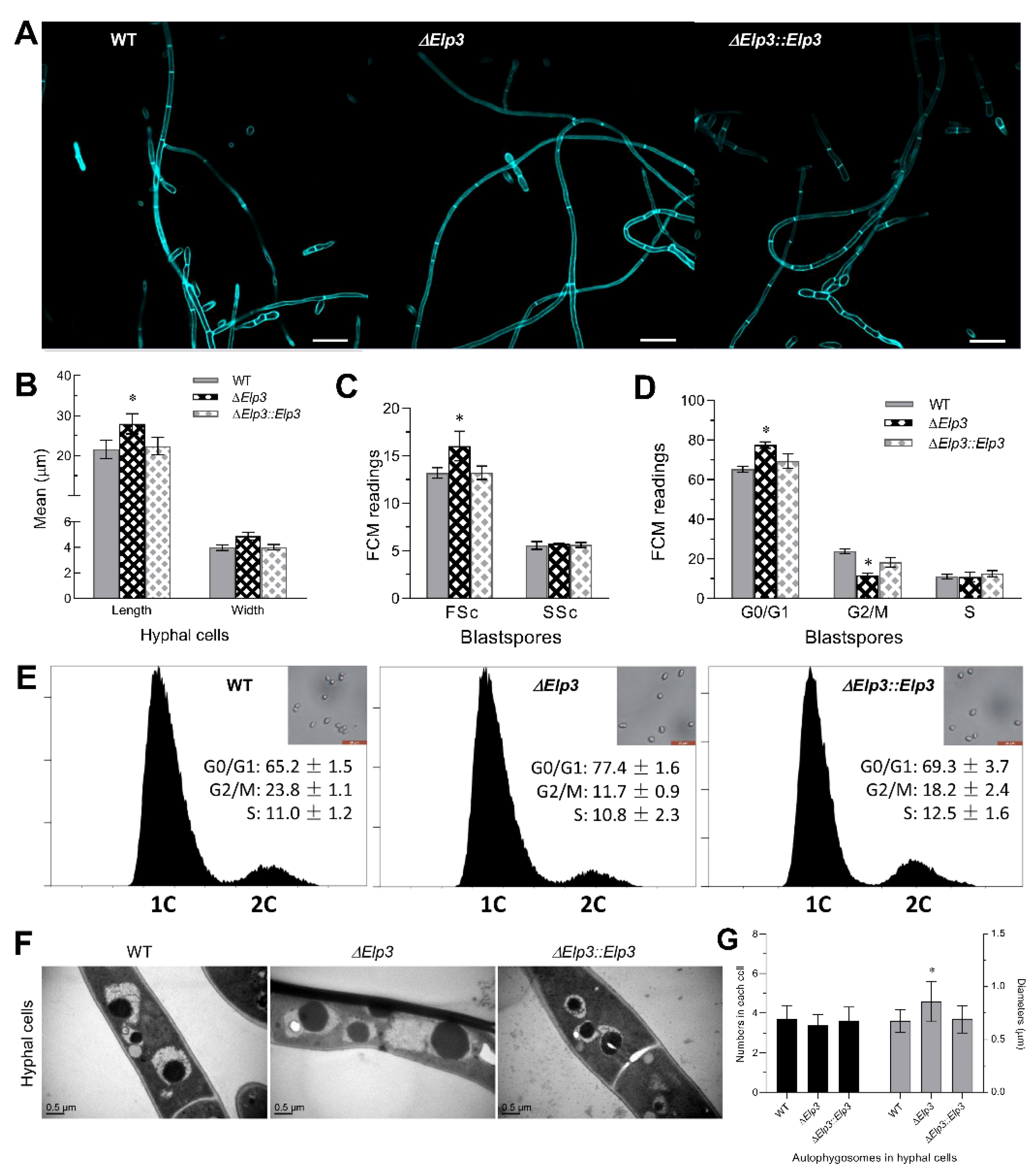
3.4. Influence of BbElp3 Deletion on Cell Cycle Progression and Hyphal Septation
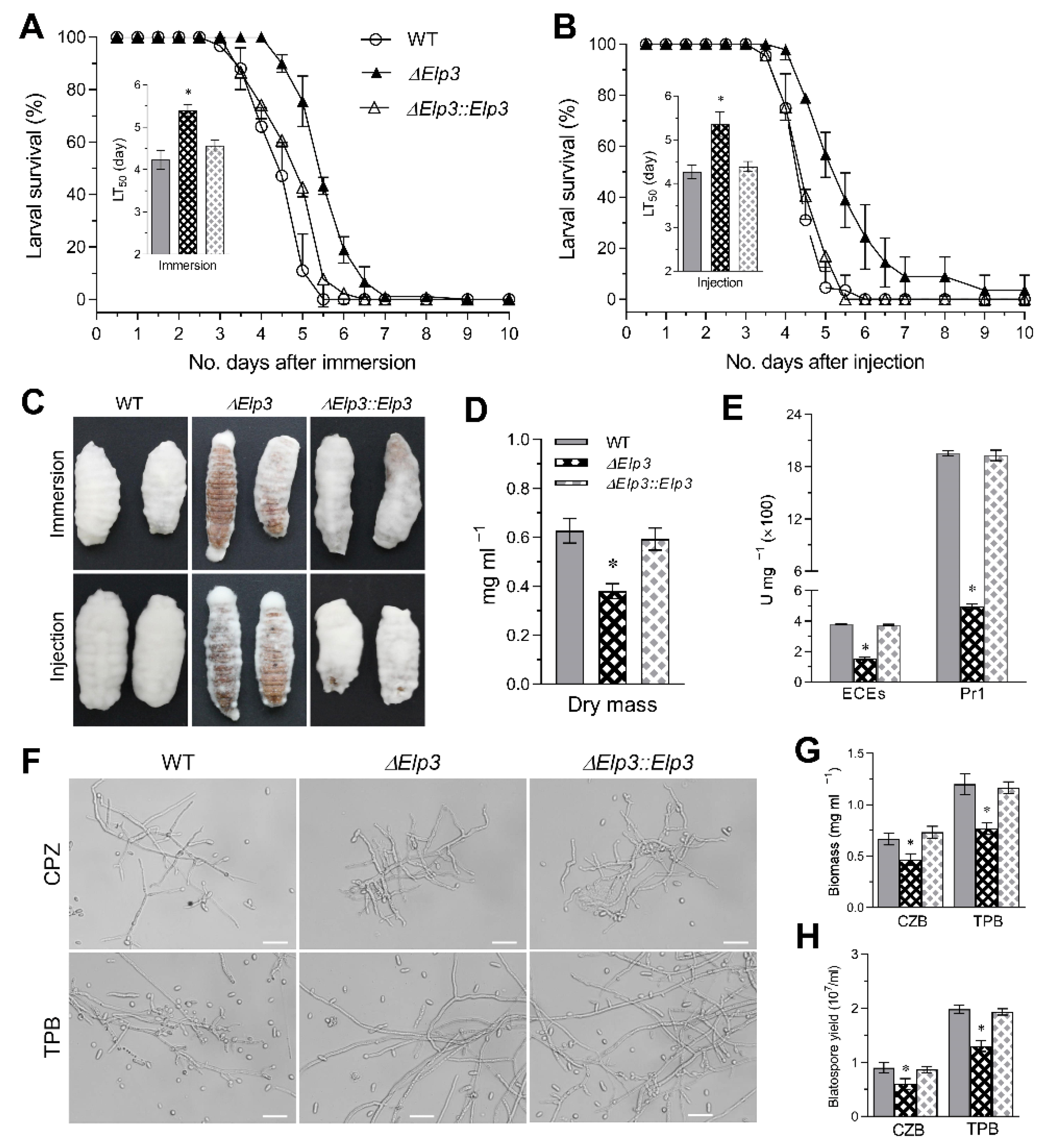
3.5. BbElp3 Deletion Attenuates B. bassiana Virulence
3.6. Global Regulatory Role of BbElp3 in Gene Transcription
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gregory, P.D.; Wagner, K.; Hörz, W. Histone acetylation and chromatin remodeling. Exp. Cell. Res. 2001, 265, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Sterner, D.E.; Berger, S.L. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 2000, 64, 435–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millar, C.B.; Grunstein, M. Genome-wide patterns of histone modifications in yeast. Nat. Rev. Mol. Cell. Biol. 2006, 7, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Carey, M.; Workman, J.L. The role of chromatin during transcription. Cell 2007, 128, 707–719. [Google Scholar] [CrossRef] [Green Version]
- Groth, A.; Rocha, W.; Verreault, A.; Almouzni, G. Chromatin challenges during DNA replication and repair. Cell 2007, 128, 721–733. [Google Scholar] [CrossRef] [Green Version]
- Otero, G.; Fellows, J.; Li, Y.; de Bizemont, T.; Dirac, A.M.; Gustafsson, C.M.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J.Q. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 1999, 3, 109–118. [Google Scholar] [CrossRef]
- Wittschieben, B.O.; Otero, G.; de Bizemont, T.; Fellows, J.; Erdjument-Bromage, H.; Ohba, R.; Li, Y.; Allis, C.D.; Tempst, P.; Svejstrup, J.Q.; et al. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 1999, 4, 123–128. [Google Scholar] [CrossRef]
- Hawkes, N.A.; Otero, G.; Winkler, G.S.; Marshall, N.; Dahmus, M.E.; Krappmann, D.; Scheidereit, C.; Thomas, C.L.; Schiavo, G.; Erdjument-Bromage, H.; et al. Purification and characterization of the human elongator complex. J. Biol. Chem. 2002, 277, 3047–3052. [Google Scholar] [CrossRef] [Green Version]
- Nelissen, H.; De Groeve, S.; Fleury, D.; Neyt, P.; Bruno, L.; Bitonti, M.B.; Vandenbussche, F.; Van Der Straeten, D.; Yamaguchi, T.; Tsukaya, H.; et al. Plant Elongator regulates auxin-related genes during RNA polymerase II transcription elongation. Proc. Natl. Acad. Sci. USA 2010, 107, 1678–1683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svejstrup, J.Q. Elongator complex: How many roles does it play? Curr. Opin. Cell Biol. 2007, 19, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Winkler, G.S.; Kristjuhan, A.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J.Q. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. USA 2002, 99, 3517–3522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, B.; Johansson, M.J.; Bystrom, A.S. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 2005, 11, 424–436. [Google Scholar] [CrossRef] [Green Version]
- Creppe, C.; Malinouskaya, L.; Volvert, M.L.; Gillard, M.; Close, P.; Malaise, O.; Laguesse, S.; Cornez, I.; Rahmouni, S.; Ormenese, S.; et al. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell 2009, 136, 551–564. [Google Scholar] [CrossRef] [Green Version]
- Okada, Y.; Yamagata, K.; Hong, K.; Wakayama, T.; Zhang, Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature 2010, 463, 554–558. [Google Scholar] [CrossRef]
- Wittschieben, B.O.; Fellows, J.; Du, W.; Stillman, D.J.; Svejstrup, J.Q. Overlapping roles for the histone acetyltransferase activities of SAGA and Elongator in vivo. EMBO J. 2000, 19, 3060–3068. [Google Scholar] [CrossRef]
- Rezgui, V.A.N.; Tyagi, K.; Ranjan, N.; Konevega, A.L.; Mittelstaet, J.; Rodnina, M.V.; Peter, M.; Pedrioli, P.G.A. tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc. Natl. Acad. Sci. USA 2013, 110, 12289–12294. [Google Scholar] [CrossRef] [Green Version]
- Creppe, C.; Buschbeck, M. Elongator: An ancestral complex driving transcription and migration through protein acetylation. J. Biomed. Biotechnol. 2011, 2011, 924898. [Google Scholar] [CrossRef] [Green Version]
- Han, Q.; Hou, X.; Su, D.; Pan, L.; Duan, J.; Cui, L.; Huang, B.; Lu, J. HELP3 subunit of the Elongator complex regulates the transcription of HSP70 gene in human cells. Acta. Biochim. Biophys. Sin. 2007, 39, 453–461. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Zhang, H.; Jablonowski, D.; Zhou, X.; Ren, X.; Hong, X.; Schaffrath, R.; Zhu, J.K.; Gong, Z. Mutations in ABO1/ELO2, a subunit of holo-Elongator, increase abscisic acid sensitivity and drought tolerance in Arabidopsis thaliana. Mol. Cell. Biol. 2006, 26, 6902–6912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stilger, K.L.; Sullivan, W.J. Elongator protein 3 (Elp3) lysine acetyltransferase is a tail-anchored mitochondrial protein in Toxoplasma gondii. J. Biol. Chem. 2013, 288, 25318–25329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barton, D.; Braet, F.; Marc, J.; Overall, R.; Gardiner, J. ELP3 localises to mitochondria and actin-rich domains at edges of HeLa cells. Neurosci. Lett. 2009, 455, 60–64. [Google Scholar] [CrossRef]
- Lee, Y.; Min, K.; Son, H.; Park, A.R.; Kim, J.C.; Choi, G.J.; Lee, Y.W. ELP3 is involved in sexual and asexual development, virulence, and the oxidative stress response in Fusarium graminearum. Mol. Plant. Microbe Interact. 2014, 27, 1344–1355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.M.; Chen, S.T.; Qi, M.; Cao, X.Q.; Liang, N.; LI, Q.; Tang, W.; Lu, G.-D.; Zhou, J.; Yu, W.-Y.; et al. The putative elongator complex protein Elp3 is involved in asexual development and pathogenicity by regulating autophagy in the rice blast fungus. J. Integr. Agric. 2021, 20, 2944–2956. [Google Scholar] [CrossRef]
- De Faria, M.R.; Wraight, S.P. Mycoinsecticides and mycoacaricides: A comprehensive list with worldwide coverage and international classification of formulation types. Biol. Control 2007, 43, 237–256. [Google Scholar] [CrossRef]
- Glare, T.; Caradus, J.; Gelernter, W.; Jackson, T.; Keyhani, N.; Köhl, J.; Marrone, P.; Morin, L.; Stewart, A. Have biopesticides come of age? Trends Biotechnol. 2012, 30, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Urquiza, A.; Keyhani, N.O. Action on the Surface: Entomopathogenic Fungi versus the Insect Cuticle. Insects 2013, 4, 357–374. [Google Scholar] [CrossRef]
- Pendland, J.C.; Hung, S.Y.; Boucias, D.G. Evasion of host defense by in vivo-produced protoplast-like cells of the insect mycopathogen Beauveria bassiana. J. Bacteriol. 1993, 175, 5962–5969. [Google Scholar] [CrossRef] [Green Version]
- Wanchoo, A.; Lewis, M.W.; Keyhani, N.O. Lectin mapping reveals stage-specific display of surface carbohydrates in in vitro and haemolymph-derived cells of the entomopathogenic fungus Beauveria bassiana. Microbiology 2009, 155, 3121–3133. [Google Scholar] [CrossRef] [Green Version]
- Lewis, M.W.; Robalino, I.V.; Keyhani, N.O. Uptake of the fluorescent probe FM4-64 by hyphae and haemolymph-derived in vivo hyphal bodies of the entomopathogenic fungus Beauveria bassiana. Microbiology 2009, 155, 3110–3120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holder, D.J.; Keyhani, N.O. Adhesion of the entomopathogenic fungus Beauveria (Cordyceps) bassiana to substrata. Appl. Environ. Microbiol. 2005, 71, 5260–5266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Liu, Y.; Keyhani, N.O.; Zhu, S.; Wang, J.; Wang, J.; Jin, D.; Fan, Y. A Perilipin Affects Lipid Droplet Homeostasis and Aerial Hyphal Growth, but Has Only Small Effects on Virulence in the Insect Pathogenic Fungus Beauveria bassiana. J. Fungi 2022, 8, 634. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Liu, X.; Keyhani, N.O.; Tang, G.; Pei, Y.; Zhang, W.; Tong, S. Regulatory cascade and biological activity of Beauveria bassiana oosporein that limits bacterial growth after host death. Proc. Natl. Acad. Sci. USA 2017, 114, E1578–E1586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortiz-Urquiza, A.; Luo, Z.B.; Keyhani, N.O. Improving mycoinsecticides for insect biological control. Appl. Microbiol. Biotechnol. 2015, 99, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Urquiza, A.; Keyhani, N.O. Stress response signaling and virulence: Insights from entomopathogenic fungi. Curr. Genet. 2015, 61, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Urquiza, A.; Keyhani, N.O. Molecular genetics of Beauveria bassiana infection of insects. Adv. Genet. 2016, 94, 165–249. [Google Scholar]
- Cai, Q.; Wang, J.J.; Fu, B.; Ying, S.H.; Feng, M.G. Gcn5-dependent histone H3 acetylation and gene activity is required for the asexual development and virulence of Beauveria bassiana. Environ. Microbiol. 2018, 20, 1484–1497. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Wang, J.J.; Xie, J.T.; Jiang, D.H.; Keyhani, N.O. The Spt10 GNAT Superfamily Protein Modulates Development, Cell Cycle Progression and Virulence in the Fungal Insect Pathogen, Beauveria bassiana. J. Fungi 2021, 7, 905. [Google Scholar] [CrossRef]
- Wang, J.J.; Cai, Q.; Qiu, L.; Ying, S.H.; Feng, M.G. The histone acetyltransferase Mst2 sustains the biological control potential of a fungal insect pathogen through transcriptional regulation. Appl. Microbiol. Biotechnol. 2018, 102, 1343–1355. [Google Scholar] [CrossRef]
- Cai, Q.; Wang, J.J.; Shao, W.; Ying, S.H.; Feng, M.G. Rtt109-dependent histone H3 K56 acetylation and gene activity are essential for the biological control potential of Beauveria bassiana. Pest. Manag. Sci. 2018, 74, 2626–2635. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.H.; Ying, S.H.; Zheng, P.; Wang, Z.L.; Zhang, S.W.; Zhang, S.; Xie, X.Q.; Shang, Y.; Leger, R.J.; Zhao, G.P.; et al. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci. Rep. 2012, 2, 483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, Y.; Zhang, S.; Kruer, N.; Keyhani, N.O. High-throughput insertion mutagenesis and functional screening in the entomopathogenic fungus Beauveria bassiana. J. Invertebr. Pathol. 2011, 106, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Tong, S.M.; Shao, W.; Ying, S.H.; Feng, M.G. Pleiotropic effects of the histone deacetylase Hos2 linked to H4-K16 deacetylation, H3-K56 acetylation and H2A-S129 phosphorylation in Beauveria bassiana. Cell. Microbiol. 2018, 20, e12839. [Google Scholar] [CrossRef] [PubMed]
- Holder, D.J.; Kirkland, B.H.; Lewis, M.W.; Keyhani, N.O. Surface characteristics of the entomopathogenic fungus Beauveria (Cordyceps) bassiana. Microbiology 2007, 153, 3448–3457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Ying, S.H.; Hu, Y.; Feng, M.G. Mas5, a homologue of bacterial DnaJ, is indispensable for the host infection and environmental adaptation of a filamentous fungal insect pathogen. Environ. Microbiol. 2016, 18, 1037–1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hercyk, B.; Das, M. Rho Family GTPases in Fission Yeast Cytokinesis. Commun. Integr. Biol. 2019, 12, 171–180. [Google Scholar] [CrossRef] [Green Version]
- Cen, K.; Li, B.; Lu, Y.; Zhang, S.; Wang, C. Divergent LysM effectors contribute to the virulence of Beauveria bassiana by evasion of insect immune defenses. PLoS Pathog. 2017, 13, e1006604. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Liu, Q.; Zeng, L.; Huang, X. Protein Acetylation/Deacetylation: A Potential Strategy for Fungal Infection Control. Front. Microbiol. 2020, 11, 574736. [Google Scholar] [CrossRef]
- Wang, X.; Chang, P.; Ding, J.; Chen, J. Distinct and redundant roles of the two MYST histone acetyltransferases Esa1 and Sas2 in cell growth and morphogenesis of Candida albicans. Eukaryot. Cell 2013, 12, 43–449. [Google Scholar] [CrossRef] [Green Version]
- Lan, H.; Sun, R.; Fan, K.; Yang, K.; Zhang, F.; Nie, X.Y.; Wang, X.; Zhuang, Z.; Wang, S. The Aspergillus flavus histone acetyltransferase AflGcnE regulates morphogenesis, aflatoxin biosynthesis, and pathogenicity. Front. Microbiol. 2016, 7, 1324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Meara, T.R.; Hay, C.; Price, M.S.; Giles, S.; Alspaugh, J.A. Cryptococcus neoformans histone acetyltransferase Gcn5 regulates fungal adaptation to the host. Eukaryot. Cell 2010, 9, 1193–1202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niehaus, E.M.; Rindermann, L.; Janevska, S.; Münsterkötter, M.; Güldener, U.; Tudzynski, B. Analysis of the global regulator Lae1 uncovers a connection between Lae1 and the histone acetyltransferase HAT1 in Fusarium fujikuroi. Appl. Microbiol. Biotechnol. 2018, 102, 279–295. [Google Scholar] [CrossRef]
- González-Prieto, J.M.; Rosas-Quijano, R.; Domínguez, A.; Ruiz-Herrera, J. The UmGcn5 gene encoding histone acetyltransferase from Ustilago maydis is involved in dimorphism and virulence. Fungal Genet. Biol. 2014, 71, 86–95. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhao, X.; Gao, Y.; Keyhani, N.O.; Wang, H.; Deng, J.; Lu, Z.; Kan, Y.; Luo, Z.; Zhang, Y. The fungal mitochondrial membrane protein, BbOhmm, antagonistically controls hypoxia tolerance. Environ. Microbiol. 2020, 22, 2514–2535. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.; Li, M.; Keyhani, N.O.; Liu, Y.; Yuan, M.; Lin, D.; Li, X.; Pei, Y.; Fan, Y. Characterization of a fungal competition factor: Production of a conidial cell-wall associated antifungal peptide. PLoS pathog. 2020, 16, e1008518. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Keyhani, N.O.; Zhao, X.; Zhang, Y. The Thm1 Zn(II)2 Cys6 transcription factor contributes to heat, membrane integrity and virulence in the insect pathogenic fungus Beauveria bassiana. Environ. Microbiol. 2019, 21, 3153–3171. [Google Scholar] [CrossRef]
- Luo, Z.; Zhang, T.; Liu, P.; Bai, Y.; Chen, Q.; Zhang, Y.; Keyhani, N.O. The Beauveria bassiana Gas3 β-Glucanosyltransferase Contributes to Fungal Adaptation to Extreme Alkaline Conditions. Appl. Environ. Microbiol. 2018, 84, e01086-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Q.; Wang, Z.K.; Shao, W.; Ying, S.H.; Feng, M.G. Essential role of Rpd3-dependent lysine modification in the growth, development and virulence of Beauveria bassiana. Environ. Microbiol. 2018, 20, 1590–1606. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Tian, L.; Xie, J.T.; Huang, Q.Y.; Feng, M.G.; Keyhani, N.O. A fungal sirtuin modulates development and virulence in the insect pathogen, Beauveria bassiana. Environ. Microbiol. 2021, 23, 5164–5183. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Tian, L.; Xie, J.T.; Jiang, D.H.; Keyhani, N.O. Contributions of a Histone Deacetylase (SirT2/Hst2) to Beauveria bassiana Growth, Development, and Virulence. J. Fungi 2022, 8, 236. [Google Scholar] [CrossRef]
- Nugent, R.L.; Johnsson, A.; Fleharty, B.; Gogol, M.; Xue-Franzén, Y.; Seidel, C.; Wright, A.P.; Forsburg, S.L. Expression profiling of S. pombe acetyltransferase mutants identifies redundant pathways of gene regulation. BMC Genom. 2010, 11, 59. [Google Scholar] [CrossRef] [Green Version]
- Ortiz-Urquiza, A.; Fan, Y.; Garrett, T.; Keyhani, N.O. Growth substrates and caleosin-mediated functions affect conidial virulence in the insect pathogenic fungus Beauveria bassiana. Microbiology 2016, 162, 1913–1921. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Ortiz-Urquiza, A.; Garrett, T.; Pei, Y.; Keyhani, N.O. Involvement of a caleosin in lipid storage, spore dispersal, and virulence in the entomopathogenic filamentous fungus, Beauveria Bassiana. Environ. Microbiol. 2015, 17, 4600–4614. [Google Scholar] [CrossRef] [Green Version]
- Qin, Y.; Ortiz-Urquiza, A.; Keyhani, N.O. A putative methyltransferase, mtrA, contributes to development, spore viability, protein secretion and virulence in the entomopathogenic fungus Beauveria bassiana. Microbiology 2014, 160, 2526–2537. [Google Scholar] [CrossRef]
- Qiu, L.; Wang, J.J.; Ying, S.H.; Feng, M.G. Wee1 and Cdc25 control morphogenesis, virulence and multistress tolerance of Beauveria bassiana by balancing cell cycle-required cyclin-dependent kinase 1 activity. Environ. Microbiol. 2015, 17, 1119–1133. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Qiu, L.; Cai, Q.; Ying, S.H.; Feng, M.G. Transcriptional control of fungal cell cycle and cellular events by Fkh2, a forkhead transcription factor in an insect pathogen. Sci. Rep. 2015, 5, 10108. [Google Scholar] [CrossRef] [Green Version]
- Ying, S.H.; Ji, X.P.; Wang, X.X.; Feng, M.G.; Keyhani, N.O. The transcriptional co-activator multiprotein bridging factor 1 from the fungal insect pathogen, Beauveria bassiana, mediates regulation of hyphal morphogenesis, stress tolerance and virulence. Environ. Microbiol. 2014, 16, 1879–1897. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhang, S.; Keyhani, N.O.; Song, Y.; Huang, S.; Pei, Y.; Zhang, Y. A novel mitochondrial membrane protein, Ohmm, limits fungal oxidative stress resistance and virulence in the insect fungal pathogen Beauveria bassiana. Environ. Microbiol. 2015, 17, 4213–4238. [Google Scholar] [CrossRef]
- Moonjely, S.; Keyhani, N.O.; Bidochka, M.J. Hydrophobins contribute to root colonization and stress responses in the rhizosphere-competent insect pathogenic fungus Beauveria bassiana. Microbiology 2018, 164, 517–528. [Google Scholar] [CrossRef]
- Mou, Y.N.; Gao, B.J.; Ren, K.; Tong, S.M.; Ying, S.H.; Feng, M.G. P-type Na+/K+ ATPases essential and nonessential for cellular homeostasis and insect pathogenicity of Beauveria bassiana. Virulence 2020, 11, 1415–1431. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, Z.K.; Sun, H.H.; Ying, S.H.; Feng, M.G. Characterization of the Hog1 MAPK pathway in the entomopathogenic fungus Beauveria bassiana. Environ. Microbiol. 2017, 19, 1808–1821. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.J.; Mou, Y.N.; Tong, S.M.; Ying, S.H.; Feng, M.G. Subtilisin-like Pr1 proteases marking the evolution of pathogenicity in a wide-spectrum insect-pathogenic fungus. Virulence 2020, 11, 365–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.Z.; Xia, Y.X.; Kim, B.; Keyhani, N.O. Two hydrophobins are involved in fungal spore coat rodlet layer assembly and each play distinct roles in surface interactions, development and pathogenesis in the entomopathogenic fungus Beauveria bassiana. Mol. Microbiol. 2011, 80, 811–826. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, Q.; Wang, J.; Xie, J.; Jiang, D.; Keyhani, N.O. The Elongator Subunit Elp3 Regulates Development, Stress Tolerance, Cell Cycle, and Virulence in the Entomopathogenic Fungus Beauveria bassiana. J. Fungi 2022, 8, 834. https://doi.org/10.3390/jof8080834
Cai Q, Wang J, Xie J, Jiang D, Keyhani NO. The Elongator Subunit Elp3 Regulates Development, Stress Tolerance, Cell Cycle, and Virulence in the Entomopathogenic Fungus Beauveria bassiana. Journal of Fungi. 2022; 8(8):834. https://doi.org/10.3390/jof8080834
Chicago/Turabian StyleCai, Qing, Juanjuan Wang, Jiatao Xie, Daohong Jiang, and Nemat O. Keyhani. 2022. "The Elongator Subunit Elp3 Regulates Development, Stress Tolerance, Cell Cycle, and Virulence in the Entomopathogenic Fungus Beauveria bassiana" Journal of Fungi 8, no. 8: 834. https://doi.org/10.3390/jof8080834
APA StyleCai, Q., Wang, J., Xie, J., Jiang, D., & Keyhani, N. O. (2022). The Elongator Subunit Elp3 Regulates Development, Stress Tolerance, Cell Cycle, and Virulence in the Entomopathogenic Fungus Beauveria bassiana. Journal of Fungi, 8(8), 834. https://doi.org/10.3390/jof8080834






