RNA-Seq Provides Insights into the Mechanisms Underlying Ilyonectria robusta Responding to Secondary Metabolites of Bacillus methylotrophicus NJ13
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Culture Conditions of Strains
2.2. Preparation of CBLJ-3’s Conidia and Fermentation Filtrates without NJ13’s Cells
2.3. Antifungal Activity Assay and Microscopic Observation
2.4. Effect of NJ13’s SMs on the Germination of I. robusta Conidia
2.5. Measurement of Mycelial Extracellular Conductivity
2.6. Detection of Malondialdehyde (MDA) in CBLJ-3’s Mycelia in the Presence of NJ13’s SMs
2.7. Measurement of the Glucose Produced by CBLJ-3 in the Presence of NJ13’s SMs
2.8. Control Effect on Ginseng Rusty Roots Infected by I. robusta Using NJ13’s SMs
2.9. RNA Extraction and Sequencing
2.10. Data Analysis
2.11. QRT-PCR Validation
2.12. Statistical Analysis
3. Results
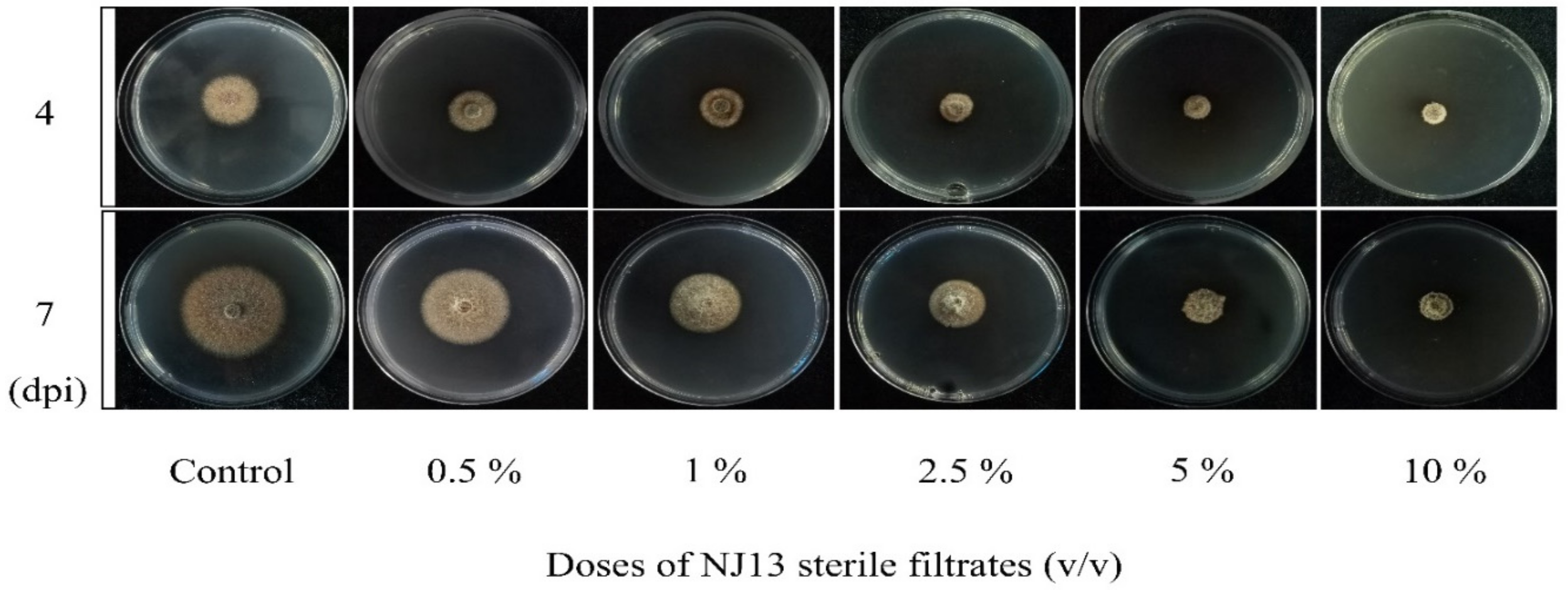
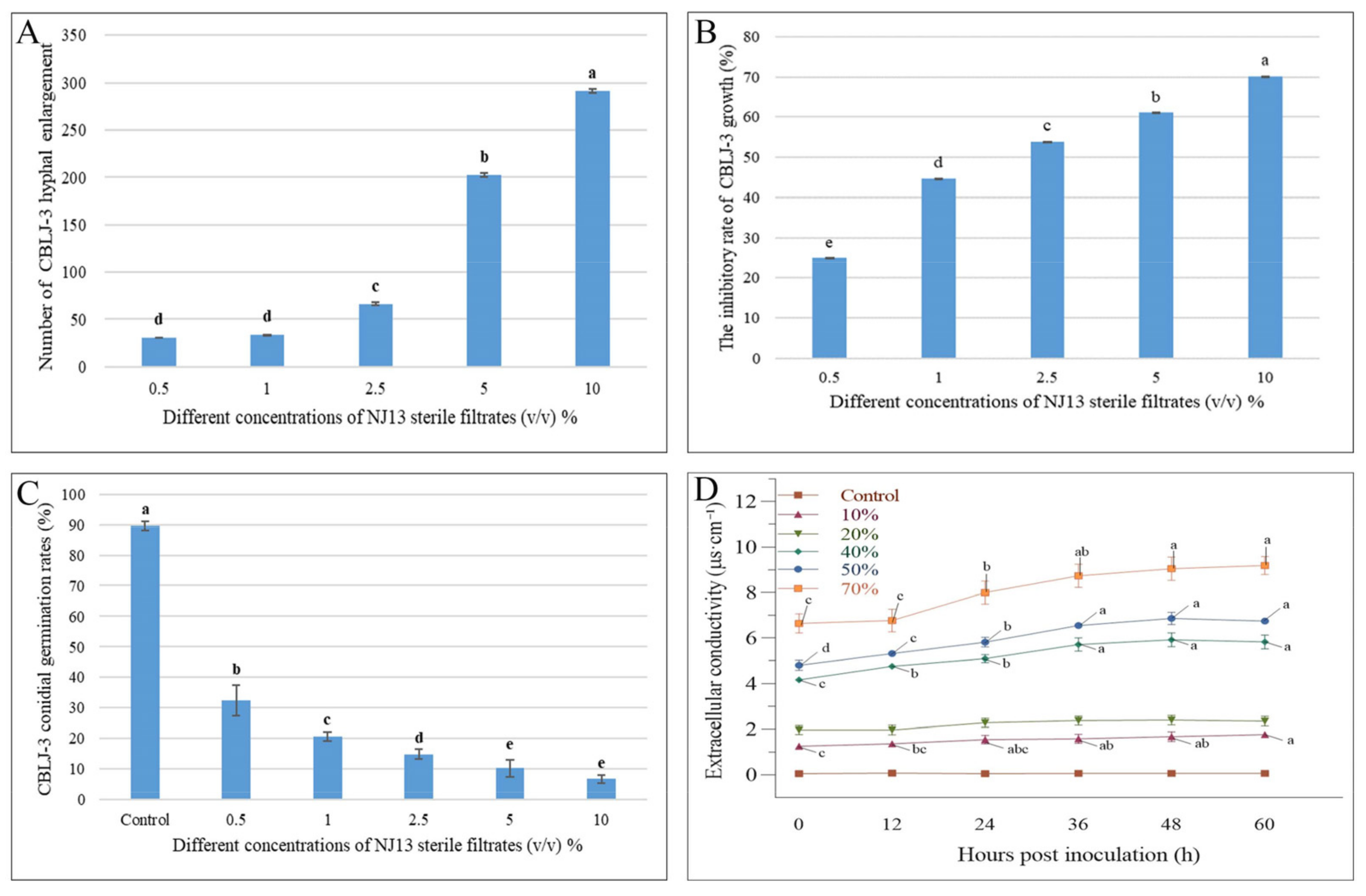
3.1. SMs from B. methylotrophicus NJ13 Inhibited CBLJ-3
3.2. Effect of NJ13’s SMs on the Germination of CBLJ-3’s Conidia
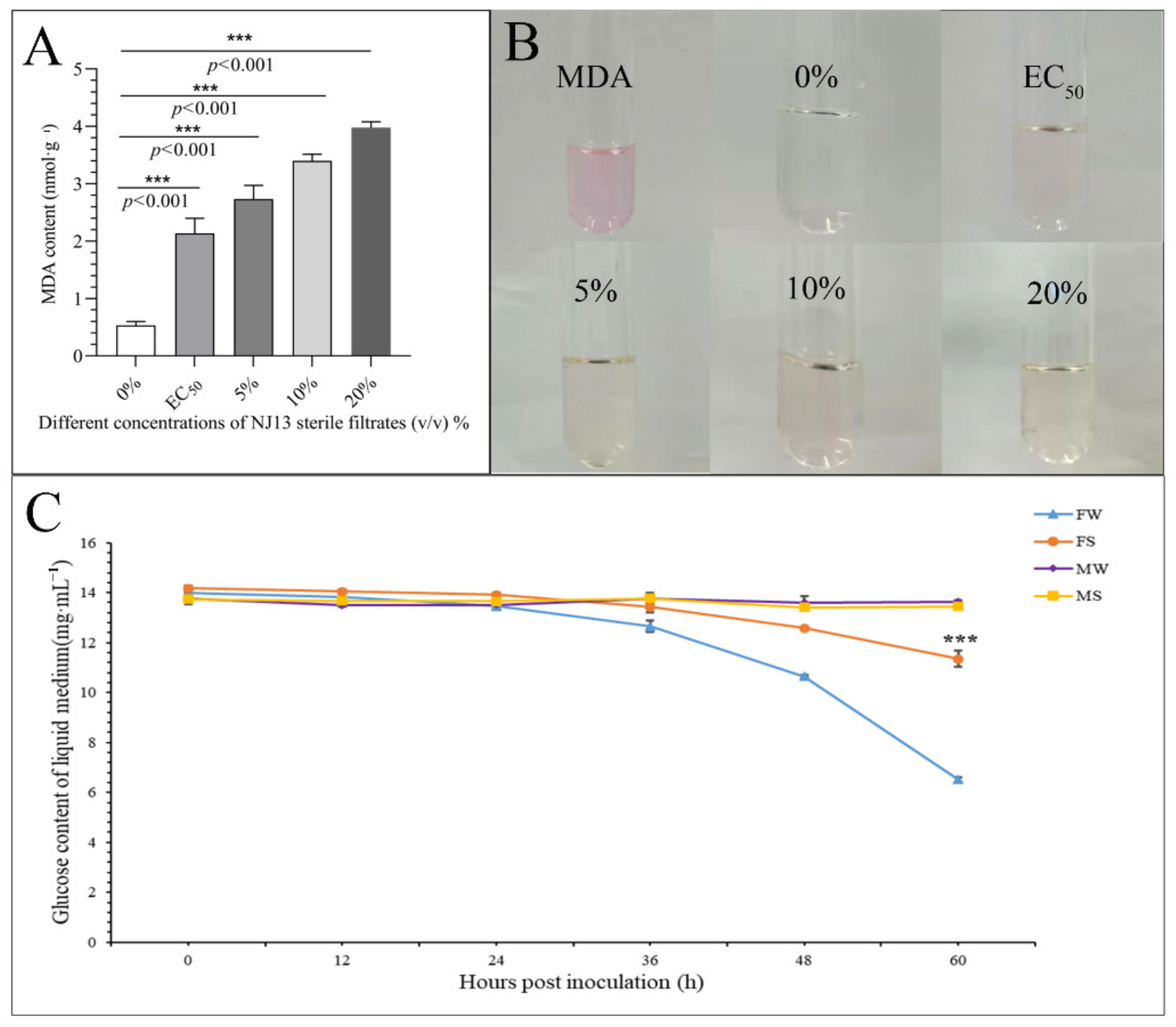
3.3. Effect of NJ13’s SMs on the Cell Membrane of CBLJ-3
3.4. Glucose Absorption of Strain CBLJ-3 Was Affected by NJ13 Sterile Filtrates
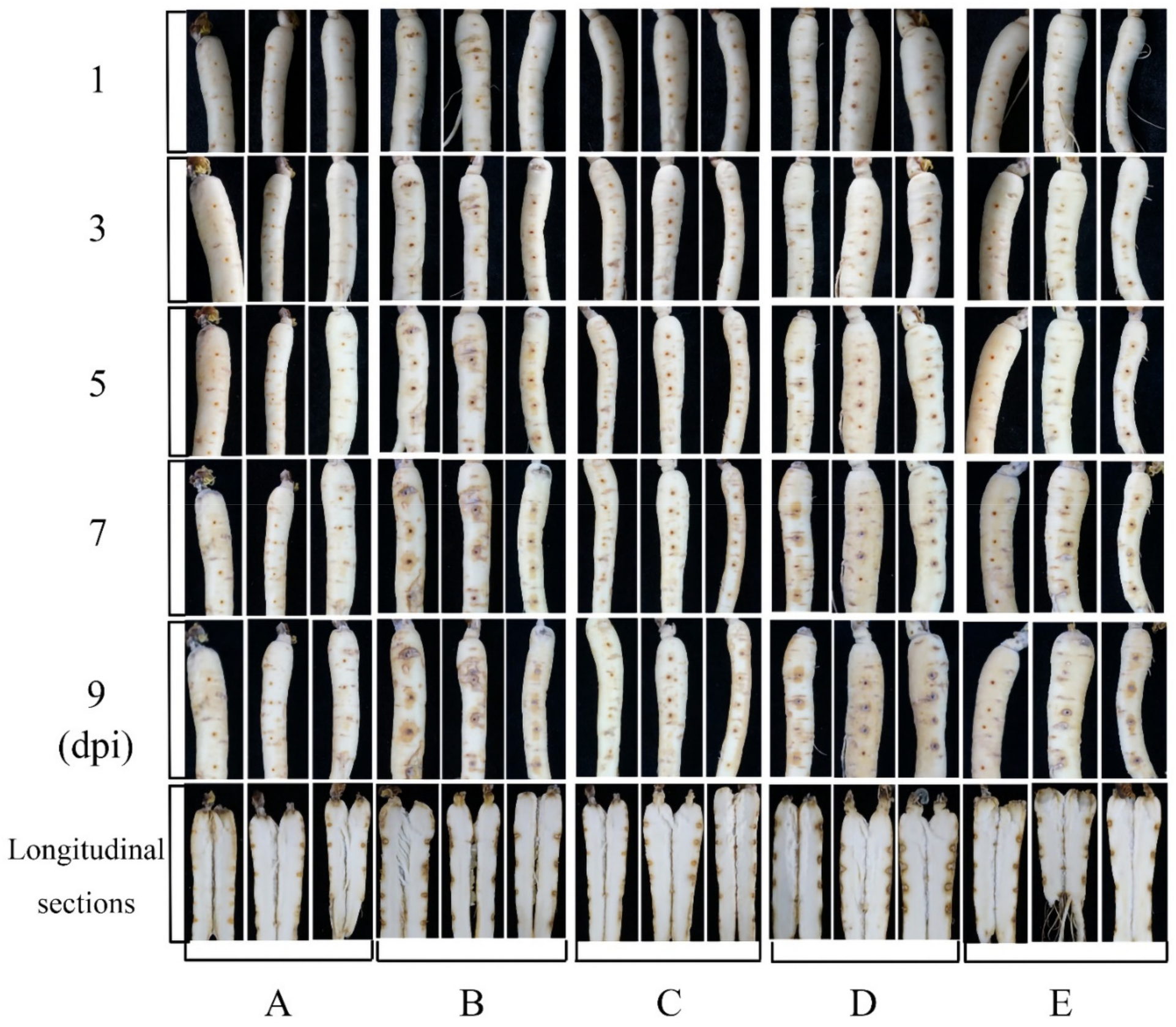
3.5. In Vivo Test of Antifungal Activity in Panax ginseng Roots
3.6. Quality Control and Quantification of Gene Expression Levels
3.7. Overall Analysis of Differentially Expressed Genes (DEGs)
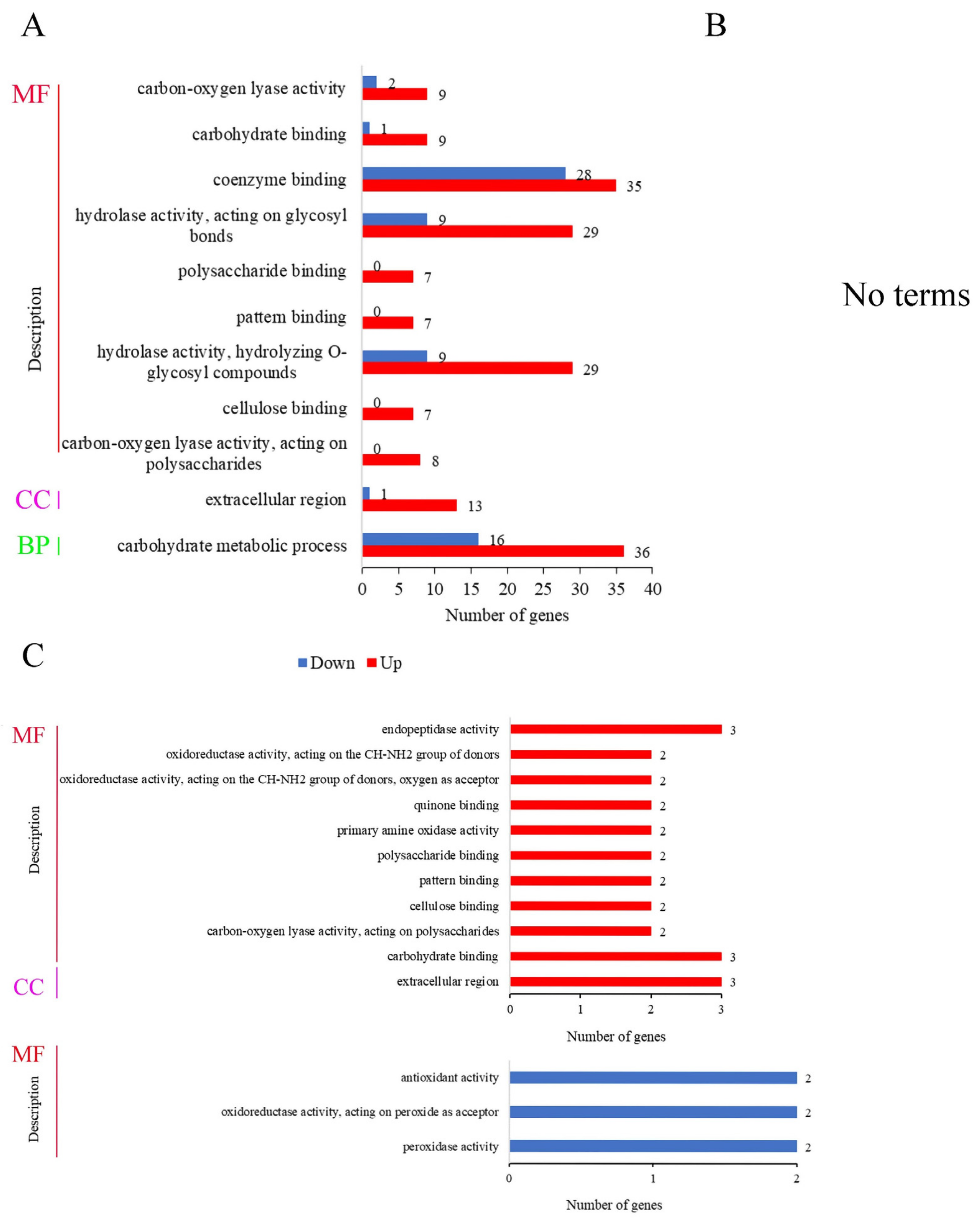
3.8. GO Enrichment Analysis of DEGs
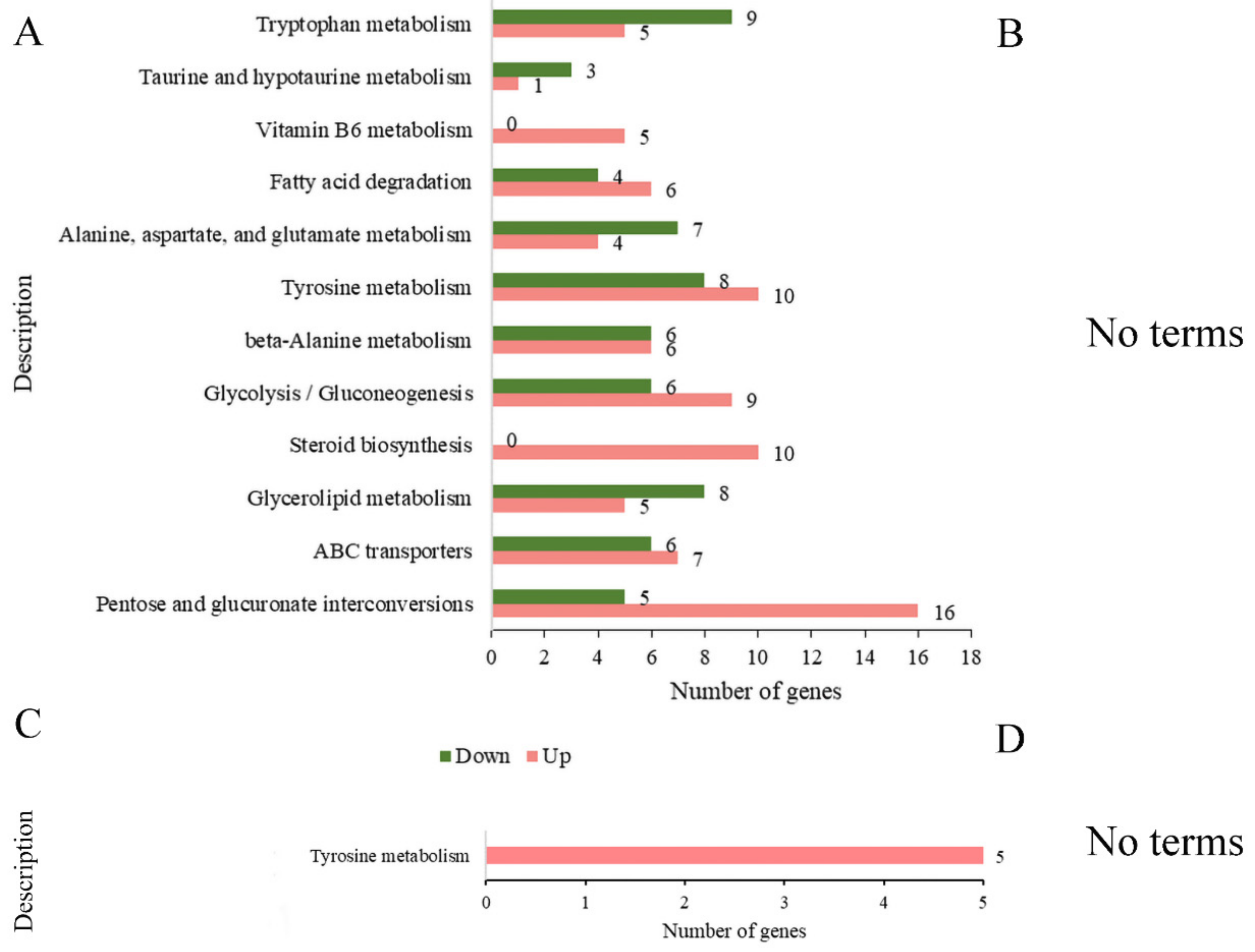
3.9. KEGG Enrichment Analysis of DEGs
3.10. Analysis of DEGs in Response to Stress Caused by B. methylotrophicus NJ13’s SMs
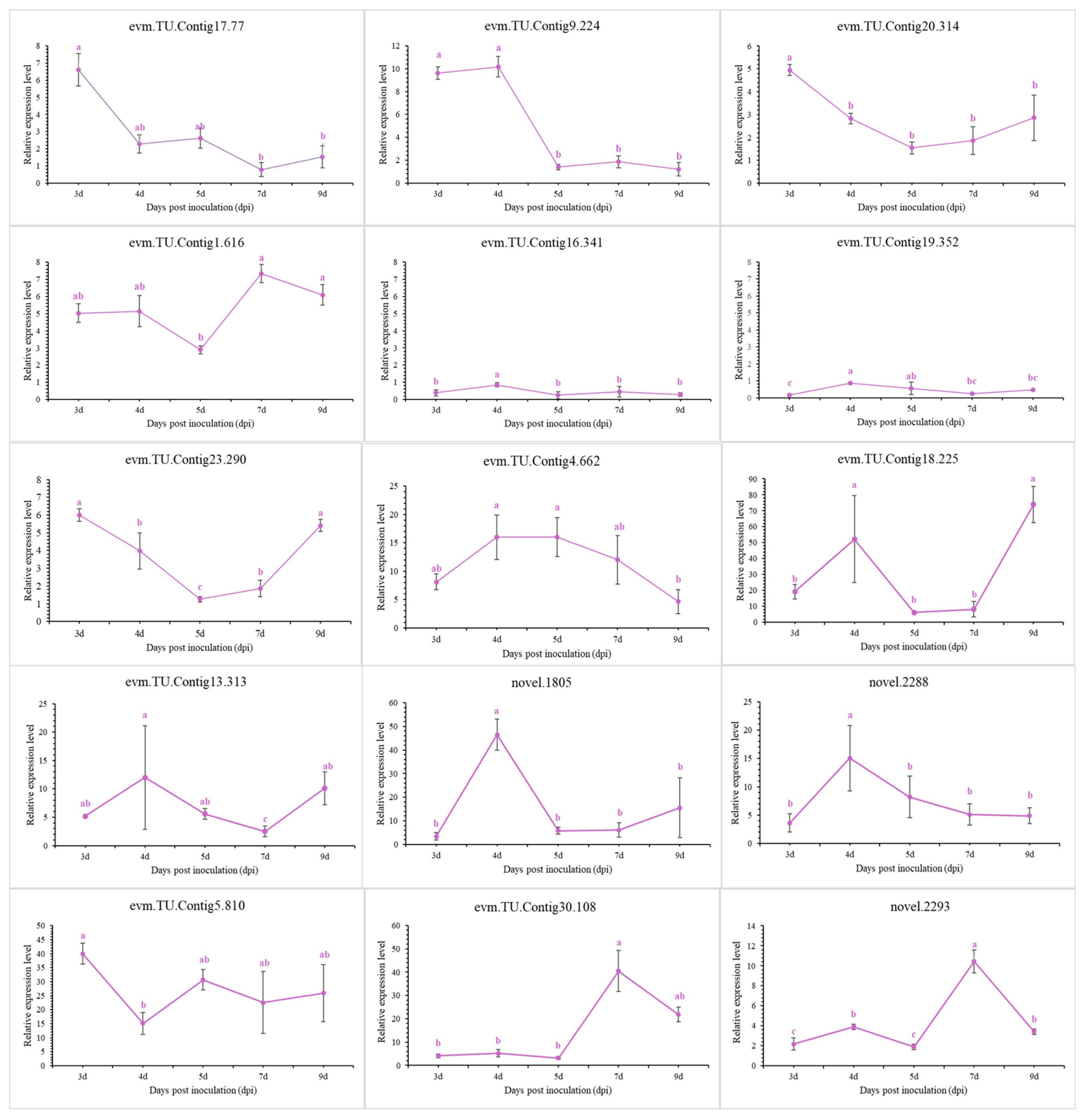
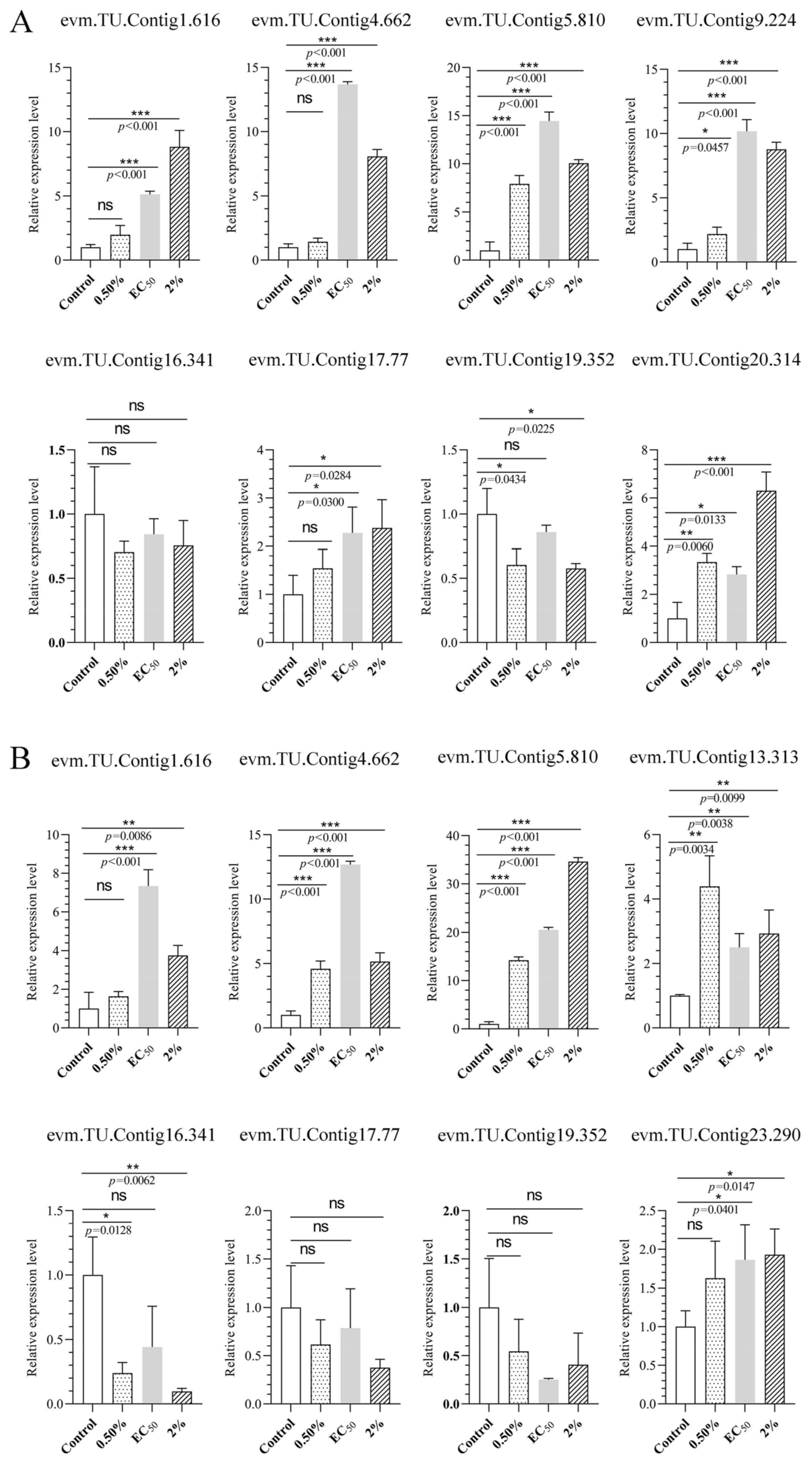
3.11. Validation of Transcriptomic Results Using QRT-PCR
4. Discussion
4.1. Effect of the Antifungal Activity of B. methylotrophicus NJ13’s SMs on CBLJ-3
4.2. Effects of B. methylotrophicus NJ13’s SMs on the Cellular Structure of CBLJ-3
4.3. Analysis of the Effect of B. methylotrophicus NJ13’s SMs on CBLJ-3’s Metabolism
4.4. Unanswered Questions and Future Possibilities
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ernst, E. Panax ginseng: An overview of the clinical evidence. J. Ginseng Res. 2010, 34, 259–263. [Google Scholar] [CrossRef] [Green Version]
- Park, J.D.; Rhee, D.K.; Lee, Y.H. Biological activities and chemistry of saponins from Panax ginseng C. A. Meyer. Phytochem. Rev. 2005, 4, 159–175. [Google Scholar] [CrossRef] [Green Version]
- Cho, K.M.; Hong, S.Y.; Lee, S.M.; Kim, Y.H.; Kahng, G.G.; Lim, Y.P.; Kim, H.; Yun, H.D. Endophytic bacterial communities in ginseng and their antifungal activity against pathogens. Microb. Ecol. 2007, 54, 341–351. [Google Scholar] [CrossRef]
- Sathiyaraj, G.; Srinivasan, S.; Subramanium, S.; Kim, Y.-J.; Kim, Y.-J.; Kwon, W.-S.; Yang, D.-C. Polygalacturonase inhibiting protein: Isolation, developmental regulation and pathogen related expression in Panax ginseng C.A. Meyer. Mol. Biol. Rep. 2010, 37, 3445–3454. [Google Scholar] [CrossRef]
- Lu, X.H.; Jiao, X.L.; Chen, A.J.; Luo, Y.; Gao, W.W. First report of Ilyonectria robusta causing rusty root of Asian ginseng in China. Plant Dis. 2015, 99, 156. [Google Scholar] [CrossRef]
- Halleen, F.; Schroers, H.-J.; Groenewald, J.Z.; Rego, C.; Oliveira, H.; Crous, P.W. Neonectria liriodendri sp. nov., the main causal agent of black foot disease of grapevines. Stud. Mycol. 2006, 55, 227–234. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Diz, M.; Díaz-Losada, E.; Armengol, J.; León, M.; Berlanas, C.; Andrés-Sodupe, M.; Gramaje, D. First report of Ilyonectria robusta causing black foot disease of grapevine in Spain. Plant Dis. 2018, 102, 2381. [Google Scholar] [CrossRef]
- Tewoldemedhin, Y.T.; Mazzola, M.; Mostert, L.; McLeod, A. Cylindrocarpon species associated with apple tree roots in South Africa and their quantification using real-time PCR. Eur. J. Plant Pathol. 2011, 129, 637–651. [Google Scholar] [CrossRef]
- Castlebury, L.A.; Rossman, A.Y.; Hyten, A.S. Phylogenetic relationships of Neonectria/Cylindrocarpon on fagus in North America. Botany 2006, 84, 1417–1433. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.; Cao, J.; Li, Y.; Luo, T.; Zhu, T.; Li, S.; Liu, Y.; Qiao, T.; Yang, C.; Qin, G. Root rot of Codonopsis tangshen caused by Ilyonectria robusta in Chongqing, China. Plant Dis. 2021, 106, 1989. [Google Scholar] [CrossRef]
- Karthikeyan, V.; Gnanamanickam, S. Biological control of Setaria blast (Magnaporthe grisea) with bacterial strains. Crop Prot. 2008, 27, 263–267. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, H.; Xu, S.; Liu, K.; Qi, H.; Wang, M.; Chen, X.; Berg, G.; Ma, Z.; Cernava, T.; et al. Bacterial-fungal interactions under agricultural settings: From physical to chemical interactions. Stress Biol. 2022, 2, 1–17. [Google Scholar] [CrossRef]
- Ongena, M.; Jacques, P. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 2008, 16, 115–125. [Google Scholar] [CrossRef]
- Xie, Y.; Peng, Q.; Ji, Y.; Xie, A.; Yang, L.; Mu, S.; Li, Z.; He, T.; Xiao, Y.; Zhao, J. Isolation and identification of antibacterial bioactive compounds from Bacillus megaterium L2. Front. Microbiol. 2021, 12, 645484. [Google Scholar] [CrossRef]
- Falardeau, J.; Wise, C.; Novitsky, L.; Avis, T.J. Ecological and mechanistic insights into the direct and indirect antimicrobial properties of Bacillus subtilis lipopeptides on plant pathogens. J. Chem. Ecol. 2013, 39, 869–878. [Google Scholar] [CrossRef]
- Park, Y.-H.; Chandra Mishra, R.; Yoon, S.; Kim, H.; Park, C.; Seo, S.-T.; Bae, H. Endophytic Trichoderma citrinoviride isolated from mountain-cultivated ginseng (Panax ginseng) has great potential as a biocontrol agent against ginseng pathogens. J. Ginseng Res. 2019, 43, 408–420. [Google Scholar] [CrossRef]
- Tumbarski, Y.; Deseva, I.; Mihaylova, D.; Stoyanova, M.; Krastev, L.; Nikolova, R.; Yanakieva, V.; Ivanov, I. Isolation, characterization and amino acid composition of a bacteriocin produced by Bacillus methylotrophicus strain BM47. Food Technol. Biotechnol. 2018, 56, 546–552. [Google Scholar] [CrossRef]
- Im, S.M.; Yu, N.H.; Joen, H.W.; Kim, S.O.; Park, H.W.; Park, A.R.; Kim, J.-C. Biological control of tomato bacterial wilt by oxydifficidin and difficidin-producing Bacillus methylotrophicus DR-08. Pestic. Biochem. Physiol. 2020, 163, 130–137. [Google Scholar] [CrossRef]
- Deleu, M.; Paquot, M.; Nylander, T. Fengycin interaction with lipid monolayers at the air–aqueous interface—implications for the effect of fengycin on biological membranes. J. Colloid Interface Sci. 2005, 283, 358–365. [Google Scholar] [CrossRef]
- Rodriguez, R.; Redman, R. Balancing the generation and elimination of reactive oxygen species. Proc. Natl. Acad. Sci. USA 2005, 102, 3175–3176. [Google Scholar] [CrossRef] [Green Version]
- Han, Q.; Wu, F.; Wang, X.; Qi, H.; Shi, L.; Ren, A.; Liu, Q.; Zhao, M.; Tang, C. The bacterial lipopeptide iturins induce V erticillium dahliae cell death by affecting fungal signalling pathways and mediate plant defence responses involved in pathogen-associated molecular pattern-triggered immunity. Environ. Microbiol. 2015, 17, 1166–1188. [Google Scholar] [CrossRef]
- Gu, Q.; Yang, Y.; Yuan, Q.; Shi, G.; Wu, L.; Lou, Z.; Huo, R.; Wu, H.; Borriss, R.; Gao, X. Bacillomycin D produced by Bacillus amyloliquefaciens is involved in the antagonistic interaction with the plant-pathogenic fungus Fusarium graminearum. Appl. Environ. Microbiol. 2017, 83, e01075-17. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Kuipers, O.P. Identification and classification of known and putative antimicrobial compounds produced by a wide variety of Bacillales species. BMC Genom. 2016, 17, 882. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Zhang, S.-w.; Li, K.-t. Antagonistic activity and mechanism of an isolated Streptomyces corchorusii stain AUH-1 against phytopathogenic fungi. World J. Microbiol. Biotechnol. 2019, 35, 145. [Google Scholar] [CrossRef]
- Miethke, M.; Klotz, O.; Linne, U.; May, J.J.; Beckering, C.L.; Marahiel, M.A. Ferri-bacillibactin uptake and hydrolysis in Bacillus subtilis. Mol. Microbiol. 2006, 61, 1413–1427. [Google Scholar] [CrossRef]
- Niehus, R.; Picot, A.; Oliveira, N.M.; Mitri, S.; Foster, K.R. The evolution of siderophore production as a competitive trait. Evolution 2017, 71, 1443–1455. [Google Scholar] [CrossRef]
- Barakat, A.; DiLoreto, D.S.; Zhang, Y.; Smith, C.; Baier, K.; Powell, W.A.; Wheeler, N.; Sederoff, R.; Carlson, J.E. Comparison of the transcriptomes of American chestnut (Castanea dentata) and Chinese chestnut (Castanea mollissima) in response to the chestnut blight infection. BMC Plant Biol. 2009, 9, 51. [Google Scholar] [CrossRef] [Green Version]
- Dassanayake, M.; Haas, J.S.; Bohnert, H.J.; Cheeseman, J.M. Shedding light on an extremophile lifestyle through transcriptomics. New Phytol. 2009, 183, 764–775. [Google Scholar] [CrossRef]
- Gkarmiri, K.; Finlay, R.D.; Alström, S.; Thomas, E.; Cubeta, M.A.; Högberg, N. Transcriptomic changes in the plant pathogenic fungus Rhizoctonia solani AG-3 in response to the antagonistic bacteria Serratia proteamaculans and Serratia plymuthica. BMC Genom. 2015, 16, 630. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Yin, W.; Chen, C.; Tian, L.; Xu, P. Identification and optimized fermentation condition of an endophyte antagonistic Bacteria NJ13. Agrochemicals 2013, 52, 97–101. [Google Scholar] [CrossRef]
- Chen, C.-Q.; Yan, D.; Jiang, Y.; Xu, P.; Chu, Y.-X.; Gao, J. Inhibition effect of biocontrol bacteria NJ13 and its mixture with chemical fungicides against ginseng root rot caused by Fusarium solani. China J. Chin. Mater. Med. 2019, 44, 2015–2019. [Google Scholar] [CrossRef]
- Chen, C.-Q.; Jin, H.; Jiang, Y.; Xu, P.; Yan, D.; Gao, J. Co-toxicity and field control effect of biocontrol strain NJ13 and its mixture with chemical fungicides against Alternaria panax. Agrochemicals 2019, 58, 381–384. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Y.-L.; Li, J.; Gao, J.; Jiang, Y.; Chen, C.-Q. Genomic and transcriptomic analyses of Bacillus methylotrophicus NJ13 reveal a molecular response strategy combating Ilyonectria robusta causing ginseng rusty root rot. Biol. Control 2022, 172, 104972. [Google Scholar] [CrossRef]
- Jiang, Y.; Ran, C.; Chen, L.; Yin, W.; Liu, Y.; Chen, C.; Gao, J. Purification and characterization of a novel antifungal flagellin protein from endophyte Bacillus methylotrophicus NJ13 against Ilyonectria robusta. Microorganisms 2019, 7, 605. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Diao, Y.; Wang, W.; Hao, J.; Imran, M.; Duan, H.; Liu, X. Assessing the risk for resistance and elucidating the genetics of Colletotrichum truncatum that is only sensitive to some DMI fungicides. Front. Microbiol. 2017, 8, 1779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [Green Version]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [Green Version]
- Mering, C.V.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.-L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2013, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, G. Hyphal growth: A tale of motors, lipids, and the spitzenkörper. Eukaryot. Cell 2007, 6, 351–360. [Google Scholar] [CrossRef] [Green Version]
- Sha, Y.; Wang, Q.; Li, Y. Suppression of Magnaporthe oryzae and interaction between Bacillus subtilis and rice plants in the control of rice blast. SpringerPlus 2016, 5, 1238. [Google Scholar] [CrossRef] [Green Version]
- Ge, B.; Liu, B.; Nwet, T.T.; Zhao, W.; Shi, L.; Zhang, K. Bacillus methylotrophicus strain NKG-1, isolated from Changbai Mountain, China, has potential applications as a biofertilizer or biocontrol agent. PLoS ONE 2016, 11, e0166079. [Google Scholar] [CrossRef] [Green Version]
- Villa-Rodríguez, E.; Parra-Cota, F.; Castro-Longoria, E.; López-Cervantes, J.; de los Santos-Villalobos, S. Bacillus subtilis TE3: A promising biological control agent against Bipolaris sorokiniana, the causal agent of spot blotch in wheat (Triticum turgidum L. subsp. durum). Biol. Control 2019, 132, 135–143. [Google Scholar] [CrossRef]
- Chen, Y.; Gao, X.; Chen, Y.; Qin, H.; Huang, L.; Han, Q. Inhibitory efficacy of endophytic Bacillus subtilis EDR4 against Sclerotinia sclerotiorum on rapeseed. Biol. Control 2014, 78, 67–76. [Google Scholar] [CrossRef]
- Tang, Q.; Bie, X.; Lu, Z.; Lv, F.; Tao, Y.; Qu, X. Effects of fengycin from Bacillus subtilis fmbJ on apoptosis and necrosis in Rhizopus stolonifer. J. Microbiol. 2014, 52, 675–680. [Google Scholar] [CrossRef]
- Kim, K.; Lee, Y.; Ha, A.; Kim, J.I.; Park, A.R.; Yu, N.H.; Son, H.; Choi, G.J.; Park, H.W.; Lee, C.W.; et al. Chemosensitization of Fusarium graminearum to chemical fungicides using cyclic lipopeptides produced by Bacillus amyloliquefaciens strain JCK-12. Front. Plant Sci. 2017, 8, 2010. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Le, K.D.; Yu, N.H.; Kim, J.I.; Kim, J.-C.; Lee, C.W. Structure and antifungal activity of pelgipeptins from Paenibacillus elgii against phytopathogenic fungi. Pestic. Biochem. Physiol. 2020, 163, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Hasim, S.; Coleman, J.J. Targeting the fungal cell wall: Current therapies and implications for development of alternative antifungal agents. Future Med. Chem. 2019, 11, 869–883. [Google Scholar] [CrossRef] [PubMed]
- Gow Neil, A.R.; Latge, J.-P.; Munro Carol, A.; Heitman, J. The fungal cell wall: Structure, biosynthesis, and function. Microbiol. Spectr. 2017, 5, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Gui, Y.-J.; Chen, J.-Y.; Zhang, D.-D.; Li, N.-Y.; Li, T.-G.; Zhang, W.-Q.; Wang, X.-Y.; Short, D.P.G.; Li, L.; Guo, W.; et al. Verticillium dahliae manipulates plant immunity by glycoside hydrolase 12 proteins in conjunction with carbohydrate-binding module 1. Environ. Microbiol. 2017, 19, 1914–1932. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.X.; Li, B.H.; Zhou, S.Y. A novel glycoside hydrolase 74 xyloglucanase CvGH74A is a virulence factor in Coniella vitis. J. Integr. Agric. 2020, 19, 2725–2735. [Google Scholar] [CrossRef]
- Abubaker, K.S.; Sjaarda, C.; Castle, A.J. Regulation of three genes encoding cell-wall-degrading enzymes of Trichoderma aggressivum during interaction with Agaricus bisporus. Can. J. Microbiol. 2013, 59, 417–424. [Google Scholar] [CrossRef]
- Abdelmoteleb, A.; Troncoso-Rojas, R.; Gonzalez-Soto, T.; González-Mendoza, D. Antifungical activity of autochthonous Bacillus subtilis isolated from Prosopis juliflora against phytopathogenic fungi. Mycobiology 2017, 45, 385–391. [Google Scholar] [CrossRef] [Green Version]
- Sant, D.G.; Tupe, S.G.; Ramana, C.V.; Deshpande, M.V. Fungal cell membrane-promising drug target for antifungal therapy. J. Appl. Microbiol. 2016, 121, 1498–1510. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, S.; Esquivel Brooke, D.; White Theodore, C.; Lorenz, M. Overexpression or deletion of ergosterol biosynthesis genes alters doubling time, response to stress agents, and drug susceptibility in Saccharomyces cerevisiae. MBio 2018, 9, e01291-18. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Tong, J.; Lee, C.W.; Ha, S.; Eom, S.H.; Im, Y.J. Structural mechanism of ergosterol regulation by fungal sterol transcription factor Upc2. Nat. Commun. 2015, 6, 6129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zavrel, M.; Hoot Sam, J.; White Theodore, C. Comparison of sterol import under aerobic and anaerobic conditions in three fungal species, Candida albicans, Candida glabrata, and Saccharomyces cerevisiae. Eukaryot. Cell 2013, 12, 725–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warnecke, D.; Erdmann, R.; Fahl, A.; Hube, B.; Müller, F.; Zank, T.; Zähringer, U.; Heinz, E. Cloning and functional expression of UGT genes encoding sterol glucosyltransferases from Saccharomyces cerevisiae, Candida albicans, Pichia pastoris, and Dictyostelium discoideum. J. Biol. Chem. 1999, 274, 13048–13059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Li, J.; Gao, J.; Cai, P.; Han, X.; Tian, C. Identification and characterization of the glucose dual-affinity transport system in Neurospora crassa: Pleiotropic roles in nutrient transport, signaling, and carbon catabolite repression. Biotechnol. Biofuels 2017, 10, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Rodriguez, R. Glucose regulation of the paralogous glucose sensing receptors Rgt2 and Snf3 of the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129881. [Google Scholar] [CrossRef]
- Maier, A.; Volker, B.; Boles, E.; Fuhrmann, G.F. Characterisation of glucose transport in Saccharomyces cerevisiae with plasma membrane vesicles (countertransport) and intact cells (initial uptake) with single Hxt1, Hxt2, Hxt3, Hxt4, Hxt6, Hxt7 or Gal2 transporters. FEMS Yeast Res. 2002, 2, 539–550. [Google Scholar] [CrossRef] [Green Version]
- Krishnan, N.; Velramar, B.; Velu, R.K. Investigation of antifungal activity of surfactin against mycotoxigenic phytopathogenic fungus Fusarium moniliforme and its impact in seed germination and mycotoxicosis. Pestic. Biochem. Physiol. 2019, 155, 101–107. [Google Scholar] [CrossRef]
- Desjardins, A.E.; Proctor, R.H. Molecular biology of Fusarium mycotoxins. Int. J. Food Microbiol. 2007, 119, 47–50. [Google Scholar] [CrossRef]
- Niehaus, E.M.; Kleigrewe, K.; Wiemann, P.; Studt, L.; Sieber, C.M.; Connolly, L.R.; Freitag, M.; Guldener, U.; Tudzynski, B.; Humpf, H.U. Genetic manipulation of the Fusarium fujikuroi fusarin gene cluster yields insight into the complex regulation and fusarin biosynthetic pathway. Chem. Biol. 2013, 20, 1055–1066. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, M.; Mahmud, N.U.; Gupta, D.R.; Tareq, F.S.; Shin, H.J.; Islam, T. Inhibitory effects of linear lipopeptides from a marine Bacillus subtilis on the wheat blast fungus Magnaporthe oryzae Triticum. Front. Microbiol. 2020, 11, 665. [Google Scholar] [CrossRef]
- Biesenthal, C.J.; Zaharia, I.L. Comparison of the phytotoxic activity of the phytotoxin destruxin B and four natural analogs. Plant Sci. Int. J. Exp. Plant Biol. 2000, 156, 185–192. [Google Scholar] [CrossRef]
- Wang, B.; Kang, Q.; Lu, Y.; Bai, L.; Wang, C. Unveiling the biosynthetic puzzle of destruxins in Metarhizium species. Proc. Natl. Acad. Sci. USA 2016, 113, E4578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morschhauser, J. Regulation of multidrug resistance in pathogenic fungi. Fungal Genet. Biol. 2010, 47, 94–106. [Google Scholar] [CrossRef]
- Duffy, B.; Schouten, A.; Raaijmakers, J.M. Pathogen self-defense: Mechanisms to counteract microbial antagonism. Annu. Rev. Phytopathol. 2003, 41, 501–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasad, R.; Banerjee, A.; Khandelwal, N.K.; Dhamgaye, S. The ABCs of Candida albicans multidrug transporter Cdr1. Eukaryot. Cell 2015, 14, 1154–1164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.-H.; Ji, Y.-P.; Qu, Y.-Y.; Qi, Y.-K.; Li, D.-W.; Liu, Z.-Y.; Wu, X.-Q. The response strategies of Colletotrichum gloeosporioides s.s. due to the stress caused by biological control agent Bacillus amyloliquefaciens deciphered by transcriptome analyses. Biol. Control 2020, 150, 104372. [Google Scholar] [CrossRef]
- Omrane, S.; Sghyer, H.; Audeon, C.; Lanen, C.; Duplaix, C.; Walker, A.S.; Fillinger, S. Fungicide efflux and the MgMFS1 transporter contribute to the multidrug resistance phenotype in Zymoseptoria tritici field isolates. Environ. Microbiol. 2015, 17, 2805–2823. [Google Scholar] [CrossRef]
- Viglas, J.; Olejnikova, P. An update on ABC transporters of filamentous fungi—from physiological substrates to xenobiotics. Microbiol. Res. 2021, 246, 126684. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, Y.; Li, B.; Chen, T.; Tian, S. Reactive oxygen species: A generalist in regulating development and pathogenicity of phytopathogenic fungi. Comput. Struct. Biotechnol. J. 2020, 18, 3344–3349. [Google Scholar] [CrossRef]
- Zhang, L.; Sun, C.; Master Emma, R. Fengycins, cyclic lipopeptides from marine Bacillus subtilis strains, kill the plant-pathogenic fungus Magnaporthe grisea by inducing reactive oxygen species production and chromatin condensation. Appl. Environ. Microbiol. 2018, 84, e00445-18. [Google Scholar] [CrossRef] [Green Version]
- Ma, D.; Cui, X.; Zhang, Z.; Li, B.; Xu, Y.; Tian, S.; Chen, T. Honokiol suppresses mycelial growth and reduces virulence of Botrytis cinerea by inducing autophagic activities and apoptosis. Food Microbiol. 2020, 88, 103411. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-H.; Xu, F.; Snyder, J.H.; Shi, H.-B.; Lu, J.-P.; Lin, F.-C. Autophagy in plant pathogenic fungi. Semin. Cell Dev. Biol. 2016, 57, 128–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reggiori, F.; Klionsky Daniel, J. Autophagy in the eukaryotic cell. Eukaryot. Cell 2002, 1, 11–21. [Google Scholar] [CrossRef] [Green Version]
- Pollack, J.K.; Li, Z.J.; Marten, M.R. Fungal mycelia show lag time before re-growth on endogenous carbon. Biotechnol. Bioeng. 2008, 100, 458–465. [Google Scholar] [CrossRef]
- Jin, X.; Guo, L.; Jin, B.; Zhu, S.; Mei, X.; Wu, J.; Liu, T.; He, X. Inhibitory mechanism of 6-Pentyl-2H-pyran-2-one secreted by Trichoderma atroviride T2 against Cylindrocarpon destructans. Pestic. Biochem. Physiol. 2020, 170, 104683. [Google Scholar] [CrossRef]
- Mariño, G.; Niso-Santano, M.; Baehrecke, E.H.; Kroemer, G. Self-consumption: The interplay of autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2014, 15, 81–94. [Google Scholar] [CrossRef] [Green Version]
- Chu, L.L.; Bae, H. Bacterial endophytes from ginseng and their biotechnological application. J. Ginseng Res. 2022, 46, 1–10. [Google Scholar] [CrossRef]
- Kim, D.-S.; Cook, R.J.; Weller, D.M. Bacillus sp. L324-92 for biological control of three root diseases of wheat grown with reduced tillage. Phytopathology 1997, 87, 551–558. [Google Scholar] [CrossRef] [Green Version]
- Fira, D.; Dimkić, I.; Berić, T.; Lozo, J.; Stanković, S. Biological control of plant pathogens by Bacillus species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar] [CrossRef]
- Jangir, M.; Pathak, R.; Sharma, S.; Sharma, S. Biocontrol mechanisms of Bacillus sp., isolated from tomato rhizosphere, against Fusarium oxysporum f. sp. lycopersici. Biol. Control 2018, 123, 60–70. [Google Scholar] [CrossRef]
- Zhou, L.; Yuen, G.; Wang, Y.; Wei, L.; Ji, G. Evaluation of bacterial biological control agents for control of root-knot nematode disease on tomato. Crop Prot. 2016, 84, 8–13. [Google Scholar] [CrossRef]
- Frikha-Gargouri, O.; Ben Abdallah, D.; Ghorbel, I.; Charfeddine, I.; Jlaiel, L.; Triki, M.A.; Tounsi, S. Lipopeptides from a novel Bacillus methylotrophicus 39b strain suppress Agrobacterium crown gall tumours on tomato plants. Pest Manag. Sci. 2017, 73, 568–574. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Li, J.; Meng, Z.; Zhang, S.; Dong, B.; Qiao, K. Synergistic effect of combined application of a new fungicide fluopimomide with a biocontrol agent Bacillus methylotrophicus TA-1 for management of gray mold in tomato. Plant Dis. 2019, 103, 1991–1997. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Li, M.; Liu, X.; Jiang, Y.; Zhao, D.; Gao, J.; Wang, Z.; Jiang, Y.; Chen, C. RNA-Seq Provides Insights into the Mechanisms Underlying Ilyonectria robusta Responding to Secondary Metabolites of Bacillus methylotrophicus NJ13. J. Fungi 2022, 8, 779. https://doi.org/10.3390/jof8080779
Li X, Li M, Liu X, Jiang Y, Zhao D, Gao J, Wang Z, Jiang Y, Chen C. RNA-Seq Provides Insights into the Mechanisms Underlying Ilyonectria robusta Responding to Secondary Metabolites of Bacillus methylotrophicus NJ13. Journal of Fungi. 2022; 8(8):779. https://doi.org/10.3390/jof8080779
Chicago/Turabian StyleLi, Xiang, Mengtao Li, Xiangkai Liu, Yilin Jiang, Dongfang Zhao, Jie Gao, Zhenhui Wang, Yun Jiang, and Changqing Chen. 2022. "RNA-Seq Provides Insights into the Mechanisms Underlying Ilyonectria robusta Responding to Secondary Metabolites of Bacillus methylotrophicus NJ13" Journal of Fungi 8, no. 8: 779. https://doi.org/10.3390/jof8080779
APA StyleLi, X., Li, M., Liu, X., Jiang, Y., Zhao, D., Gao, J., Wang, Z., Jiang, Y., & Chen, C. (2022). RNA-Seq Provides Insights into the Mechanisms Underlying Ilyonectria robusta Responding to Secondary Metabolites of Bacillus methylotrophicus NJ13. Journal of Fungi, 8(8), 779. https://doi.org/10.3390/jof8080779






