Integrated Transcriptomics and Nontargeted Metabolomics Analysis Reveal Key Metabolic Pathways in Ganoderma lucidum in Response to Ethylene
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
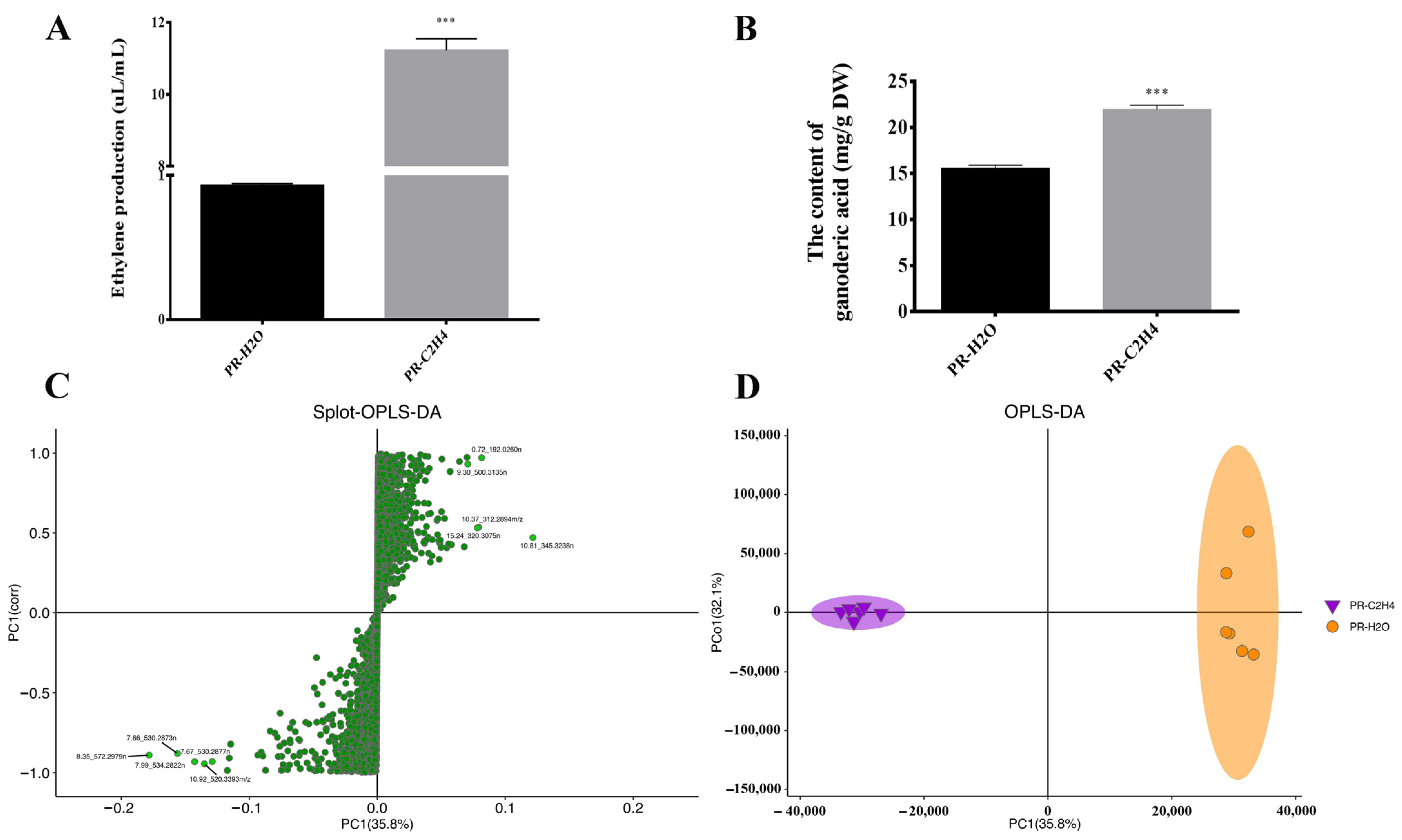
2.2. Measurement of Ethylene Production and GA Content
2.3. Sample Preparation and LC-MS Analysis
2.4. Metabolite Identification and Quantification
2.5. Transcriptome Sequencing Analysis
2.6. Integrative Analysis of Metabolome and Transcriptome
2.7. WGCNA
3. Results
3.1. The Metabolome Profiling of G. lucidum in Response to Ethylene
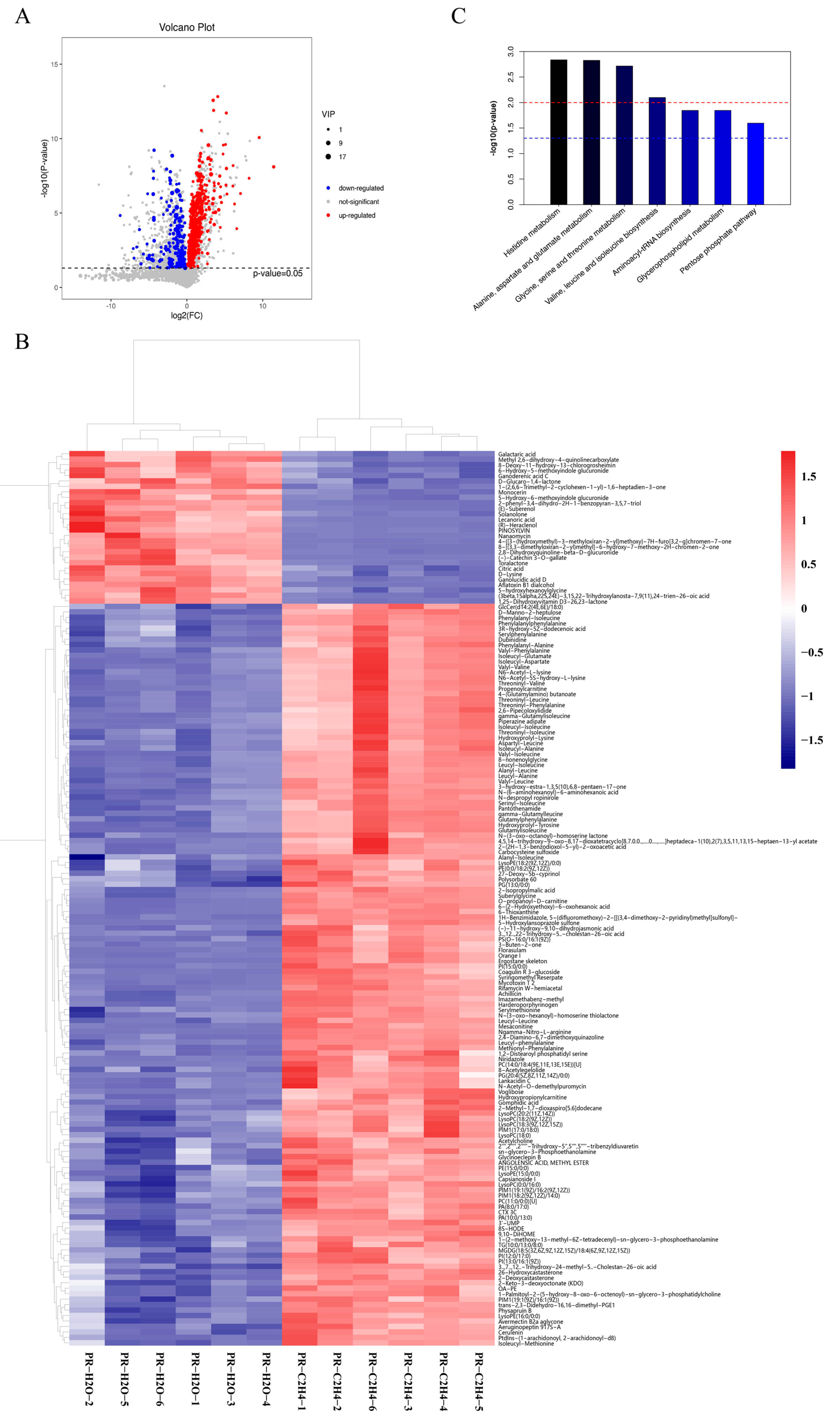
3.2. Differentially Accumulated Metabolites in Response to Ethylene
3.3. Transcriptome Profiling of G. lucidum in Response to Ethylene
3.4. Integrated Analysis of Transcriptomics and Metabolomics Data
4. Discussion
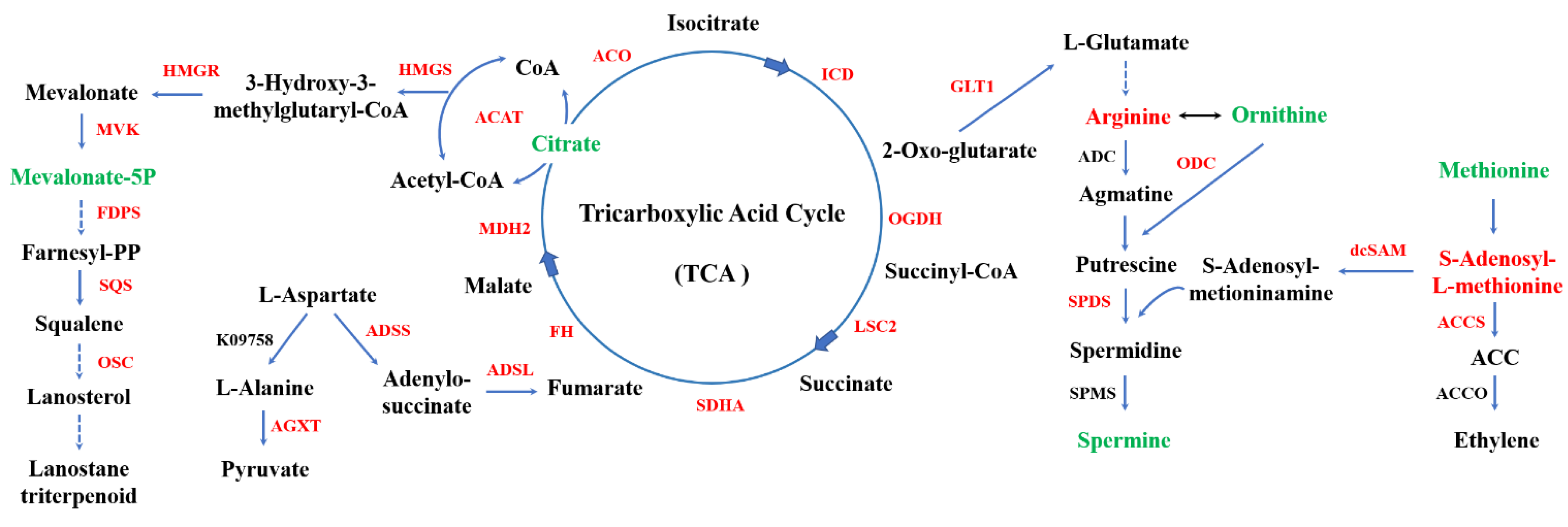
4.1. Terpenoid Backbone Biosynthesis Pathway
4.2. Polyamine Metabolism Pathway
4.3. Ethylene Signaling Pathway
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Guo, J.; Bao, J.; Li, X.; Chen, X.; Lu, J.; Wang, Y. Anti-cancer properties of triterpenoids isolated from Ganoderma lucidum-a review. Expert Opin. Investig. Drugs 2013, 22, 981–992. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.Q.; Zhang, Y.J.; Tian, Z.K. Anti-oxidant, anti-inflammatory and anti-fibrosis effects of ganoderic acid A on carbon tetrachloride induced nephrotoxicity by regulating the Trx/TrxR and JAK/ROCK pathway. Chem.-Biol. Interact. 2021, 344, 109529. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Shi, K.; Dong, J.; Jin, Z.; Wu, Y.; Cai, Y.; Lin, T.; Cai, Q.; Liu, L.; Zhang, Y. Ganoderic acid A attenuates high-fat-diet-induced liver injury in rats by regulating the lipid oxidation and liver inflammation. Arch. Pharmacal Res. 2020, 43, 744–754. [Google Scholar] [CrossRef] [PubMed]
- Ren, A.; Shi, L.; Zhu, J.; Yu, H.; Jiang, A.; Zheng, H.; Zhao, M. Shedding light on the mechanisms underlying the environmental regulation of secondary metabolite ganoderic acid in Ganoderma lucidum using physiological and genetic methods. Fungal Genet. Biol. 2019, 128, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Ren, A.; Qin, L.; Shi, L.; Dong, X.; Mu, D.S.; Li, Y.X.; Zhao, M.W. Methyl jasmonate induces ganoderic acid biosynthesis in the basidiomycetous fungus Ganoderma lucidum. Bioresour. Technol. 2010, 101, 6785–6790. [Google Scholar] [CrossRef]
- Jiang, A.; Liu, Y.; Liu, R.; Ren, A.; Ma, H.; Shu, L.; Shi, L.; Zhu, J.; Zhao, M. Integrated proteomics and metabolomics analysis provides insights into ganoderic acid biosynthesis in response to methyl jasmonate in Ganoderma lucidum. Int. J. Mol. Sci. 2019, 20, 6116. [Google Scholar] [CrossRef] [Green Version]
- Cao, P.F.; Wu, C.; Dang, Z.; Shi, L.; Jiang, A.; Ren, A.; Zhao, M. Effects of exogenous salicylic acid on ganoderic acid biosynthesis and the expression of key genes in the ganoderic acid biosynthesis pathway in the lingzhi or reishi medicinal mushroom, Ganoderma lucidum (Agaricomycetes). Int. J. Med. Mushrooms 2017, 19, 65–73. [Google Scholar] [CrossRef]
- Zhang, X.; Ren, A.; Li, M.J.; Cao, P.F.; Chen, T.X.; Zhang, G.; Shi, L.; Jiang, A.L.; Zhao, M.W. Heat stress modulates mycelium growth, heat shock protein expression, ganoderic acid biosynthesis, and hyphal branching of Ganoderma lucidum via cytosolic Ca2+. Appl. Environ. Microbiol. 2016, 82, 4112–4125. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.L.; Zhang, G.; Ren, A.; Dang, Z.H.; Shi, L.; Jiang, A.L.; Zhao, M.W. The pH-responsive transcription factor PacC regulates mycelial growth, fruiting body development, and ganoderic acid biosynthesis in Ganoderma lucidum. Mycologia 2016, 108, 1104–1113. [Google Scholar]
- Zhao, W.; Xu, J.W.; Zhong, J.J. Enhanced production of ganoderic acids in static liquid culture of Ganoderma lucidum under nitrogen-limiting conditions. Bioresour. Technol. 2011, 102, 8185–8190. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Ren, A.; Wu, F.; Yu, H.; Shi, L.; Zhao, M. Ethylene promotes mycelial growth and ganoderic acid biosynthesis in Ganoderma lucidum. Biotechnol. Lett. 2017, 39, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Papon, N.; Binder, B.M. An Evolutionary Perspective on Ethylene Sensing in Microorganisms. Trends Microbiol. 2019, 27, 193–196. [Google Scholar] [CrossRef] [PubMed]
- Ke, S.W.; Chen, G.H.; Chen, C.T.; Taken, J.T.C.; Yang, C. Ethylene signaling modulates contents of catechin and ability of antioxidant in Camellia sinensis. Bot. Stud. 2018, 59, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, W.; Xu, L.; Gao, S.; Lyu, X.; Cao, X.; Yao, Y. Melatonin alters the secondary metabolite profile of grape berry skin by promoting VvMYB14-mediated ethylene biosynthesis. Hortic. Res. 2021, 8, 43. [Google Scholar] [CrossRef]
- Choi, H.; Baek, M.W.; Cheol, L.H.; Jeong, C.S.; Tilahun, S. Changes in metabolites and antioxidant activities of green ‘Hayward’ and gold ‘Haegeum’ kiwifruits during ripening with ethylene treatment. Food Chem. 2022, 384, 132490. [Google Scholar] [CrossRef]
- Binder, B.M. Ethylene signaling in plants. J. Biol. Chem. 2020, 295, 7710–7725. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Zhang, J.; Gao, X.; Chen, J.; Zheng, Y.; Gao, Y.; Qiu, L. The molecular mechanism for the ethylene regulation of postharvest button mushrooms maturation and senescence. Postharv. Biol. Technol. 2019, 156, 110930. [Google Scholar] [CrossRef]
- Zhang, C.; Huang, T.; Shen, C.; Wang, X.; Qi, Y.; Shen, J.; Song, A.; Qiu, L.; Ai, Y. Downregulation of ethylene production increases mycelial growth and primordia formation in the button culinary-medicinal mushroom, Agaricus bisporus (Agaricomycetes). Int. J. Med. Mushrooms 2016, 18, 1131–1140. [Google Scholar] [CrossRef]
- Chagué, V. Ethylene Production by Fungi: Biological Questions and Future Developments towards a Sustainable Polymers Industry. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar] [CrossRef]
- Meng, L.; Bai, X.; Zhang, S.; Zhang, M.; Zhou, S.; Mukhtar, I.; Wang, L.; Li, Z.; Wang, W. Enhanced ganoderic acids accumulation and transcriptional responses of biosynthetic genes in Ganoderma lucidum fruiting bodies by elicitation supplementation. Int. J. Mol. Sci. 2019, 20, 2830. [Google Scholar] [CrossRef] [Green Version]
- Meng, L.; Zhang, S.; Chen, B.; Bai, X.; Li, Y.; Yang, J.; Wang, W.; Li, C.; Li, Y.; Li, Z. The MADS-box transcription factor GlMADS1 regulates secondary metabolism in Ganoderma lucidum. Mycologia 2021, 113, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Haug, K.; Cochrane, K.; Nainala, V.C.; Williams, M.; Chang, J.; Jayaseelan, K.V.; O’Donovan, C. MetaboLights: A resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2019, 48, D440–D444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Xu, J.; Liu, C.; Zhu, Y.; Nelson, D.R.; Zhou, S.; Li, C.; Wang, L.; Guo, X.; Sun, Y.; et al. Genome sequence of the model medicinal mushroom Ganoderma lucidum. Nat. Commun. 2012, 3, 913. [Google Scholar] [CrossRef] [Green Version]
- Shi, Q.Q.; Huang, Y.J.; Su, H.G.; Gao, Y.; Lu, S.Y.; Peng, X.R.; Li, X.N.; Zhou, L.; Qiu, M.H. Structurally diverse lanostane triterpenoids from medicinal and edible mushroom Ganoderma resinaceum Boud. Bioorg. Chem. 2020, 100, 103871. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Wang, W.; Wu, H.; Gong, X.; Moriguchi, T. Polyamines function in stress tolerance: From synthesis to regulation. Front. Plant Sci. 2015, 6, 827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, R.; Zhang, X.; Ren, A.; Shi, D.K.; Shi, L.; Zhu, J.; Yu, H.S.; Zhao, M.W. Heat stress-induced reactive oxygen species participate in the regulation of HSP expression, hyphal branching and ganoderic acid biosynthesis in Ganoderma lucidum. Microbiol. Res. 2018, 209, 43–54. [Google Scholar] [CrossRef]
- Rocha, R.O.; Wilson, R.A. Essential, deadly, enigmatic: Polyamine metabolism and roles in fungal cells. Fungal Biol. Rev. 2019, 33, 47–57. [Google Scholar] [CrossRef]
- Wu, C.G.; Tian, J.L.; Liu, R.; Cao, P.F.; Zhang, T.J.; Ren, A.; Shi, L.; Zhao, M.W. Ornithine decarboxylase-mediated production of putrescine influences ganoderic acid biosynthesis by regulating reactive oxygen species in Ganoderma lucidum. Appl. Environ. Microbiol. 2017, 83, e01289-17. [Google Scholar] [CrossRef] [Green Version]
- Tao, Y.; Han, X.; Ren, A.; Li, J.; Song, H.; Xie, B.; Zhao, M. Heat stress promotes the conversion of putrescine to spermidine and plays an important role in regulating ganoderic acid biosynthesis in Ganoderma lucidum. Appl. Microbiol. Biotechnol. 2021, 105, 5039–5051. [Google Scholar] [CrossRef]
- Alonso, J.M.; Stepanova, A.N. The ethylene signaling pathway. Science 2004, 306, 1513–1515. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Yin, C.C.; Ma, B.; Chen, S.; Zhang, J. Ethylene signaling in rice and Arabidopsis: New regulators and mechanisms. J. Integr. Plant Biol. 2021, 63, 102–125. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Umuhoza Karemera, N.J.; Wu, T.; Yang, Y.; Zhang, X.; Xu, X.; Wang, Y.; Han, Z. The ethylene response factor AtERF4 negatively regulates the iron deficiency response in Arabidopsis thaliana. PLoS ONE 2017, 12, e0186580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Zhang, Y.; Ren, J.; Jia, F.; Zeng, H.; Li, G.; Yang, X. Ethylene-responsive factor ERF114 mediates fungal pathogen effector PevD1-induced disease resistance in Arabidopsis thaliana. Mol. Plant Pathol. 2022, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, S.Y.; Ohta, M.; Usui, A.; Shinshi, H.; Ohme-Takagi, M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 2000, 12, 393–404. [Google Scholar]
- Cary, J.W.; Harris-Coward, P.; Scharfenstein, L.; Mack, B.M.; Chan, P.; Wei, Q.; Lebar, M.; Carter-Wientjes, C.; Majumdar, R.; Mitra, C.; et al. The Aspergillus flavus homeobox gene, hbx1, is required for development and aflatoxin production. Toxins 2017, 9, 315. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, L.; Zhou, R.; Lin, J.; Wang, Q.; Wang, P.; Wang, W.; Wang, L.; Li, Z. Integrated Transcriptomics and Nontargeted Metabolomics Analysis Reveal Key Metabolic Pathways in Ganoderma lucidum in Response to Ethylene. J. Fungi 2022, 8, 456. https://doi.org/10.3390/jof8050456
Meng L, Zhou R, Lin J, Wang Q, Wang P, Wang W, Wang L, Li Z. Integrated Transcriptomics and Nontargeted Metabolomics Analysis Reveal Key Metabolic Pathways in Ganoderma lucidum in Response to Ethylene. Journal of Fungi. 2022; 8(5):456. https://doi.org/10.3390/jof8050456
Chicago/Turabian StyleMeng, Li, Ruyue Zhou, Jialong Lin, Qingji Wang, Panmeng Wang, Wei Wang, Li Wang, and Zhuang Li. 2022. "Integrated Transcriptomics and Nontargeted Metabolomics Analysis Reveal Key Metabolic Pathways in Ganoderma lucidum in Response to Ethylene" Journal of Fungi 8, no. 5: 456. https://doi.org/10.3390/jof8050456
APA StyleMeng, L., Zhou, R., Lin, J., Wang, Q., Wang, P., Wang, W., Wang, L., & Li, Z. (2022). Integrated Transcriptomics and Nontargeted Metabolomics Analysis Reveal Key Metabolic Pathways in Ganoderma lucidum in Response to Ethylene. Journal of Fungi, 8(5), 456. https://doi.org/10.3390/jof8050456





