Two Cladosporium Fungi with Opposite Functions to the Chinese White Wax Scale Insect Have Different Genome Characters
Abstract
1. Introduction
2. Materials and Methods
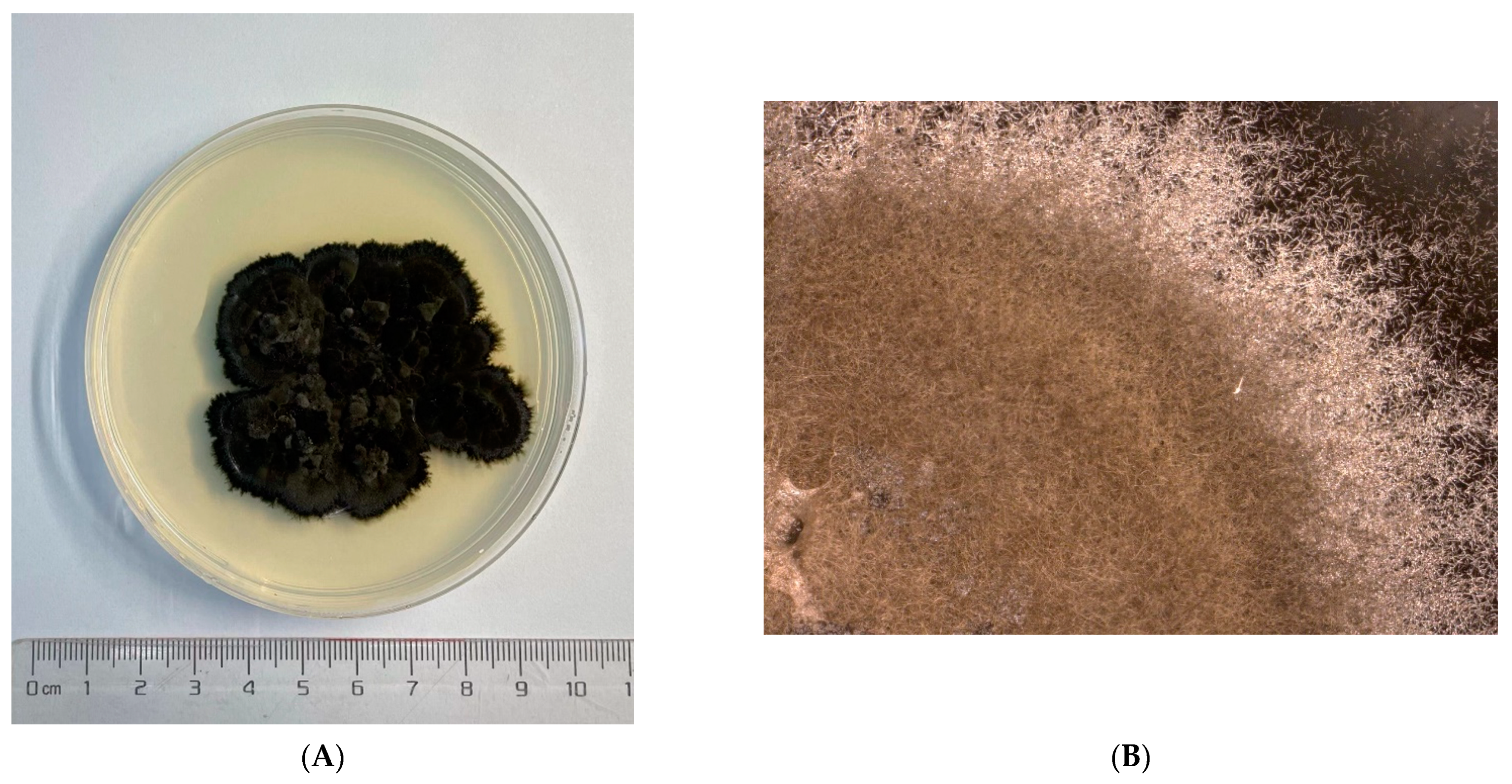
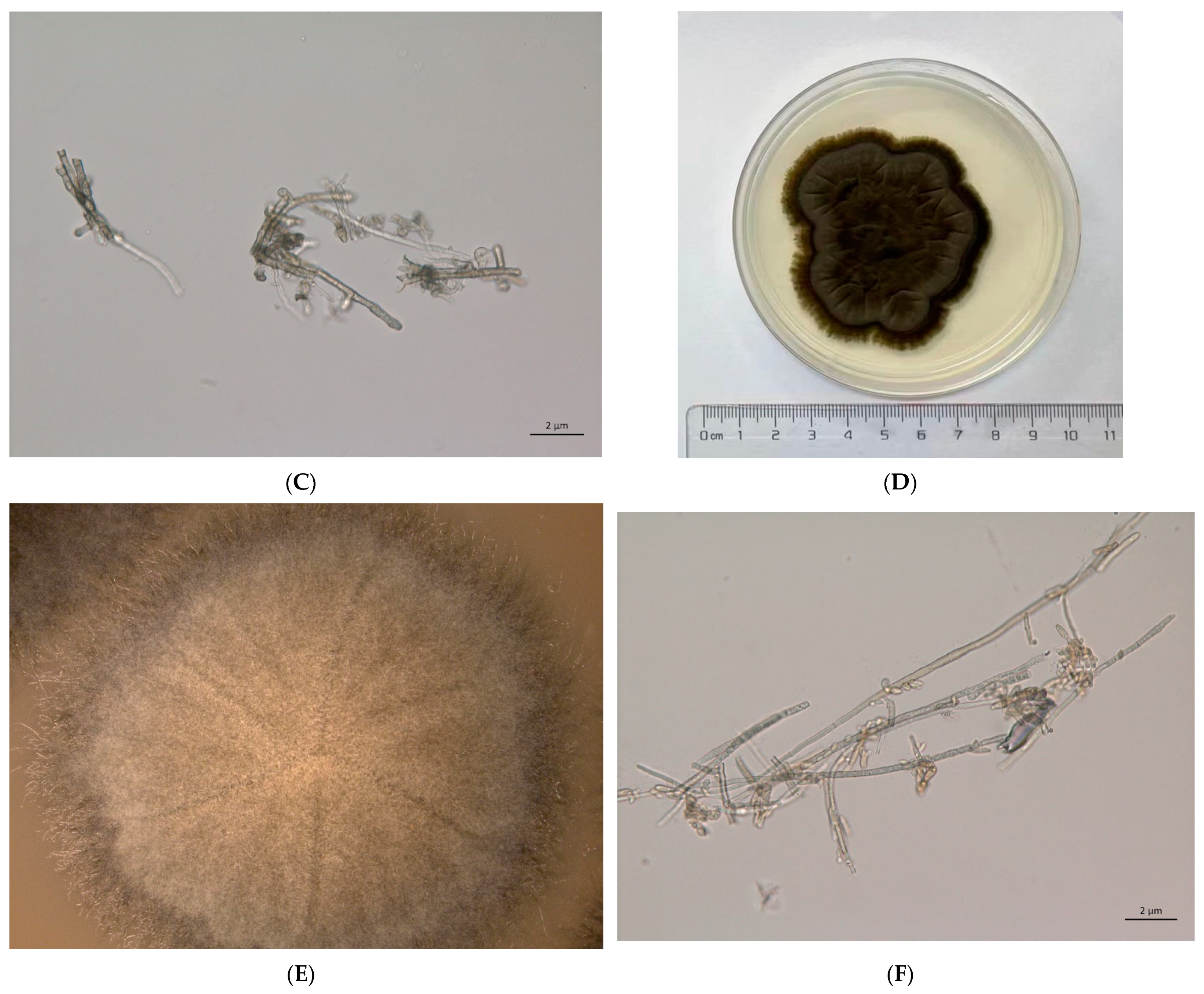
2.1. Fungi Cultivation
2.2. DNA Isolation
2.3. Library Construction and Sequence Filter
2.4. Genome Assembly
2.5. Gene Prediction
2.6. Gene Functional Annotation
2.7. Core Gene and Specific Gene Analyses
2.8. Phylogenetic Tree Construction
2.9. Synteny Analyses
2.10. Secondary Metabolite Biosynthetic Gene Clusters Prediction
2.11. Cytotoxicity Assay of the Two Cladosporium Species
3. Results
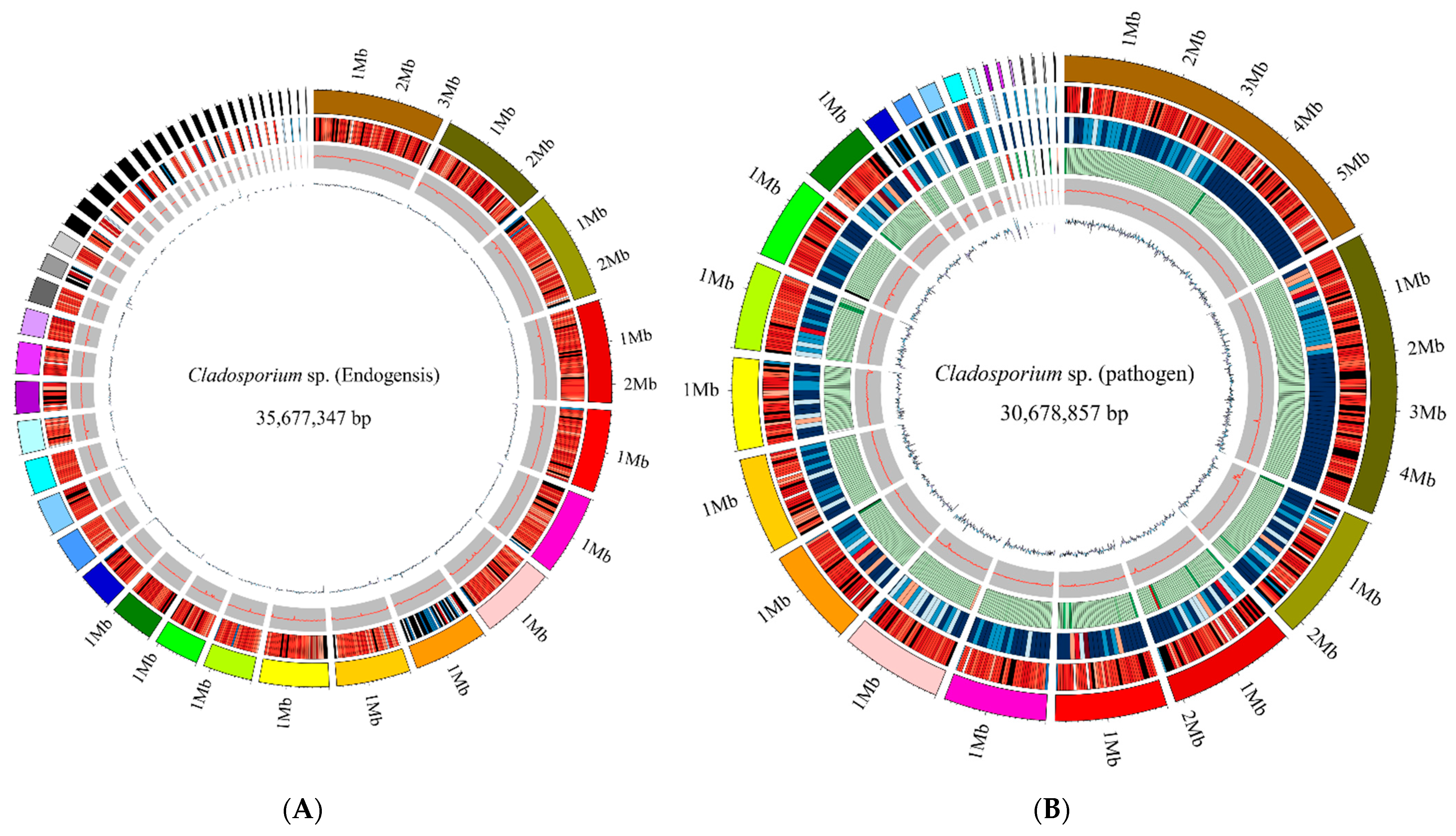
3.1. Genome Assemble Results
3.2. Genome Component Analyses
3.3. Gene Annotation and Functional Analyses
3.4. Pathogenic Analyses
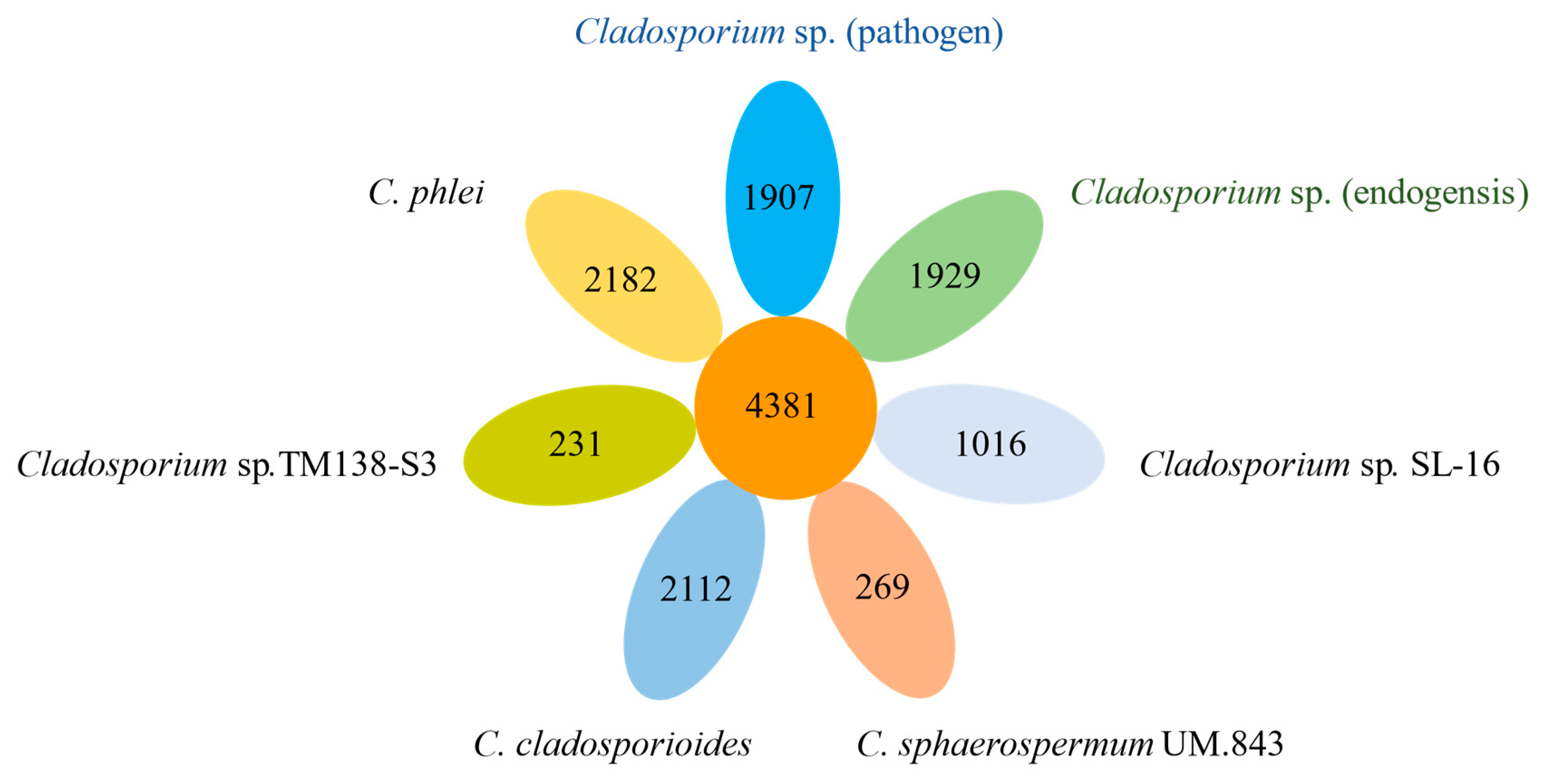
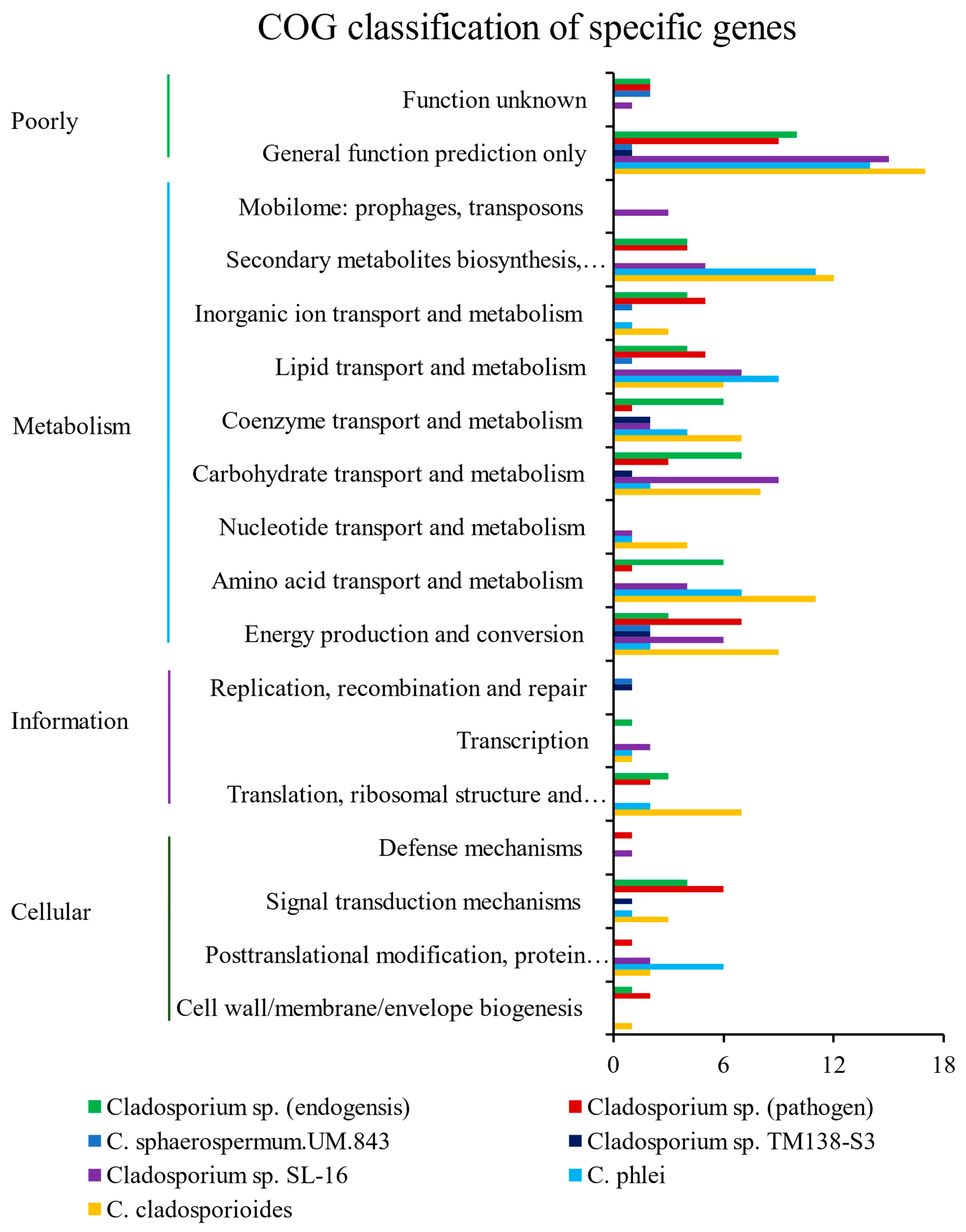
3.5. Specific Gene Analyses
3.6. Secondary Metabolites Analyses
3.7. Phylogenetic Analyses
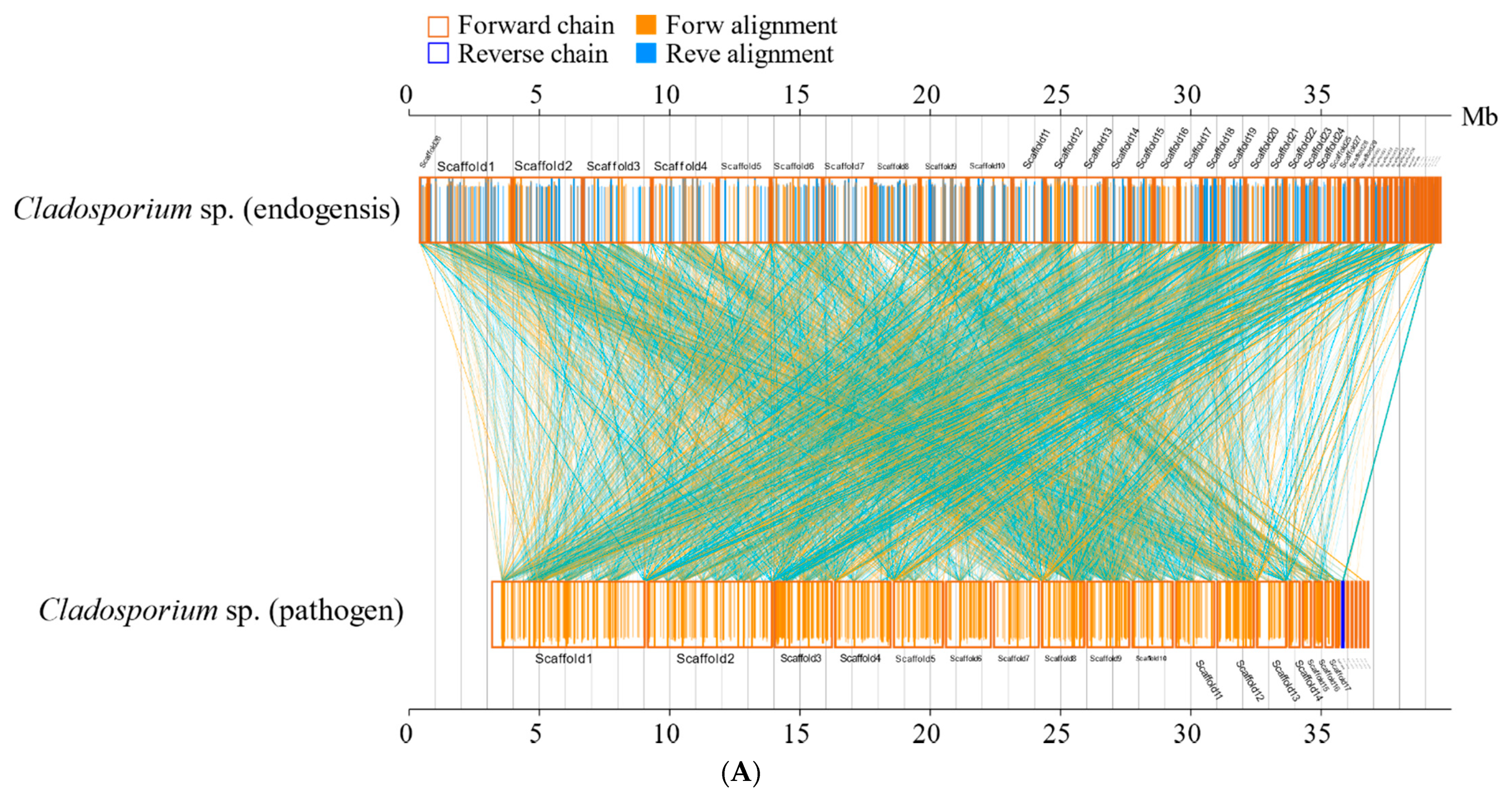
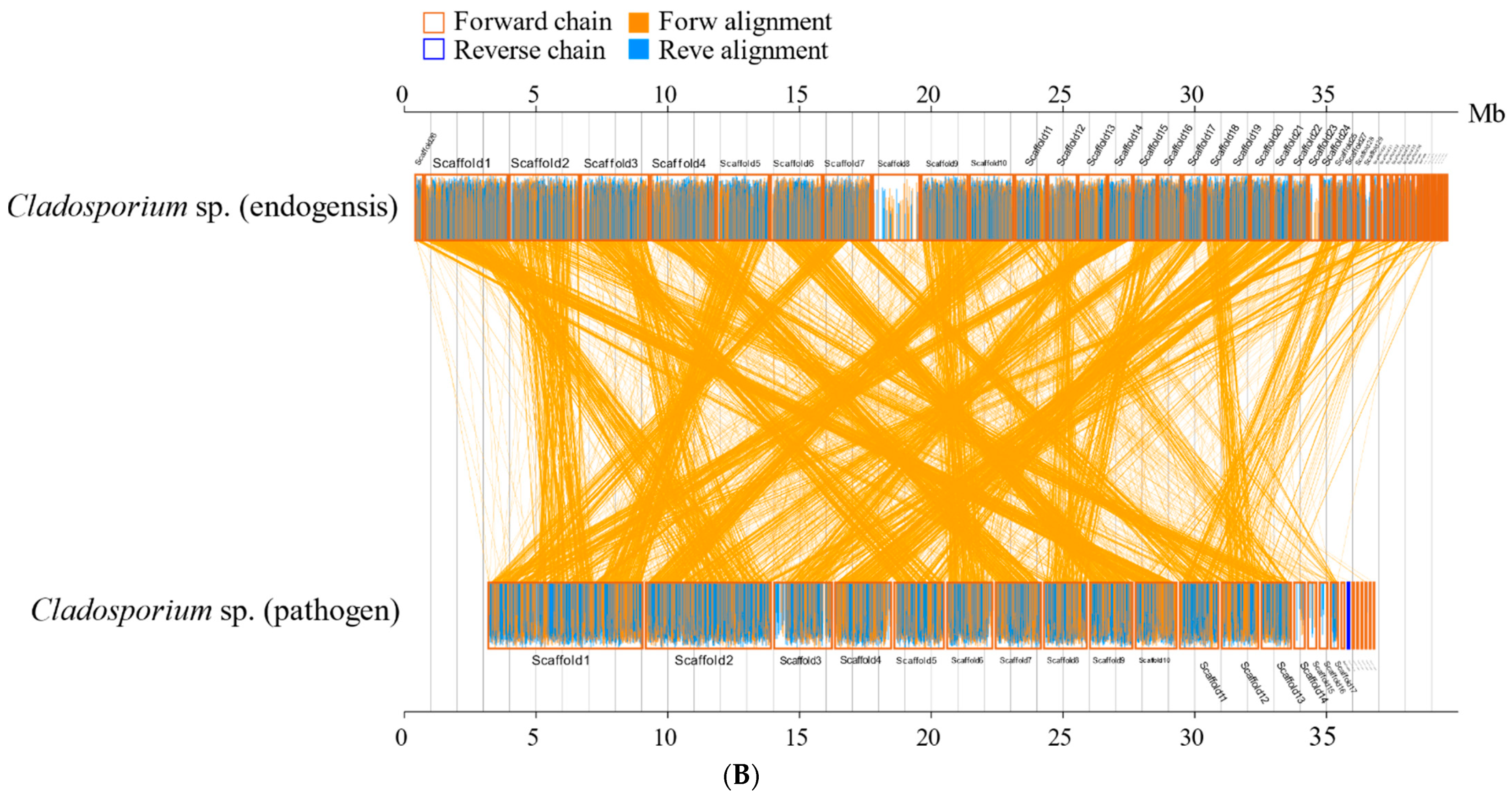
3.8. Synteny Analyses
3.9. Cytotoxicity Analyses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xie, S.; Lan, Y.; Sun, C.; Shao, Y. Insect microbial symbionts as a novel source for biotechnology. World J. Microbiol. Biotechnol. 2019, 35, 25. [Google Scholar] [CrossRef] [PubMed]
- Masson, F.; Lemaitre, B. Growing ungrowable bacteria: Overview and perspectives on insect symbiont culturability. Microbiol. Mol. Biol. Rev. 2020, 84, e00089-20. [Google Scholar] [CrossRef] [PubMed]
- Stone, L.B.L.; Bidochka, M.J. The multifunctional lifestyles of Metarhizium: Evolution and applications. Appl. Microbiol. Biotechnol. 2020, 104, 9935–9945. [Google Scholar] [CrossRef] [PubMed]
- Joseph, R.; Keyhani, N.O. Fungal mutualisms and pathosystems: Life and death in the ambrosia beetle mycangia. Appl. Microbiol. Biotechnol. 2021, 105, 3393–3410. [Google Scholar] [CrossRef] [PubMed]
- Teoh, M.C.; Furusawa, G.; Singham, G.V. Multifaceted interactions between the pseudomonads and insects: Mechanisms and prospects. Arch. Microbiol. 2021, 203, 1891–1915. [Google Scholar] [CrossRef] [PubMed]
- Attardo, G.M.; Scolari, F.; Malacrida, A. Bacterial symbionts of tsetse flies: Relationships and functional interactions between tsetse flies and their symbionts. Results Probl. Cell Differ. 2020, 69, 497–536. [Google Scholar]
- Sun, T.; Wang, X.Q.; Zhao, Z.L.; Yu, S.H.; Yang, P.; Chen, X.M. A lethal fungus infects the Chinese white wax scale insect and causes dramatic changes in the host microbiota. Sci. Rep. 2018, 8, 5324. [Google Scholar] [CrossRef]
- Yang, P.; Zhu, J.Y.; Gong, Z.J.; Xu, D.L.; Chen, X.M.; Liu, W.W.; Lin, X.D.; Li, Y.F. Transcriptome analysis of the Chinese white wax scale Ericerus pela with focus on genes involved in wax biosynthesis. PLoS ONE 2012, 7, e35719. [Google Scholar] [CrossRef][Green Version]
- Yang, P.; Chen, X.M.; Liu, W.W.; Feng, Y.; Sun, T. Transcriptome analysis of sexually dimorphic Chinese white wax scale insects reveals key differences in developmental programs and transcription factor expression. Sci. Rep. 2015, 5, 8141. [Google Scholar] [CrossRef]
- Yang, P.; Yu, S.; Hao, J.; Liu, W.; Zhao, Z.; Zhu, Z.; Sun, T.; Wang, X.; Song, Q. Genome sequence of the Chinese white wax scale insect Ericerus pela: The first draft genome for the Coccidae family of scale insects. Gigascience 2019, 8, giz113. [Google Scholar] [CrossRef]
- Zhao, Z.L.; Yu, S.H.; Zhou, W.W.; Zhu, Z.R.; Guo, Z.J.; Yang, P. The composition and function of the honeydew secreted by Ericerus pela as deduced from metabolome and transcriptome data. J. Appl. Entomol. 2018, 5, 1007–1018. [Google Scholar]
- Zhan, Y.; Tian, H.; Ji, X.; Liu, Y. Myzus persicae (Hemiptera: Aphididae) infestation increases the risk of bacterial contamination and alters nutritional content in storage Chinese cabbage. J. Sci. Food Agric. 2020, 100, 3007–3012. [Google Scholar] [CrossRef] [PubMed]
- Salvatore, M.M.; Andolfi, A.; Nicoletti, R. The genus Cladosporium: A rich source of diverse and bioactive natural compounds. Molecules 2021, 26, 3959. [Google Scholar] [CrossRef] [PubMed]
- Iturrieta-González, I.; García, D.; Gené, J. Novel species of Cladosporium from environmental sources in Spain. MycoKeys 2021, 77, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Zimowska, B.; Becchimanzi, A.; Krol, E.D.; Furmanczyk, A.; Bensch, K.; Nicoletti, R. New Cladosporium species from normal and galled flowers of Lamiaceae. Pathogens 2021, 10, 369. [Google Scholar] [CrossRef] [PubMed]
- Kratzsch, D.; Nenoff, P.; Uhrlaß, S.; Wendt, S.; Simon, J.C.; Treudler, R. Phaeohyphomycosis caused by Cladosporium cladosporioides in an immunosuppressed boy with Ewing sarcoma. J. Dtsch. Dermatol. Ges. 2021, 19, 1204–1206. [Google Scholar] [CrossRef]
- Ma, X.; Hu, J.; Yu, Y.; Wang, C.; Gu, Y.; Cao, S.; Huang, X.; Wen, Y.; Zhao, Q.; Wu, R.; et al. Assessment of the pulmonary adaptive immune response to Cladosporium cladosporioides infection using an experimental mouse model. Sci. Rep. 2021, 11, 909. [Google Scholar] [CrossRef]
- Li, C.; Cao, P.; Du, C.; Xu, X.; Xiang, W.; Wang, X.; Zhao, J. First report of leaf spot caused by Cladosporium tenuissimum on panicle hydrangea (Hydrangea paniculate) in China. Plant. Dis. 2021, 105, 2240. [Google Scholar] [CrossRef]
- Mukhtar, I.; Ashraf, H.J.; Khokhar, I.; Huang, Q.; Chen, B.; Xie, B. First report of Cladosporium blossom blight caused by Cladosporium cladosporioides on Calliandra haematocephala in China. Plant. Dis. 2020, 105, 1570. [Google Scholar] [CrossRef]
- Kumar, A.; Verma, V.C.; Gond, S.K.; Kumar, V.; Kharwar, R.N. Bio-control potential of Cladosporium sp. (MCPL-461), against a noxious weed Parthenium hysterophorus L. J. Environ. Biol. 2009, 30, 307–312. [Google Scholar]
- Singh, B.; Kaur, T.; Kaur, S.; Manhas, R.K.; Kaur, A. Insecticidal potential of an endophytic Cladosporium velox against Spodoptera litura mediated through inhibition of alpha glycosidases. Pestic. Biochem. Physiol. 2016, 131, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Gange, A.C.; Eschen, R.; Wearn, J.A.; Thawer, A.; Sutton, B.C. Differential effects of foliar endophytic fungi on insect herbivores attacking a herbaceous plant. Oecologia 2012, 168, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- Thakur, A.; Kaur, S.; Kaur, A.; Singh, V. Enhanced resistance to Spodoptera litura in endophyte infected cauliflower plants. Environ. Entomol 2013, 42, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Renoz, F.; Foray, V.; Ambroise, J.; Baa-Puyoulet, P.; Bearzatto, B.; Mendez, G.L.; Grigorescu, A.S.; Mahillon, J.; Mardulyn, P.; Gala, J.L.; et al. At the gate of mutualism: Identification of genomic traits predisposing to insect-bacterial symbiosis in pathogenic strains of the aphid symbiont Serratia symbiotica. Front. Cell Infect. Microbiol. 2021, 11, 660007. [Google Scholar] [CrossRef] [PubMed]
- Nakabachi, A.; Piel, J.; Malenovský, I.; Hirose, Y. Comparative genomics underlines multiple roles of Profftella, an obligate symbiont of psyllids: Providing toxins, vitamins, and carotenoids. Genome Biol. Evol. 2020, 12, 1975–1987. [Google Scholar] [CrossRef] [PubMed]
- Nakabachi, A.; Ueoka, R.; Oshima, K.; Teta, R.; Mangoni, A.; Gurgui, M.; Oldham, N.J.; van Echten-Deckert, G.; Okamura, K.; Yamamoto, K.; et al. Defensive bacteriome symbiont with a drastically reduced genome. Curr. Biol. 2013, 23, 1478–1484. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, Y.; Moriyama, M.; Łukasik, P.; Vanderpool, D.; Tanahashi, M.; Meng, X.Y.; McCutcheon, J.; Fukatsu, T.P. Recurrent symbiont recruitment from fungal parasites in cicadas. Proc. Natl. Acad. Sci. USA 2018, 115, E5970–E5979. [Google Scholar] [CrossRef] [PubMed]
- Szabó, G.; Schulz, F.; Manzano-Marín, A.; Toenshoff, E.R.; Horn, M. Evolutionarily recent dual obligatory symbiosis among adelgids indicates a transition between fungus- and insect-associated lifestyles. ISME J. 2022, 16, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Gullan, P.J.; Kosztarab, M. Adaptations in scale insects. Ann. Rev. Entomol. 1997, 42, 23–50. [Google Scholar] [CrossRef]
- Yang, A.; Troup, M.; Lin, P.; Ho, J.W.K. Falco: A quick and flexible single-cell RNA-seq processing framework on the cloud. Bioinformatics 2017, 33, 767–769. [Google Scholar] [CrossRef]
- Hackl, T.; Hedrich, R.; Schultz, J.; Förster, F. proovread: Large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics 2014, 30, 3004–3011. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar] [CrossRef] [PubMed]
- English, A.C.; Richards, S.; Han, Y.; Wang, M.; Vee, V.; Qu, J.; Qin, X.; Muzny, D.M.; Reid, J.G.; Worley, K.C.; et al. Mind the gap: Upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS ONE 2012, 7, e47768. [Google Scholar] [CrossRef] [PubMed]
- Tarailo-Graovac, M.; Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2009, 4, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Staerfeldt, H.-H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Gardner, P.P.; Daub, J.; Tate, J.G.; Nawrocki, E.P.; Kolbe, D.L.; Lindgreen, S.; Wilkinson, A.C.; Finn, R.D.; Griffiths-Jones, S.; Eddy, S.R.; et al. Rfam: Updates to the RNA families database. Nucleic Acids Res. 2009, 37, D136–D140. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef]
- Birney, E.; Clamp, M.; Durbin, R. GeneWise and genomewise. Genome Res. 2004, 14, 988–995. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Diekhans, M.; Baertsch, R.; Haussler, D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 2008, 24, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Ter-Hovhannisyan, V.; Lomsadze, A.; Chernoff, Y.O.; Borodovsky, M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 2008, 18, 1979–1990. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Niu, B.; Gao, Y.; Fu, L.; Li, W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics 2010, 26, 680–682. [Google Scholar] [CrossRef] [PubMed]
- Niu, B.; Fu, L.; Sun, S.; Li, W. Artificial and natural duplicates in pyrosequencing reads of metagenomic data. BMC Bioinform. 2010, 11, 187. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Vilella, A.J.; Severin, J.; Ureta-Vidal, A.; Heng, L.; Durbin, R.; Birney, E. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009, 19, 327–335. [Google Scholar] [CrossRef]
- Bensch, K.; Braun, U.; Groenewald, J.Z.; Crous, P.W. The Genus Cladosporium. In Studies in Mycologyi; Elsevier: Amsterdam, The Netherlands, 2012; Volume 72, pp. 1–401. [Google Scholar]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Comp. Study Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Russell, J.A.; Oliver, K.M.; Hansen, A.K. Band-aids for Buchnera and B vitamins for all. Mol. Ecol. 2017, 26, 2199–2203. [Google Scholar] [CrossRef]
- Biely, P. Microbial carbohydrate esterases deacetylating plant polysaccharides. Biotechnol. Adv. 2012, 30, 1575–1588. [Google Scholar] [CrossRef] [PubMed]
- Rafiei, V.; Vélëz, H.; Tzelepis, G. The role of glycoside hydrolases in phytopathogenic fungi and oomycetes virulence. Int. J. Mol. Sci. 2021, 22, 9359. [Google Scholar] [CrossRef]
- Staats, C.C.; Junges, A.; Guedes, R.L.; Thompson, C.E.; de Morais, G.L.; Boldo, J.T.; Paula de Almeida, L.G.; Andreis, F.C.; Gerber, A.L.; Sbaraini, N.; et al. Comparative genome analysis of entomopathogenic fungi reveals a complex set of secreted proteins. BMC Genom. 2014, 15, 822. [Google Scholar] [CrossRef]
- Kang, Y.-N.; So, K.-K.; Kim, D.-W.; Kim, D.-H.; Lee, T.-H. Draft genome sequencing of the pathogenic fungus Cladosporium phlei ATCC 36193 identifies candidates of novel polyketide synthase genes involved in perylenequinone-group pigment production. Evol. Bioinform. Online 2019, 15, 1176934319831306. [Google Scholar] [CrossRef]
- Seto, Y.; Kogami, Y.; Shimanuki, T.; Takahashi, K.; Matsuura, H.; Yoshihara, T. Production of phleichrome by Cladosporium phlei as stimulated by diketopiperadines of Epichloe typhina. Biosci. Biotechnol. Biochem. 2005, 69, 1515–1519. [Google Scholar] [CrossRef]
- Cline, A.R.; Skelley, P.E.; Kinnee, S.A.; Rooney-Latham, S.; Winterton, S.L.; Borkent, C.J.; Audisio, P. Interactions between a sap beetle, sabal palm, scale insect, filamentous fungi and yeast, with discovery of potential antifungal compounds. PLoS ONE 2014, 9, e89295. [Google Scholar] [CrossRef] [PubMed]
- Yun, T.S.; Park, S.Y.; Yu, J.; Hwang, Y.; Hong, K.J. Isolation and identification of fungal species from the insect pest Tribolium castaneum in rice processing complexes in Korea. Plant. Pathol. J. 2018, 34, 356–366. [Google Scholar] [CrossRef]
- Liu, W.-W.; Yang, P.; Chen, X.-M.; Xu, D.-L.; Hu, Y.-H. Cloning and expression analysis of four heat shock protein genes in Ericerus pela (Homoptera: Coccidae). J. Insect Sci. 2014, 14, 142. [Google Scholar] [CrossRef]
- Yu, S.-H.; Yang, P.; Sun, T.; Qi, Q.; Wang, X.-Q.; Xu, D.-L.; Chen, X.-M. Identification and evaluation of reference genes in the Chinese white wax scale insect Ericerus pela. Springerplus 2016, 5, 791. [Google Scholar] [CrossRef]
- Zhang, H.; Ye, L.; Liu, Q.; Huang, X. Genome assembly of Scorias spongiosa and comparative genomics provide insights into ecological adaptation of honeydew-dependent sooty mould fungi. Genomics 2021, 113, 2189–2198. [Google Scholar] [CrossRef] [PubMed]
- Spanu, P.D. The genomics of obligate (and nonobligate) biotrophs. Ann. Rev. Phytopathol. 2012, 50, 91–109. [Google Scholar] [CrossRef] [PubMed]


| Species Name | Accession Number in GenBank |
|---|---|
| Cladosporium phlei | QZFA00000000 |
| Cladosporium sphaerospermum.UM.843 | AIIA02 |
| Cladosporium sp.SL-16 | PEGC00000000.1 |
| Cladosporium sp.TM138-S3 | JAAQHG000000000.1 |
| Cladosporium cladosporioides | NOXB01 |
| Sequence Type | Sample | Total Number | Total Length (bp) | N50 Length (bp) | N90 Length (bp) | Max Length (bp) | Min Length (bp) | Gap Number (bp) | GC Content |
|---|---|---|---|---|---|---|---|---|---|
| Scaffold | Cladosporium sp. (pathogen) | 25 | 30,678,857 | 1,877,121 | 1,391,070 | 5,850,328 | 22,622 | 89,418 | 53.05% |
| Cladosporium sp. (endogensis) | 46 | 35,677,347 | 1,764,234 | 4,247,99 | 3,136,047 | 11,843 | 23,760 | 52.58% | |
| Contig | Cladosporium sp. (pathogen) | 49 | 30,589,439 | 1,610,294 | 428,466 | 3,437,199 | 22,329 | - | 53.05% |
| Cladosporium sp. (endogensis) | 55 | 35,653,587 | 1,749,682 | 37,981,6 | 25,907,13 | 11,843 | - | 52.58% |
| Method | Sample | Repeat Size (bp) | Percent in Genome |
|---|---|---|---|
| Repbase | Cladosporium sp. (pathogen) | 240,641 | 0.7844% |
| Cladosporium sp. (endogensis) | 252,517 | 0.7078% | |
| ProMask | Cladosporium sp. (pathogen) | 490,596 | 1.5991% |
| Cladosporium sp. (endogensis) | 525,112 | 1.4718% | |
| denovo | Cladosporium sp. (pathogen) | 758,713 | 2.4731% |
| Cladosporium sp. (endogensis) | 1,150,874 | 3.2258% | |
| TRF | Cladosporium sp. (pathogen) | 155,625 | 0.5073% |
| Cladosporium sp. (endogensis) | 190746 | 0.5346% | |
| Total | Cladosporium sp. (pathogen) | 1,125,929 | 3.67% |
| Cladosporium sp. (endogensis) | 1,572,493 | 4.4075% |
| Type | Sample | Copy | Average Length | Total Length | Percent in Genome |
|---|---|---|---|---|---|
| tRNA | Cladosporium sp. (pathogen) | 221 | 92 | 20,406 | 0.0665% |
| Cladosporium sp. (endogensis) | 274 | 91 | 25,030 | 0.0702% | |
| rRNA (by denovo prediction) | Cladosporium sp. (pathogen) | 82 | 1237 | 101,498 | 0.3308% |
| Cladosporium sp. (endogensis) | 80 | 465 | 37,235 | 0.1044% | |
| sRNA | Cladosporium sp. (pathogen) | 10 | 58 | 581 | 0.0019% |
| Cladosporium sp. (endogensis) | 46 | 88 | 4044 | 0.0113% | |
| snRNA | Cladosporium sp. (pathogen) | 4 | 58 | 232 | 0.0008% |
| Cladosporium sp. (endogensis) | 34 | 113 | 3848 | 0.0108% | |
| miRNA | Cladosporium sp. (pathogen) | 0 | 0 | 0 | 0 |
| Cladosporium sp. (endogensis) | 64 | 60 | 3877 | 0.0109% |
| Type | Sample | Total Number | Total Length (bp) | Average Length (bp) | Percent in Genome |
|---|---|---|---|---|---|
| Gene | Cladosporium sp. (pathogen) | 10,930 | 17,633,154 | 1613 | 57.48% |
| Cladosporium sp. (endogensis) | 13,522 | 21,235,259 | 1570 | 59.52% | |
| Exons | Cladosporium sp. (pathogen) | 25,247 | 16,208,934 | 642 | 52.83% |
| Cladosporium sp. (endogensis) | 31,128 | 19,653,616 | 631 | 55.09% | |
| CDS | Cladosporium sp. (pathogen) | 10,930 | 16,208,934 | 1483 | 52.83% |
| Cladosporium sp. (endogensis) | 13,522 | 19,653,616 | 1453 | 55.09% | |
| Introne | Cladosporium sp. (pathogen) | 14,317 | 1,424,220 | 99 | 4.64% |
| Cladosporium sp. (endogensis) | 17,606 | 1,581,643 | 90 | 4.43% |
| Number | Percent | |||
|---|---|---|---|---|
| Annotation Database | Cladosporium sp. (Pathogen) | Cladosporium sp. (Endogensis) | Cladosporium sp. (Pathogen) | Cladosporium sp. (Endogensis) |
| Total | 10,930 | 13,522 | ||
| NR | 9255 | 10,627 | 84.67% | 78.59% |
| SWISSPROT | 3187 | 3456 | 29.15% | 25.55% |
| GO | 6185 | 7331 | 56.58% | 54.21% |
| KEGG | 4202 | 4569 | 38.44% | 33.78% |
| KOG | 2151 | 2237 | 19.67% | 16.54% |
| COG | 1337 | 1506 | 12.23% | 11.13% |
| NOG | 7799 | 8835 | 71.35% | 65.33% |
| IPR | 8405 | 10,064 | 76.89% | 74.42% |
| CAZY | 433 | 544 | 3.96% | 4.02% |
| DBCAN | 583 | 725 | 5.33% | 5.36% |
| PHI | 1143 | 1202 | 10.45% | 8.89% |
| P450 | 1762 | 2202 | 16.12% | 16.28% |
| KINASE | 115 | 133 | 1.05% | 0.98% |
| CARD | 0 | 1 | 0.00% | 0.00% |
| TCDB | 565 | 614 | 5.16% | 4.54% |
| PHOSPHATASE | 34 | 36 | 0.31% | 0.26% |
| Over All | 9702 | 11,566 | 88.76% | 85.53% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Yu, S.-H.; Zhang, H.-P.; Fu, Z.-Y.; An, J.-Q.; Zhang, J.-Y.; Yang, P. Two Cladosporium Fungi with Opposite Functions to the Chinese White Wax Scale Insect Have Different Genome Characters. J. Fungi 2022, 8, 286. https://doi.org/10.3390/jof8030286
Liu W, Yu S-H, Zhang H-P, Fu Z-Y, An J-Q, Zhang J-Y, Yang P. Two Cladosporium Fungi with Opposite Functions to the Chinese White Wax Scale Insect Have Different Genome Characters. Journal of Fungi. 2022; 8(3):286. https://doi.org/10.3390/jof8030286
Chicago/Turabian StyleLiu, Wei, Shu-Hui Yu, Hong-Ping Zhang, Zuo-Yi Fu, Jia-Qi An, Jin-Yang Zhang, and Pu Yang. 2022. "Two Cladosporium Fungi with Opposite Functions to the Chinese White Wax Scale Insect Have Different Genome Characters" Journal of Fungi 8, no. 3: 286. https://doi.org/10.3390/jof8030286
APA StyleLiu, W., Yu, S.-H., Zhang, H.-P., Fu, Z.-Y., An, J.-Q., Zhang, J.-Y., & Yang, P. (2022). Two Cladosporium Fungi with Opposite Functions to the Chinese White Wax Scale Insect Have Different Genome Characters. Journal of Fungi, 8(3), 286. https://doi.org/10.3390/jof8030286






