Establishment of High-Efficiency Screening System for Gene Deletion in Fusarium venenatum TB01
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of F. venenatum and E. coli DH5α Cultures for Transformation
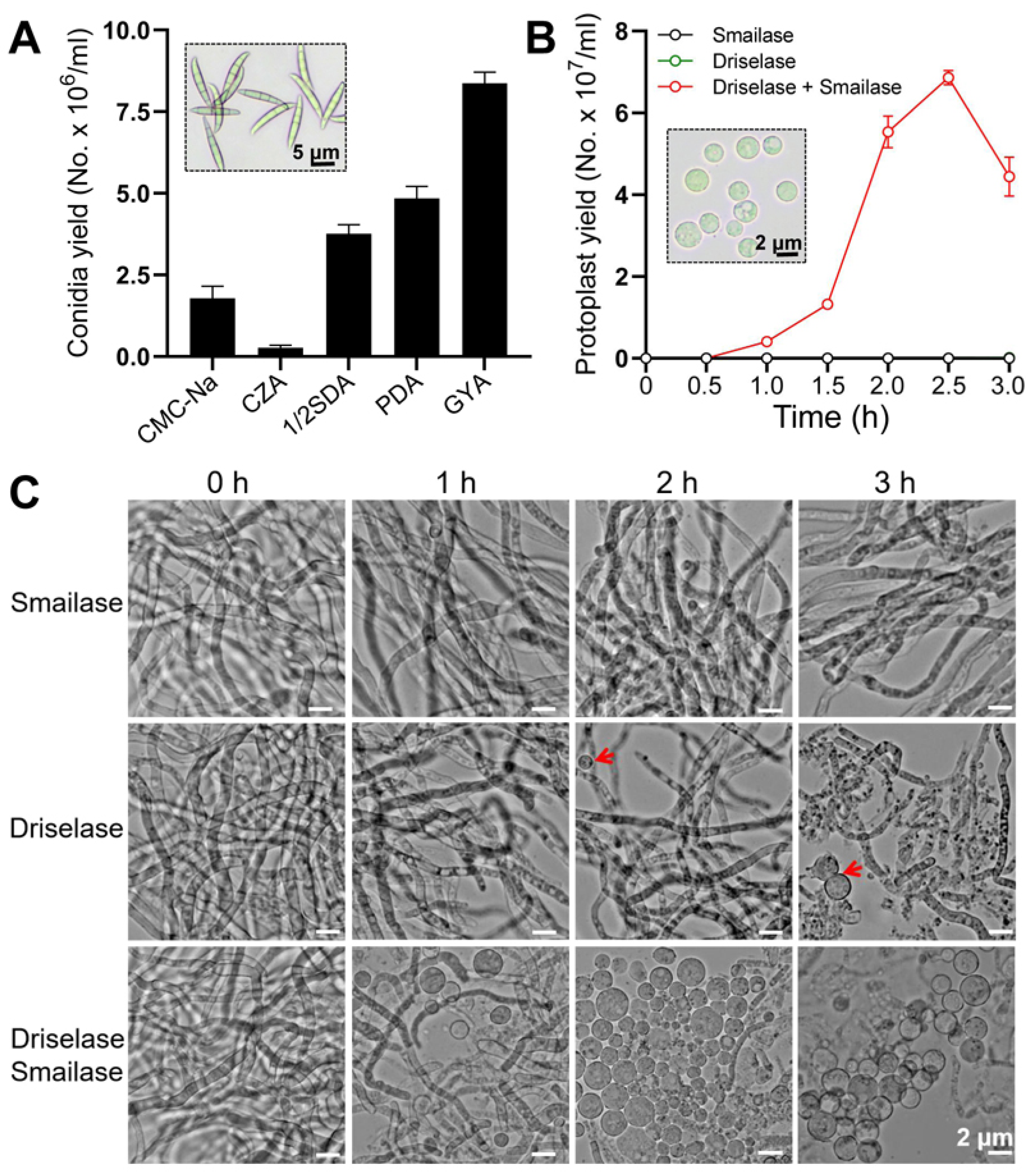
2.2. Quantification of Conidia Yield
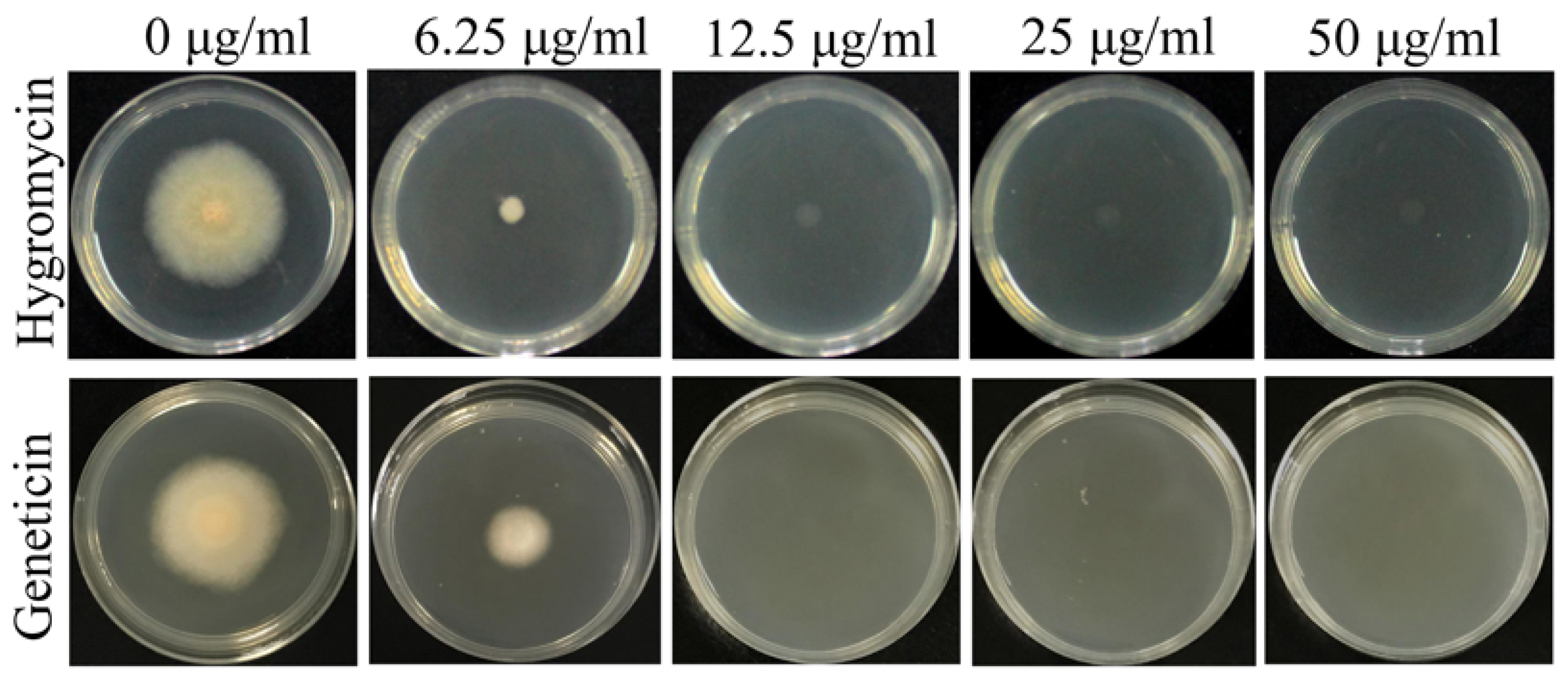
2.3. Antibiotic Sensivity Test for F. venenatum
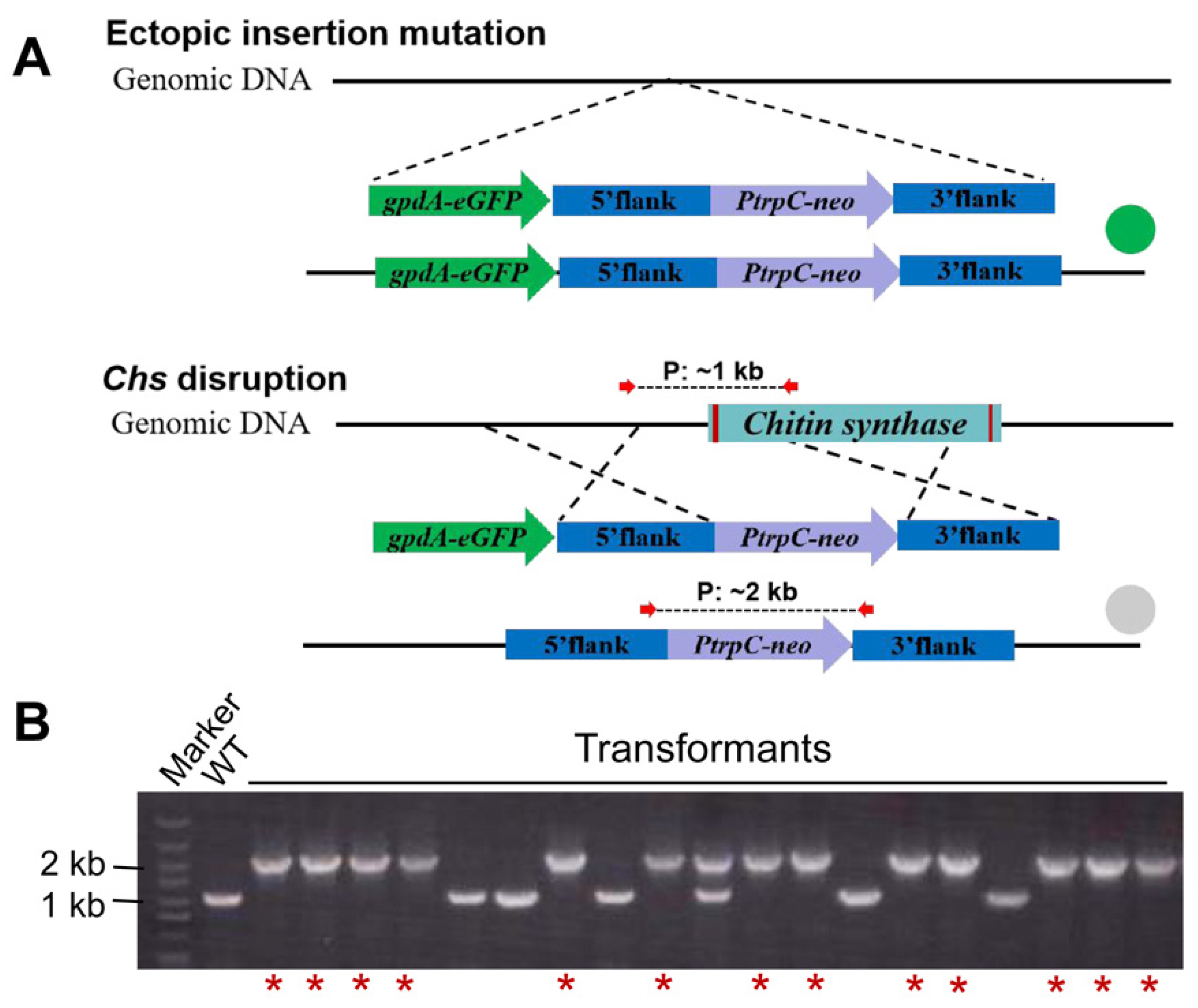
2.4. Construction of Targeted Gene Knockout and eGFP Gene Overexpression Vector
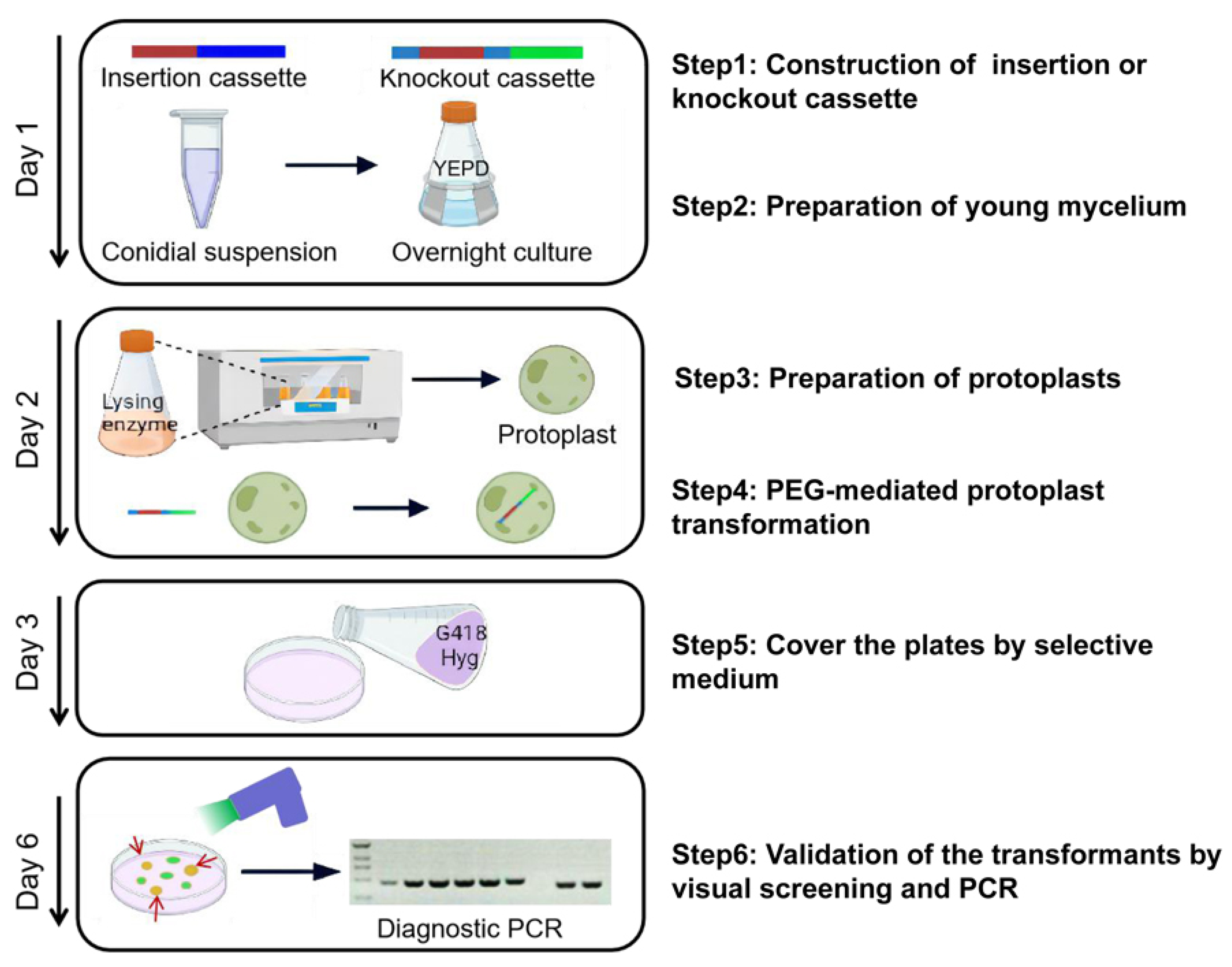
2.5. PEG-Mediated Protoplast Transformation
2.6. Fluorescence of Fungus Strains
2.7. Validation of the Transformants by PCR
3. Results
3.1. Optimization of the Protocol for Protoplast Transformation
3.2. Rate of Homologus Recombination in F. venenatum TB01
3.3. Screening the Strong Promoter for the Visualized Gene Deletion System
3.4. Visualized Gene Deletion System Significantly Improves the Screening Efficiency of Gene Deletion Transformants
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Derbyshire, E.; Ayoob, K.T. Mycoprotein: Nutritional and health properties. Nutr. Today 2019, 54, 7–15. [Google Scholar] [CrossRef]
- Finnigan, T.J.A.; Wall, B.T.; Wilde, P.J.; Stephens, F.B.; Taylor, S.L.; Freedman, M.R. Mycoprotein: The future of nutritious nonmeat protein, a symposium review. Curr. Dev. Nutr. 2019, 3, nzz021. [Google Scholar] [CrossRef]
- Colosimo, R.; Mulet-Cabero, A.I.; Warren, F.J.; Edwards, C.H.; Finnigan, T.J.A.; Wilde, P.J. Mycoprotein ingredient structure reduces lipolysis and binds bile salts during simulated gastrointestinal digestion. Food Funct. 2020, 11, 10896–10906. [Google Scholar] [CrossRef]
- Thomas, A.B.; Shetane, T.D.; Singha, R.G.; Nanda, R.K.; Poddar, S.S.; Shirsat, A. Employing central composite design for evaluation of biomass production by Fusarium venenatum: In vivo antioxidant and antihyperlipidemic properties. Appl. Biochem. Biotech. 2017, 183, 91–109. [Google Scholar] [CrossRef]
- Spalvins, K.; Raita, S.; Valters, K.; Blumberga, D. Improving single cell protein yields and amino acid profile via mutagenesis: Review of applicable amino acid inhibitors for mutant selection. Agron. Res. 2021, 19, 1285–1307. [Google Scholar]
- Finnigan, T.; Needham, L.; Abbott, C. Chapter 19-Mycoprotein: A healthy new protein with a low environmental impact. In Sustainable Protein Sources; Nadathus, S.R., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 305–325. [Google Scholar]
- Jacobson, M.F.; Deporter, J. Self-reported adverse reactions associated with mycoprotein (Quorn-brand) containing foods. Ann. Allergy Asthma Immunol. 2018, 120, 626–630. [Google Scholar] [CrossRef]
- Awange, J.; Kiema, J. Environmental pollution. In Environmental Geoinformatics; Forstner, U., Ed.; Springer: Berlin/Heidelberg, Germany, 2019; pp. 579–595. [Google Scholar]
- Hashempour-Baltor, F.; Khosravi-Darani, K.; Hosseini, H.; Farshi, P.; Reihani, S.F. Mycoproteins as safe meat substitutes. J. Clean. Prod. 2020, 253, 119958. [Google Scholar] [CrossRef]
- Lv, X.; Wu, Y.; Gong, M.; Deng, J.; Gu, Y.; Liu, Y.; Li, J.; Du, G.; Ledesma-Amaro, R.; Liu, L.; et al. Synthetic biology for future food: Research progress and future directions. Future Foods 2021, 3, 100025. [Google Scholar] [CrossRef]
- Gruber, F.; Visser, J.; Kubicek, C.P.; Graaff, L.H. The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr. Genet. 1990, 18, 71–76. [Google Scholar] [CrossRef]
- Lu, S.W.; Lyngholm, L.; Yang, G.; Bronson, C.; Yoder, O.; Turgeon, B.G. Tagged mutations at the Toxl locus of Cochliobolus heterostrophus by restriction enzyme-mediated integration. Proc. Natl. Acad. Sci. USA 1995, 91, 12649–12653. [Google Scholar] [CrossRef]
- Robinson, M.; Sharon, A. Transformation of the bioherbicide Colletotrichum gloeosporioides f. sp. aeschynomene by electroporation of germinated conidia. Curr. Genet. 1999, 36, 98–104. [Google Scholar] [CrossRef]
- Fang, W.; Zhang, Y.; Yang, X.; Zheng, X.; Duan, H.; Li, Y.; Pei, Y. Agrobacterium tumefaciens-mediated transformation of Beauveria bassiana using an herbicide resistance gene as a selection marker. J. Invertebr. Pathol. 2004, 85, 18–24. [Google Scholar] [CrossRef]
- Fungaro, M.H.; Rech, E.G.; Muhlen, G.; Vainstein, M.H.; Pascon, R.C.; Queiroz, M.; Pizzirani-Kleiner, A.A.; Azevedo, J.L. Transformation of Aspergillus nidulans by microprojectile bombardment on intact conidia. FEMS Microbiol. Lett. 2010, 125, 293–297. [Google Scholar] [CrossRef][Green Version]
- Xu, J.; Li, J.; Lin, L.; Liu, Q.; Sun, W.; Huang, B.; Tian, C. Development of genetic tools for Myceliophthora thermophila. BMC Biotechnol. 2015, 15, 35. [Google Scholar] [CrossRef]
- Groot, M.J.; Bundock, P.; Hooykaas, P.J.; Beijersbergen, A.G. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 1998, 16, 839–842. [Google Scholar] [CrossRef]
- Royer, J.C.; Moyer, D.L.; Reiwitch, S.G.; Madden, M.S.; Jensen, E.B.; Brown, S.H.; Yonker, C.C.; Johnstone, J.A.; Golightly, E.J.; Yoder, W.T.; et al. Fusarium graminearum a 3/5 as a novel host for heterologous protein production. Nat. Biotechnol. 1995, 13, 1479–1483. [Google Scholar] [CrossRef]
- Royer, J.C.; Christianson, L.M.; Yoder, W.T.; Gambetta, G.A.; Klotz, A.V.; Morris, C.L.; Brody, H.; Otani, S. Deletion of the trichodiene synthase gene of Fusarium venenatum: Two systems for repeated gene deletions. Fungal Genet. Biol. 1999, 28, 68–78. [Google Scholar] [CrossRef]
- Meyer, V.; Arentshorst, M.; El-Ghezal, A.; Drews, A.C.; Kooistra, R.; Hondel, C.A.; Ram, A.F. Highly efficient gene targeting in the Aspergillus niger kusA mutant. J. Biotechnol. 2007, 128, 770–775. [Google Scholar] [CrossRef]
- Koh, C.M.; Liu, Y.; Moehninsi, N.; Du, M.; Ji, L. Molecular characterization of ku70 and ku80 homologues and exploitation of a ku70-deficient mutant for improving gene deletion frequency in Rhodosporidium toruloides. BMC Microbiol. 2014, 14, 50. [Google Scholar] [CrossRef]
- Colot, H.V.; Park, G.; Turner, G.E.; Ringelberg, C.; Crew, C.M.; Litvinkova, L.; Weiss, R.L.; Borkovich, K.A.; Dunlap, J.C. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 2006, 103, 10352–10357. [Google Scholar] [CrossRef]
- Zhang, G.; Hartl, L.; Schuster, A.; Polak, S.; Schmoll, M.; Wang, T.; Seidl, V.; Seiboth, B. Gene targeting in a nonhomologous end joining deficient Hypocrea jecorina. J. Biotechnol. 2009, 139, 146–151. [Google Scholar]
- Visser, H.; Joosten, V.; Punt, P.J.; Gusakov, A.V.; Olson, P.T.; Joosten, R.; Bartels, J.; Sinitsyn, A.P.; Emalfarb, M.A.; Verdoes, J.C.; et al. Development of a mature fungal technology and production platform for industrial enzymes based on a Myceliophthora thermophila isolate, previously known as Chrysosporium lucknowense C1. Ind. Biotechnol. 2011, 7, 214–223. [Google Scholar] [CrossRef]
- Zheng, X.; Zheng, P.; Sun, J.; Kun, Z.; Ma, Y. Heterologous and endogenous U6 snRNA promoters enable CRISPR/Cas9 mediated genome editing in Aspergillus niger. Fungal Biol. Biotechnol. 2018, 5, 1–9. [Google Scholar] [CrossRef]
- Li, C.; Zhou, J.; Rao, S.; Du, G.; Liu, S. Visualized multigene editing system for Aspergillus niger. ACS Synth. Biol. 2021, 10, 2607–2616. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, J.; Fang, W.; Zhang, J.; Luo, Z.; Zhang, M.; Fan, Y.; Pei, Y. Mitogen-activated protein kinase hog1 in the entomopathogenic fungus Beauveria bassiana regulates environmental stress responses and virulence to insects. Appl. Environ. Microb. 2009, 75, 3787–3795. [Google Scholar] [CrossRef]
- Ildefonso, C.J.; Jaime, H.; Brown, E.E.; Iwata, R.L.; Ahmed, C.M.; Massengill, M.T.; Biswal, M.R.; Boye, S.E.; Hauswirth, W.W.; Ash, J.D.; et al. Targeting the Nrf2 signaling pathway in the retina with a gene-delivered secretable and cell-penetrating peptide. Investig. Ophthalmol. Vis. Sci. 2016, 57, 372–386. [Google Scholar] [CrossRef]
- Fan, Y.; Ortiz-Urquiza, A.; Garrett, T.; Pei, Y.; Keyhani, N.O. Involvement of a caleosin in lipid storage, spore dispersal, and virulence in the entomopathogenic filamentous fungus, Beauveria bassiana. Environ. Microbiol. 2016, 17, 4600–4614. [Google Scholar] [CrossRef]
- Hou, Y.; Zhou, M. Establishing of an Economic and Efficient Genetic Transformation System of Protoplast for Fusarium graminearum. Sciencepaper Online. 2013. Available online: http://www.paper.edu.cn/releasepaper/content/201309-253 (accessed on 2 February 2022).
- Fang, W.; Pei, Y.; Bidochka, M.J. Transformation of Metarhizium anisopliae mediated by Agrobacterium tumefaciens. Can. J. Microbiol. 2006, 52, 623–626. [Google Scholar] [CrossRef]
- Zhong, Y.H.; Wang, X.L.; Wang, T.H.; Jiang, Q. Agrobacterium-mediated transformation (AMT) of Trichoderma reesei as an efficient tool for random insertional mutagenesis. Appl. Microbiol. Biot. 2007, 73, 1348–1354. [Google Scholar] [CrossRef]
- Shi, L.; Fang, X.; Li, M.; Mu, D.; Ren, A.; Tan, Q.; Zhao, M. Development of a simple and efficient transformation system for the basidiomycetous medicinal fungus Ganoderma lucidum. World J. Microb. Biot. 2012, 28, 283–291. [Google Scholar] [CrossRef]
- Sorensen, L.Q.; Lysoe, E.; Larsen, J.E.; Khorsand-Jamal, P.; Nielsen, K.F.; Frandsen, R.J. Genetic transformation of Fusarium avenaceum by Agrobacterium tumefaciens mediated transformation and the development of a USER-Brick vector construction system. BMC Mol. Biol. 2014, 15, 15. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Zhao, J.J.; Xie, M.; Peng, D.L. Agrobacterium tumefaciens-mediated transformation in the entomopathogenic fungus Lecanicillium lecanii and development of benzimidazole fungicide resistant strains. J. Microbiol. Meth. 2014, 105, 168–173. [Google Scholar] [CrossRef]
- Liao, X.; Fang, W.; Zhang, Y.; Fan, Y.; Wu, X.; Zhou, Q.; Pei, Y. Characterization of a highly active promoter, PBbgpd, in Beauveria bassiana. Curr. Microbiol. 2008, 57, 121–126. [Google Scholar] [CrossRef]
- Cao, Y.; Jiao, R.; Xia, Y. A strong promoter, PMagpd, provides a tool for high gene expression in entomopathogenic fungus, Metarhizium acridum. Biotechnol. Lett. 2012, 34, 557–562. [Google Scholar] [CrossRef]
- Zhang, H.; Yan, J.N.; Zhang, H.; Liu, T.Q.; Xu, Y.; Zhang, Y.Y.; Li, J. Effect of gpd box copy numbers in the gpda promoter of Aspergillus nidulans on its transcription efficiency in A. niger. FEMS Microbiol. Lett. 2018, 365, 15. [Google Scholar] [CrossRef]
- Wilson, F.M.; Harrison, R.J. CRISPR/Cas9 mediated editing of the Quorn fungus Fusarium venenatum A3/5 by transient expression of Cas9 and sgRNAs targeting endogenous marker gene PKS12. Fungal Biol. Biotechnol. 2021, 8, 1–8. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tong, S.; An, K.; Zhou, W.; Chen, W.; Sun, Y.; Wang, Q.; Li, D. Establishment of High-Efficiency Screening System for Gene Deletion in Fusarium venenatum TB01. J. Fungi 2022, 8, 169. https://doi.org/10.3390/jof8020169
Tong S, An K, Zhou W, Chen W, Sun Y, Wang Q, Li D. Establishment of High-Efficiency Screening System for Gene Deletion in Fusarium venenatum TB01. Journal of Fungi. 2022; 8(2):169. https://doi.org/10.3390/jof8020169
Chicago/Turabian StyleTong, Sheng, Kexin An, Wenyuan Zhou, Wuxi Chen, Yuanxia Sun, Qinhong Wang, and Demao Li. 2022. "Establishment of High-Efficiency Screening System for Gene Deletion in Fusarium venenatum TB01" Journal of Fungi 8, no. 2: 169. https://doi.org/10.3390/jof8020169
APA StyleTong, S., An, K., Zhou, W., Chen, W., Sun, Y., Wang, Q., & Li, D. (2022). Establishment of High-Efficiency Screening System for Gene Deletion in Fusarium venenatum TB01. Journal of Fungi, 8(2), 169. https://doi.org/10.3390/jof8020169






