Cryptococcus neoformans Causing Meningoencephalitis in Adults and a Child from Lima, Peru: Genotypic Diversity and Antifungal Susceptibility
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolates
2.2. Multilocus Sequence Typing
2.3. Antifungal Susceptibility Testing
3. Results
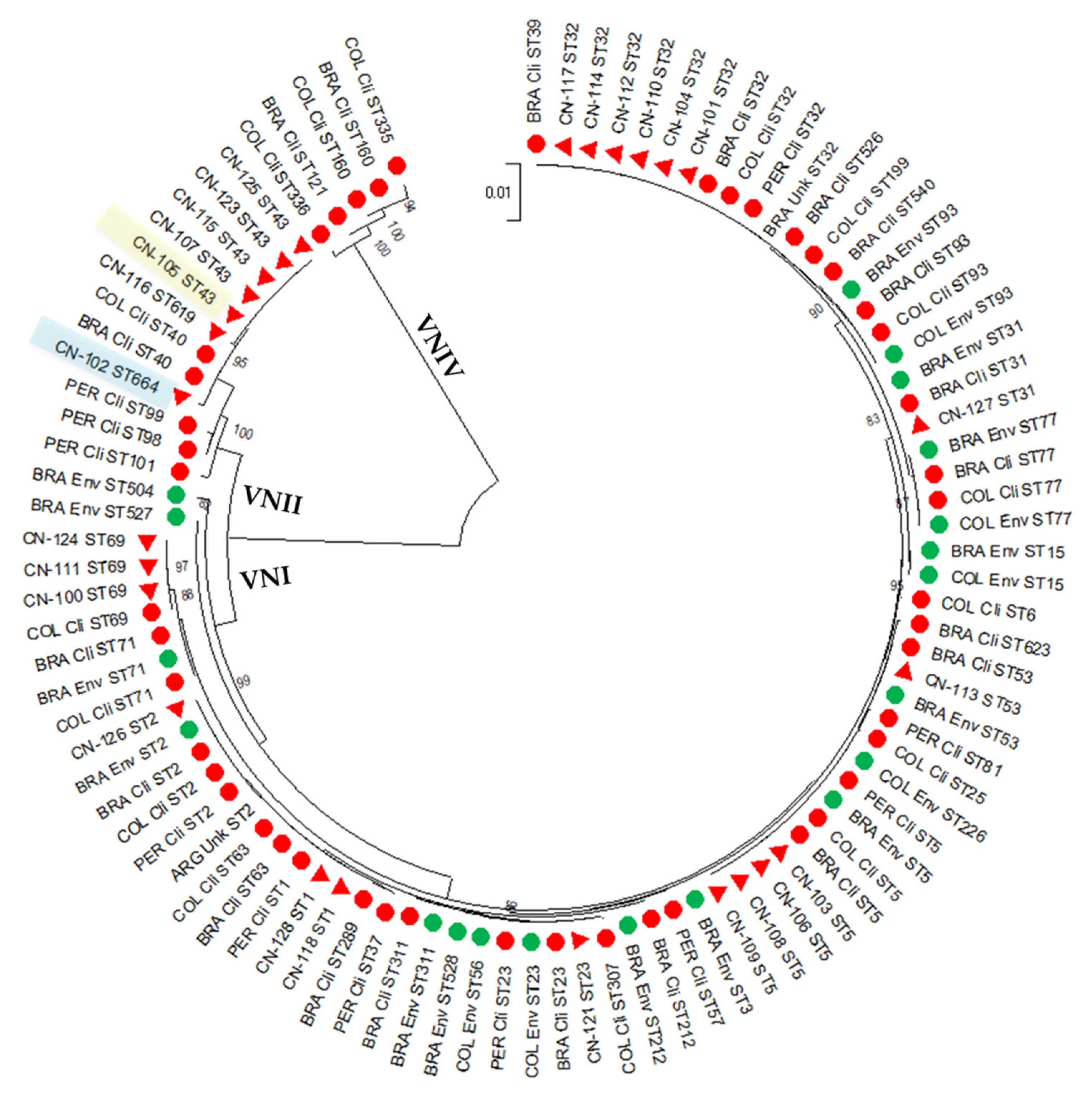
3.1. The Studied C. neoformans Population from Peru Is Highly Diverse
3.2. Most of the Studied C. neoformans Isolates from Peru Are Susceptible to Commonly Used Antifungals
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kwon-Chung, K.J.; Fraser, J.A.; Doering, T.L.; Wang, Z.; Janbon, G.; Idnurm, A.; Bahn, Y.S. Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb. Perspect. Med. 2014, 4, a019760. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, T.G.; Castañeda, E.; Nielsen, K.; Wanke, B.; Lazéra, M.S. Environmental Niches for Cryptococcus neoformans and Cryptococcus gattii. In Cryptococcus: From Human Pathogen to Model Yeast; Heitman, J., Kozel, T.R., Kwon-Chung, J.K., Perfect, J.R., Casadevall, A., Eds.; ASM Press: Washington, DC, USA, 2011; pp. 237–259. [Google Scholar]
- Mitchell, T.G.; Perfect, J.R. Cryptococcosis in the era of AIDS--100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 1995, 8, 515–548. [Google Scholar] [CrossRef] [PubMed]
- Rajasingham, R.; Govender, N.P.; Jordan, A.; Loyse, A.; Shroufi, A.; Denning, D.W.; Meya, D.B.; Chiller, T.M.; Boulware, D.R. The global burden of HIV-associated cryptococcal infection in adults in 2020: A modelling analysis. Lancet Infect. Dis. 2022, 22, 1748–1755. [Google Scholar] [CrossRef] [PubMed]
- Pappas, P.G. Cryptococcal infections in non-HIV-infected patients. Trans. Am. Clin. Climatol. Assoc. 2013, 124, 61–79. [Google Scholar]
- UNAIDS. UNAIDS. Available online: https://www.unaids.org/en/regionscountries/countries/peru (accessed on 4 February 2022).
- Eza, D.; Cerrillo, G.; Moore, D.A.; Castro, C.; Ticona, E.; Morales, D.; Cabanillas, J.; Barrantes, F.; Alfaro, A.; Benavides, A.; et al. Postmortem findings and opportunistic infections in HIV-positive patients from a public hospital in Peru. Pathol. Res. Pract. 2006, 202, 767–775. [Google Scholar] [CrossRef]
- Meyer, W.; Aanensen, D.M.; Boekhout, T.; Cogliati, M.; Diaz, M.R.; Esposto, M.C.; Fisher, M.; Gilgado, F.; Hagen, F.; Kaocharoen, S.; et al. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med. Mycol. 2009, 47, 561–570. [Google Scholar] [CrossRef]
- Bejar, V.; Tello, M.; García, R.; Guevara, J.M.; Gonzales, S.; Vergaray, G.; Valencia, E.; Abanto, E.; Ortega-Loayza, A.G.; Hagen, F.; et al. Molecular characterization and antifungal susceptibility of Cryptococcus neoformans strains collected from a single institution in Lima, Peru. Rev. Iberoam. De Micol. 2015, 32, 88–92. [Google Scholar] [CrossRef]
- Van de Wiele, N.; Neyra, E.; Firacative, C.; Gilgado, F.; Serena, C.; Bustamante, B.; Meyer, W. Molecular epidemiology reveals low genetic diversity among Cryptococcus neoformans isolates from people living with HIV in Lima, Peru, during the Pre-HAART Era. Pathogens 2020, 9, 665. [Google Scholar] [CrossRef]
- Billmyre, R.B.; Clancey, S.A.; Heitman, J. Natural mismatch repair mutations mediate phenotypic diversity and drug resistance in Cryptococcus deuterogattii. eLife 2017, 6, e28802. [Google Scholar] [CrossRef]
- Bermas, A.; Geddes-McAlister, J. Combatting the evolution of antifungal resistance in Cryptococcus neoformans. Mol. Microbiol. 2020, 114, 721–734. [Google Scholar] [CrossRef]
- Carneiro, H.C.S.; Bastos, R.W.; Ribeiro, N.Q.; Gouveia-Eufrasio, L.; Costa, M.C.; Magalhães, T.F.F.; Oliveira, L.V.N.; Paixão, T.A.; Joffe, L.S.; Rodrigues, M.L.; et al. Hypervirulence and cross-resistance to a clinical antifungal are induced by an environmental fungicide in Cryptococcus gattii. Sci. Total Environ. 2020, 740, 140135. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Alastruey-Izquierdo, A.; Berman, J.; Bicanic, T.; Bignell, E.M.; Bowyer, P.; Bromley, M.; Bruggemann, R.; Garber, G.; Cornely, O.A.; et al. Tackling the emerging threat of antifungal resistance to human health. Nat. Rev. Microbiol. 2022, 20, 557–571. [Google Scholar] [CrossRef] [PubMed]
- Firacative, C.; Meyer, W.; Castañeda, E. Cryptococcus neoformans and Cryptococcus gattii species complexes in Latin America: A map of molecular types, genotypic diversity, and antifungal susceptibility as reported by the Latin American Cryptococcal Study Group. J. Fungi 2021, 7, 282. [Google Scholar] [CrossRef] [PubMed]
- Castañeda, E.; Lizarazo, J.; Firacative, C. Criptococosis. In Fundamentos de las Micosis Humanas, 1st ed.; González, A., Gómez, B.L., Tobón, A., Restrepo, A., Eds.; CIB-Universidad de Antioquia: Medellín, Colombia, 2018; pp. 157–173. [Google Scholar]
- Franzot, S.P.; Mukherjee, J.; Cherniak, R.; Chen, L.C.; Hamdan, J.S.; Casadevall, A. Microevolution of a standard strain of Cryptococcus neoformans resulting in differences in virulence and other phenotypes. Infect. Immun. 1998, 66, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Litvintseva, A.P.; Thakur, R.; Vilgalys, R.; Mitchell, T.G. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics 2006, 172, 2223–2238. [Google Scholar] [CrossRef] [PubMed]
- Andrade-Silva, L.E.; Ferreira-Paim, K.; Ferreira, T.B.; Vilas-Boas, A.; Mora, D.J.; Manzato, V.M.; Fonseca, F.M.; Buosi, K.; Andrade-Silva, J.; Prudente, B.D.S.; et al. Genotypic analysis of clinical and environmental Cryptococcus neoformans isolates from Brazil reveals the presence of VNB isolates and a correlation with biological factors. PLoS ONE 2018, 13, e0193237. [Google Scholar] [CrossRef]
- Ferreira-Paim, K.; Andrade-Silva, L.; Fonseca, F.M.; Ferreira, T.B.; Mora, D.J.; Andrade-Silva, J.; Khan, A.; Dao, A.; Reis, E.C.; Almeida, M.T.; et al. MLST-based population genetic analysis in a global context reveals clonality amongst Cryptococcus neoformans var. grubii VNI isolates from HIV patients in Southeastern Brazil. PLoS Negl. Trop. Dis. 2017, 11, e0005223. [Google Scholar] [CrossRef]
- Rocha, D.F.S.; Cruz, K.S.; Santos, C.; Menescal, L.S.F.; Neto, J.; Pinheiro, S.B.; Silva, L.M.; Trilles, L.; Braga de Souza, J.V. MLST reveals a clonal population structure for Cryptococcus neoformans molecular type VNI isolates from clinical sources in Amazonas, Northern-Brazil. PLoS ONE 2018, 13, e0197841. [Google Scholar] [CrossRef]
- Cogliati, M.; Zani, A.; Rickerts, V.; McCormick, I.; Desnos-Ollivier, M.; Velegraki, A.; Escandon, P.; Ichikawa, T.; Ikeda, R.; Bienvenu, A.L.; et al. Multilocus sequence typing analysis reveals that Cryptococcus neoformans var. neoformans is a recombinant population. Fungal Genet. Biol. 2016, 87, 22–29. [Google Scholar] [CrossRef]
- Velez, N.; Alvarado, M.; Parra-Giraldo, C.M.; Sanchez-Quitian, Z.A.; Escandon, P.; Castaneda, E. Genotypic diversity is independent of pathogenicity in Colombian strains of Cryptococcus neoformans and Cryptococcus gattii in Galleria mellonella. J. Fungi 2018, 4, 82. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Firacative, C.; Roe, C.C.; Malik, R.; Ferreira-Paim, K.; Escandon, P.; Sykes, J.E.; Castanon-Olivares, L.R.; Contreras-Peres, C.; Samayoa, B.; Sorrell, T.C.; et al. MLST and whole-genome-based population analysis of Cryptococcus gattii VGIII links clinical, veterinary and environmental strains, and reveals divergent serotype specific sub-populations and distant ancestors. PLoS Negl. Trop. Dis. 2016, 10, e0004861. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Espinel-Ingroff, A.; Pfaller, M.; Messer, S.A.; Knapp, C.C.; Killian, S.; Norris, H.A.; Ghannoum, M.A. Multicenter comparison of the sensititre YeastOne Colorimetric Antifungal Panel with the National Committee for Clinical Laboratory standards M27-A reference method for testing clinical isolates of common and emerging Candida spp., Cryptococcus spp., and other yeasts and yeast-like organisms. J. Clin. Microbiol. 1999, 37, 591–595. [Google Scholar] [CrossRef] [PubMed]
- CLSI supplement M27M44S; Performance Standards for Antifungal Susceptibility Testing of Yeasts. CLSI: Wayne, PA, USA, 2022.
- Espinel-Ingroff, A.; Aller, A.I.; Canton, E.; Castanon-Olivares, L.R.; Chowdhary, A.; Cordoba, S.; Cuenca-Estrella, M.; Fothergill, A.; Fuller, J.; Govender, N.; et al. Cryptococcus neoformans-Cryptococcus gattii species complex: An international study of wild-type susceptibility endpoint distributions and epidemiological cutoff values for fluconazole, itraconazole, posaconazole, and voriconazole. Antimicrob. Agents Chemother. 2012, 56, 5898–5906. [Google Scholar] [CrossRef]
- Espinel-Ingroff, A.; Chowdhary, A.; Cuenca-Estrella, M.; Fothergill, A.; Fuller, J.; Hagen, F.; Govender, N.; Guarro, J.; Johnson, E.; Lass-Florl, C.; et al. Cryptococcus neoformans-Cryptococcus gattii species complex: An international study of wild-type susceptibility endpoint distributions and epidemiological cutoff values for amphotericin B and flucytosine. Antimicrob. Agents Chemother. 2012, 56, 3107–3113. [Google Scholar] [CrossRef][Green Version]
- Delma, F.Z.; Al-Hatmi, A.M.S.; Buil, J.B.; van der Lee, H.; Tehupeiory-Kooreman, M.; de Hoog, G.S.; Meletiadis, J.; Verweij, P.E. Comparison of MIC test strip and Sensititre YeastOne with the CLSI and EUCAST broth microdilution reference methods for in vitro antifungal susceptibility testing of Cryptococcus neoformans. Antimicrob. Agents Chemother. 2020, 64, e02261-19. [Google Scholar] [CrossRef]
- Meiring, S.T.; Quan, V.C.; Cohen, C.; Dawood, H.; Karstaedt, A.S.; McCarthy, K.M.; Whitelaw, A.C.; Govender, N.P.; Group for Enteric, R.; Meningeal disease Surveillance in South, A. A comparison of cases of paediatric-onset and adult-onset cryptococcosis detected through population-based surveillance, 2005–2007. AIDS 2012, 26, 2307–2314. [Google Scholar] [CrossRef]
- Lizarazo, J.; Escandón, P.; Agudelo, C.I.; Castañeda, E. Cryptococcosis in Colombian children and literature review. Mem. Inst. Oswaldo Cruz 2014, 109, 797–804. [Google Scholar] [CrossRef]
- Firacative, C.; Lizarazo, J.; Illnait-Zaragozi, M.T.; Castaneda, E.; Latin American Cryptococcal Study, G. The status of cryptococcosis in Latin America. Mem. Inst. Oswaldo Cruz 2018, 113, e170554. [Google Scholar] [CrossRef]
- Berejnoi, A.; Taverna, C.G.; Mazza, M.; Vivot, M.; Isla, G.; Cordoba, S.; Davel, G. First case report of cryptococcosis due to Cryptococcus decagattii in a pediatric patient in Argentina. Rev. Soc. Bras. Med. Trop. 2019, 52, e20180419. [Google Scholar] [CrossRef] [PubMed]
- Pinto Junior, V.L.; Pone, M.V.; Pone, S.M.; Campos, J.M.; Garrido, J.R.; de Barros, A.C.; Trilles, L.; Barbosa, G.G.; Morales, B.P.; Bezerra Cde, C.; et al. Cryptococcus gattii molecular type VGII as agent of meningitis in a healthy child in Rio de Janeiro, Brazil: Report of an autochthonous case. Rev. Soc. Bras. Med. Trop. 2010, 43, 746–748. [Google Scholar] [CrossRef]
- Martins, L.M.; Wanke, B.; Lazera Mdos, S.; Trilles, L.; Barbosa, G.G.; Macedo, R.C.; Cavalcanti Mdo, A.; Eulalio, K.D.; Castro, J.A.; Silva, A.S.; et al. Genotypes of Cryptococcus neoformans and Cryptococcus gattii as agents of endemic cryptococcosis in Teresina, Piaui (northeastern Brazil). Mem. Inst. Oswaldo Cruz 2011, 106, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Noguera, M.C.; Escandon, P.; Arevalo, M.; Piedrahita, J.; Castaneda, E. Fatal neurocryptococcosis in a Colombian underage patient. J. Infect. Dev. Ctries. 2019, 13, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Van Wyk, M.; Govender, N.P.; Mitchell, T.G.; Litvintseva, A.P.; Germs, S.A. Multilocus sequence typing of serially collected isolates of Cryptococcus from HIV-infected patients in South Africa. J. Clin. Microbiol. 2014, 52, 1921–1931. [Google Scholar] [CrossRef] [PubMed]
- ISHAM. Genotyping of Cryptococcus neoformans and C. gattii. Available online: https://www.isham.org/working-groups/genotyping-cryptococcous-neoformans-and-c-gattii (accessed on 22 November 2022).
- Desnos-Ollivier, M.; Patel, S.; Raoux-Barbot, D.; Heitman, J.; Dromer, F.; French Cryptococcosis Study, G. Cryptococcosis Serotypes Impact Outcome and Provide Evidence of Cryptococcus neoformans Speciation. mBio 2015, 6, e00311. [Google Scholar] [CrossRef]
- Sanchini, A.; Smith, I.M.; Sedlacek, L.; Schwarz, R.; Tintelnot, K.; Rickerts, V. Molecular typing of clinical Cryptococcus neoformans isolates collected in Germany from 2004 to 2010. Med. Microbiol. Immunol. 2014, 203, 333–340. [Google Scholar] [CrossRef]
- Chen, Y.H.; Yu, F.; Bian, Z.Y.; Hong, J.M.; Zhang, N.; Zhong, Q.S.; Hang, Y.P.; Xu, J.; Hu, L.H. Multilocus Sequence Typing Reveals both Shared and Unique Genotypes of Cryptococcus neoformans in Jiangxi Province, China. Sci. Rep. 2018, 8, 1495. [Google Scholar] [CrossRef]
- Mihara, T.; Izumikawa, K.; Kakeya, H.; Ngamskulrungroj, P.; Umeyama, T.; Takazono, T.; Tashiro, M.; Nakamura, S.; Imamura, Y.; Miyazaki, T.; et al. Multilocus sequence typing of Cryptococcus neoformans in non-HIV associated cryptococcosis in Nagasaki, Japan. Med. Mycol. 2013, 51, 252–260. [Google Scholar] [CrossRef]
- Umeyama, T.; Ohno, H.; Minamoto, F.; Takagi, T.; Tanamachi, C.; Tanabe, K.; Kaneko, Y.; Yamagoe, S.; Kishi, K.; Fujii, T.; et al. Determination of epidemiology of clinically isolated Cryptococcus neoformans strains in Japan by multilocus sequence typing. Jpn. J. Infect. Dis. 2013, 66, 51–55. [Google Scholar] [CrossRef][Green Version]
- Khayhan, K.; Hagen, F.; Pan, W.; Simwami, S.; Fisher, M.C.; Wahyuningsih, R.; Chakrabarti, A.; Chowdhary, A.; Ikeda, R.; Taj-Aldeen, S.J.; et al. Geographically structured populations of Cryptococcus neoformans variety grubii in Asia correlate with HIV status and show a clonal population structure. PLoS ONE 2013, 8, e72222. [Google Scholar] [CrossRef] [PubMed]
- Hatthakaroon, C.; Pharkjaksu, S.; Chongtrakool, P.; Suwannakarn, K.; Kiratisin, P.; Ngamskulrungroj, P. Molecular epidemiology of cryptococcal genotype VNIc/ST5 in Siriraj Hospital, Thailand. PLoS ONE 2017, 12, e0173744. [Google Scholar] [CrossRef] [PubMed]
- Day, J.N.; Qihui, S.; Thanh, L.T.; Trieu, P.H.; Van, A.D.; Thu, N.H.; Chau, T.T.H.; Lan, N.P.H.; Chau, N.V.V.; Ashton, P.M.; et al. Comparative genomics of Cryptococcus neoformans var. grubii associated with meningitis in HIV infected and uninfected patients in Vietnam. PLoS Negl. Trop. Dis. 2017, 11, e0005628. [Google Scholar] [CrossRef]
- Chen, Y.; Litvintseva, A.P.; Frazzitta, A.E.; Haverkamp, M.R.; Wang, L.; Fang, C.; Muthoga, C.; Mitchell, T.G.; Perfect, J.R. Comparative analyses of clinical and environmental populations of Cryptococcus neoformans in Botswana. Mol. Ecol. 2015, 24, 3559–3571. [Google Scholar] [CrossRef] [PubMed]
- Singer, L.M.; Meyer, W.; Firacative, C.; Thompson, G.R., 3rd; Samitz, E.; Sykes, J.E. Antifungal drug susceptibility and phylogenetic diversity among Cryptococcus isolates from dogs and cats in North America. J. Clin. Microbiol. 2014, 52, 2061–2070. [Google Scholar] [CrossRef]
- Cogliati, M.; Desnos-Ollivier, M.; McCormick-Smith, I.; Rickerts, V.; Ferreira-Paim, K.; Meyer, W.; Boekhout, T.; Hagen, F.; Theelen, B.; Inacio, J.; et al. Genotypes and population genetics of Cryptococcus neoformans and Cryptococcus gattii species complexes in Europe and the mediterranean area. Fungal Genet. Biol. 2019, 129, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Danesi, P.; Firacative, C.; Cogliati, M.; Otranto, D.; Capelli, G.; Meyer, W. Multilocus sequence typing (MLST) and M13 PCR fingerprinting revealed heterogeneity amongst Cryptococcus species obtained from Italian veterinary isolates. FEMS Yeast Res. 2014, 14, 897–909. [Google Scholar] [CrossRef] [PubMed]
- Prakash, A.; Sundar, G.; Sharma, B.; Hagen, F.; Meis, J.F.; Chowdhary, A. Genotypic diversity in clinical and environmental isolates of Cryptococcus neoformans from India using multilocus microsatellite and multilocus sequence typing. Mycoses 2020, 63, 284–293. [Google Scholar] [CrossRef]
- Park, S.H.; Kim, M.; Joo, S.I.; Hwang, S.M. Molecular epidemiology of clinical Cryptococcus neoformans isolates in Seoul, Korea. Mycobiology 2014, 42, 73–78. [Google Scholar] [CrossRef]
- Thanh, L.T.; Phan, T.H.; Rattanavong, S.; Nguyen, T.M.; Duong, A.V.; Dacon, C.; Hoang, T.N.; Nguyen, L.P.H.; Tran, C.T.H.; Davong, V.; et al. Multilocus sequence typing of Cryptococcus neoformans var. grubii from Laos in a regional and global context. Med. Mycol. 2018, 57, 557–565. [Google Scholar] [CrossRef]
- Park, S.H.; Choi, S.C.; Lee, K.W.; Kim, M.N.; Hwang, S.M. Genotypes of Clinical and Environmental Isolates of Cryptococcus neoformans and Cryptococcus gattii in Korea. Mycobiology 2015, 43, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Kassi, F.K.; Drakulovski, P.; Bellet, V.; Roger, F.; Chabrol, A.; Krasteva, D.; Doumbia, A.; Landman, R.; Kakou, A.; Reynes, J.; et al. Cryptococcus genetic diversity and mixed infections in Ivorian HIV patients: A follow up study. PLoS Negl. Trop. Dis. 2019, 13, e0007812. [Google Scholar] [CrossRef] [PubMed]
- Beale, M.A.; Sabiiti, W.; Robertson, E.J.; Fuentes-Cabrejo, K.M.; O’Hanlon, S.J.; Jarvis, J.N.; Loyse, A.; Meintjes, G.; Harrison, T.S.; May, R.C.; et al. Genotypic Diversity Is Associated with Clinical Outcome and Phenotype in Cryptococcal Meningitis across Southern Africa. PLoS Negl. Trop. Dis. 2015, 9, e0003847. [Google Scholar] [CrossRef]
- Fan, X.; Xiao, M.; Chen, S.; Kong, F.; Dou, H.T.; Wang, H.; Xiao, Y.L.; Kang, M.; Sun, Z.Y.; Hu, Z.D.; et al. Predominance of Cryptococcus neoformans var. grubii multilocus sequence type 5 and emergence of isolates with non-wild-type minimum inhibitory concentrations to fluconazole: A multi-centre study in China. Clin. Microbiol. Infect. 2016, 22, 887.e881–887.e889. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cogliati, M.; Zamfirova, R.R.; Tortorano, A.M.; Viviani, M.A.; Fimua Cryptococcosis, N. Molecular epidemiology of Italian clinical Cryptococcus neoformans var. grubii isolates. Med. Mycol. 2013, 51, 499–506. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wiesner, D.L.; Moskalenko, O.; Corcoran, J.M.; McDonald, T.; Rolfes, M.A.; Meya, D.B.; Kajumbula, H.; Kambugu, A.; Bohjanen, P.R.; Knight, J.F.; et al. Cryptococcal genotype influences immunologic response and human clinical outcome after meningitis. mBio 2012, 3, e00196-12. [Google Scholar] [CrossRef] [PubMed]
- Dou, H.; Wang, H.; Xie, S.; Chen, X.; Xu, Z.; Xu, Y. Molecular characterization of Cryptococcus neoformans isolated from the environment in Beijing, China. Med. Mycol. 2017, 55, 737–747. [Google Scholar] [CrossRef][Green Version]
- Simwami, S.P.; Khayhan, K.; Henk, D.A.; Aanensen, D.M.; Boekhout, T.; Hagen, F.; Brouwer, A.E.; Harrison, T.S.; Donnelly, C.A.; Fisher, M.C. Low diversity Cryptococcus neoformans variety grubii multilocus sequence types from Thailand are consistent with an ancestral African origin. PLoS Pathog. 2011, 7, e1001343. [Google Scholar] [CrossRef]
- Cogliati, M.; Andrianarivelo, M.R.; Ellabib, M.; Nnadi, E.N.; Cornet, M. Molecular-type specific multiplex PCR produces a distinct VNII PCR pattern among Cryptococcus neoformans species complex. Med. Mycol. 2019, 57, 384–386. [Google Scholar] [CrossRef]
- Nnadi, N.E.; Giosa, D.; Ayanbimpe, G.M.; D’Alessandro, E.; Aiese Cigliano, R.; Oheri, C.U.; Aguiyi, J.C.; Enweani, I.B.; Romeo, O. Whole-genome sequencing of an uncommon Cryptococcus neoformans MLST43 genotype isolated in Nigeria. Mycopathologia 2019, 184, 555–557. [Google Scholar] [CrossRef]
- Kaocharoen, S.; Ngamskulrungroj, P.; Firacative, C.; Trilles, L.; Piyabongkarn, D.; Banlunara, W.; Poonwan, N.; Chaiprasert, A.; Meyer, W.; Chindamporn, A. Molecular epidemiology reveals genetic diversity amongst isolates of the Cryptococcus neoformans/C. gattii species complex in Thailand. PLoS Negl. Trop. Dis. 2013, 7, e2297. [Google Scholar] [CrossRef] [PubMed]
- Reis, R.S.; Bonna, I.C.F.; Antonio, I.; Pereira, S.A.; Nascimento, C.; Ferraris, F.K.; Brito-Santos, F.; Ferreira Gremiao, I.D.; Trilles, L. Cryptococcus neoformans VNII as the Main Cause of Cryptococcosis in Domestic Cats from Rio de Janeiro, Brazil. J. Fungi 2021, 7, 980. [Google Scholar] [CrossRef] [PubMed]
- Dammert, P.; Bustamante, B.; Ticona, E.; Llanos-Cuentas, A.; Huaroto, L.; Chavez, V.M.; Campos, P.E. Treatment of cryptococcal meningitis in Peruvian AIDS patients using amphotericin B and fluconazole. J. Infect. 2008, 57, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Concha-Velasco, F.; Gonzalez-Lagos, E.; Seas, C.; Bustamante, B. Factors associated with early mycological clearance in HIV-associated cryptococcal meningitis. PLoS ONE 2017, 12, e0174459. [Google Scholar] [CrossRef]
- Perfect, J.R.; Cox, G.M. Drug resistance in Cryptococcus neoformans. Drug Resist. Updates 1999, 2, 259–269. [Google Scholar] [CrossRef]
- Bellet, V.; Roger, F.; Krasteva, D.; Gouveia, T.; Drakulovski, P.; Pottier, C.; Bertout, S. Multilocus sequence typing of strains from the Cryptococcus gattii species complex from different continents. Mycoses 2022, 65, 88–96. [Google Scholar] [CrossRef]
- Padda, I.S.; Parmar, M. Flucytosine. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Chong, H.S.; Dagg, R.; Malik, R.; Chen, S.; Carter, D. In vitro susceptibility of the yeast pathogen Cryptococcus to fluconazole and other azoles varies with molecular genotype. J. Clin. Microbiol. 2010, 48, 4115–4120. [Google Scholar] [CrossRef]
- Firacative, C.; Escandon, P. Antifungal susceptibility of clinical Cryptococcus gattii isolates from Colombia varies among molecular types. Med. Mycol. 2021, 59, 1122–1125. [Google Scholar] [CrossRef]
- Lockhart, S.R.; Iqbal, N.; Bolden, C.B.; DeBess, E.E.; Marsden-Haug, N.; Worhle, R.; Thakur, R.; Harris, J.R.; Cryptococcus gattii PNW Public Health Working Group. Epidemiologic cutoff values for triazole drugs in Cryptococcus gattii: Correlation of molecular type and in vitro susceptibility. Diagn. Microbiol. Infect. Dis. 2012, 73, 144–148. [Google Scholar] [CrossRef]
- Trilles, L.; Meyer, W.; Wanke, B.; Guarro, J.; Lazera, M. Correlation of antifungal susceptibility and molecular type within the Cryptococcus neoformans/C. gattii species complex. Med. Mycol. 2012, 50, 328–332. [Google Scholar] [CrossRef]
- Cogliati, M. Global molecular epidemiology of Cryptococcus neoformans and Cryptococcus gattii: An atlas of the molecular types. Scientifica 2013, 2013, 675213. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.A.; Arthur, I.; Merritt, A.; Leung, M. Molecular types of Cryptococcus neoformans and Cryptococcus gattii in Western Australia and correlation with antifungal susceptibility. Med. Mycol. 2019, 57, 1004–1010. [Google Scholar] [CrossRef] [PubMed]
- Hagen, F.; Hare Jensen, R.; Meis, J.F.; Arendrup, M.C. Molecular epidemiology and in vitro antifungal susceptibility testing of 108 clinical Cryptococcus neoformans sensu lato and Cryptococcus gattii sensu lato isolates from Denmark. Mycoses 2016, 59, 576–584. [Google Scholar] [CrossRef] [PubMed]
| Isolate | Year | Age (Years) 1 | Sex 1,2 | CAP59 | GPD1 | IGS1 | LAC1 | PLB1 | SOD1 | URA5 | ST 3 | MT 4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CN-100 | 2012 | NA | NA | 7 | 5 | 1 | 3 | 3 | 1 | 1 | 69 | VNI |
| CN-101 | 2013 | NA | NA | 1 | 1 | 10 | 3 | 4 | 1 | 1 | 32 | VNI |
| CN-102 | 2013 | NA | NA | 10 | 9 | 14 | 8 | 12 | 12 | 4 | 664 5 | VNII |
| CN-103 | 2014 | 43 | M | 1 | 3 | 1 | 5 | 2 | 1 | 1 | 5 | VNI |
| CN-104 | 2014 | 47 | M | 1 | 1 | 10 | 3 | 4 | 1 | 1 | 32 | VNI |
| CN-105 | 2014 | 9 | F | 2 | 9 | 14 | 8 | 11 | 11 | 4 | 43 | VNII |
| CN-106 | 2015 | 39 | M | 1 | 3 | 1 | 5 | 2 | 1 | 1 | 5 | VNI |
| CN-107 | 2017 | NA | NA | 2 | 9 | 14 | 8 | 11 | 11 | 4 | 43 | VNII |
| CN-108 | 2017 | NA | NA | 1 | 3 | 1 | 5 | 2 | 1 | 1 | 5 | VNI |
| CN-109 | 2018 | 36 | M | 1 | 3 | 1 | 5 | 2 | 1 | 1 | 5 | VNI |
| CN-110 | 2018 | 41 | M | 1 | 1 | 10 | 3 | 4 | 1 | 1 | 32 | VNI |
| CN-111 | 2018 | 36 | M | 7 | 5 | 1 | 3 | 3 | 1 | 1 | 69 | VNI |
| CN-112 | 2019 | 45 | M | 1 | 1 | 10 | 3 | 4 | 1 | 1 | 32 | VNI |
| CN-113 | 2019 | 54 | M | 1 | 3 | 1 | 3 | 2 | 1 | 1 | 53 | VNI |
| CN-114 6 | 2019 | 71 | F | 1 | 1 | 10 | 3 | 4 | 1 | 1 | 32 | VNI |
| CN-115 | 2019 | 35 | M | 2 | 9 | 14 | 8 | 11 | 11 | 4 | 43 | VNII |
| CN-116 | 2019 | 63 | M | 2 | 9 | 14 | 11 | 11 | 11 | 15 | 619 | VNII |
| CN-117 | 2019 | 31 | M | 1 | 1 | 10 | 3 | 4 | 1 | 1 | 32 | VNI |
| CN-118 7 | 2019 | 70 | M | 7 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | VNI |
| CN-121 | 2020 | 35 | M | 7 | 1 | 1 | 2 | 1 | 1 | 2 | 23 | VNI |
| CN-123 | 2020 | 29 | M | 2 | 9 | 14 | 8 | 11 | 11 | 4 | 43 | VNII |
| CN-124 | 2020 | 31 | M | 7 | 5 | 1 | 3 | 3 | 1 | 1 | 69 | VNI |
| CN-125 | 2020 | 25 | M | 2 | 9 | 14 | 8 | 11 | 11 | 4 | 43 | VNII |
| CN-126 | 2020 | 51 | M | 7 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | VNI |
| CN-127 | 2020 | 59 | M | 1 | 1 | 10 | 3 | 2 | 1 | 1 | 31 | VNI |
| CN-128 | 2021 | 45 | M | 7 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | VNI |
| Population | n | D 1 | SS 2 | H 3 | Hd 4 | Pi 5 |
|---|---|---|---|---|---|---|
| All | 26 | 0.892 | 72 bp | 11 | 0.892 | 0.00667 |
| VNI | 19 | 0.854 | 24 bp | 8 | 0.854 | 0.00233 |
| VNII | 7 | 0.524 | 9 bp | 3 | 0.524 | 0.00064 |
| No. of Isolates at MIC Value (µg/mL) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antifungal | MT 1 | n | GM 2 (µg/mL) | 0.008 | 0.015 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 |
| Amphotericin B | VNI | 19 | 0.4821 | 1 | 5 | 7 | 6 | ||||||||
| VNII | 7 | 0.5000 | 3 | 2 | 1 | 1 | |||||||||
| All | 26 | 0.4868 | 1 | 8 | 9 | 7 | 1 | ||||||||
| 5-flucytosine | VNI | 19 | 2.9880 | 2 | 1 | 6 | 6 | 2 | 2 | ||||||
| VNII | 7 | 1.0000 | 1 | 2 | 2 | 2 | |||||||||
| All | 26 | 2.2250 | 1 | 4 | 1 | 8 | 8 | 2 | 2 | ||||||
| Fluconazole 3 | VNI | 19 | 3.2140 | 1 | 7 | 8 | 3 | ||||||||
| VNII | 7 | 0.4529 | 1 | 5 | 1 | ||||||||||
| All | 26 | 1.8960 | 1 | 5 | 2 | 7 | 8 | 3 | |||||||
| Itraconazole 3 | VNI | 19 | 0.0485 | 2 | 8 | 4 | 5 | ||||||||
| VNII | 7 | 0.0257 | 3 | 3 | 1 | ||||||||||
| All | 26 | 0.0408 | 5 | 11 | 5 | 5 | |||||||||
| Voriconazole 3 | VNI | 19 | 0.0291 | 2 | 4 | 7 | 6 | ||||||||
| VNII | 7 | 0.0080 | 7 | ||||||||||||
| All | 26 | 0.0206 | 9 | 4 | 7 | 6 | |||||||||
| Posaconazole | VNI | 19 | 0.0629 | 1 | 5 | 6 | 6 | 1 | |||||||
| VNII | 7 | 0.0381 | 1 | 4 | 1 | 1 | |||||||||
| All | 26 | 0.0548 | 1 | 1 | 9 | 7 | 7 | 1 | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Firacative, C.; Zuluaga-Puerto, N.; Guevara, J. Cryptococcus neoformans Causing Meningoencephalitis in Adults and a Child from Lima, Peru: Genotypic Diversity and Antifungal Susceptibility. J. Fungi 2022, 8, 1306. https://doi.org/10.3390/jof8121306
Firacative C, Zuluaga-Puerto N, Guevara J. Cryptococcus neoformans Causing Meningoencephalitis in Adults and a Child from Lima, Peru: Genotypic Diversity and Antifungal Susceptibility. Journal of Fungi. 2022; 8(12):1306. https://doi.org/10.3390/jof8121306
Chicago/Turabian StyleFiracative, Carolina, Natalia Zuluaga-Puerto, and José Guevara. 2022. "Cryptococcus neoformans Causing Meningoencephalitis in Adults and a Child from Lima, Peru: Genotypic Diversity and Antifungal Susceptibility" Journal of Fungi 8, no. 12: 1306. https://doi.org/10.3390/jof8121306
APA StyleFiracative, C., Zuluaga-Puerto, N., & Guevara, J. (2022). Cryptococcus neoformans Causing Meningoencephalitis in Adults and a Child from Lima, Peru: Genotypic Diversity and Antifungal Susceptibility. Journal of Fungi, 8(12), 1306. https://doi.org/10.3390/jof8121306







