Invasion and Colonization of Pathogenic Fusarium oxysporum R1 in Crocus sativus L. during Corm Rot Disease Progression
Abstract
1. Introduction
2. Materials and Methods
2.1. Corm Sample, Fungal and Bacterial Strains and Culture Conditions
2.2. Molecular Characterization of Fox R1
2.3. Disease Assays for Fox R1 Characterization
2.4. Experimental Design to Study the Route of Infection of Fox R1 in Saffron Plant
2.5. Confirmation of Infection and Movement of Fox R1 by Semi-Quantitative PCR and q-PCR
2.5.1. Designing of Primer and Sensitivity of Primers
2.5.2. In Planta Detection and Quantification of Fox R1 in Infected Roots and Corms by Semi-Quantitative PCR and q-PCR
2.6. Calculation of Fox R1 Load at the Site of Infection
2.7. Agrobacterium Tumefaciens Mediated Transformation (ATMT) of Fox R1
2.8. Molecular Characterization of Transformed Fox R1
2.9. Microscopic Monitoring of Transformed Fox R1
2.10. Disease Progression in Saffron Infected Corms and Roots
2.11. Statistical Analysis
3. Results
3.1. Molecular Characterization and Disease Assays of Fox R1
3.2. Route of Infection of Fox R1 in Saffron
3.3. Confirmation of Infection of Fox R1 by Semi-Quantitative PCR and q-PCR
3.3.1. Semi-Quantitative PCR
3.3.2. Standard Curve Analysis and Quantification of Fox R1 Load by q-PCR
3.4. Determinations of Fox R1 Load at the Site of Infection
3.5. Tagging and Expression of EGFP in Transformed Fox R1 at Different Time Points
3.6. Conformation of Transformants
3.7. Invasion and Colonization of Fox R1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Menia, M.; Iqbal, S.; Zahida, R.; Tahir, S.; Kanth, R.H.; Saad, A.A.; Hussian, A. Production technology of saffron for enhancing productivity. J. Pharmacognos. Phytochem. 2018, 7, 1033–1039. [Google Scholar]
- Cardone, L.; Castronuovo, D.; Perniola, M.; Cicco, N.; Candido, V. Saffron (Crocus sativus L.), the king of spices: An overview. Sci. Hortic. 2020, 272, 109560. [Google Scholar] [CrossRef]
- Bhagat, N.; Sharma, S.; Ambardar, S.; Raj, S.; Trakroo, D.; Horacek, M.; Zouagui, R.; Sbabou, L.; Vakhlu, J. Microbiome Fingerprint as Biomarker for Geographical Origin and Heredity in Crocus sativus: A Feasibility Study. Front. Sustain. Food Syst. 2021, 5. [Google Scholar] [CrossRef]
- Sharma, S.; Kumar, D. Chemical Composition and Biological Uses of Crocus sativus L. (Saffron). In Edible Plants in Health and Diseases Volume II: Phytochemical and Pharmacological Properties; Springer: Singapore, 2022; p. 249. [Google Scholar]
- Shokrpour, M. Saffron (Crocus sativus L.) breeding: Opportunities and challenges. In Advances in Plant Breeding Strategies: Industrial and Food Crops; Springer: Cham, Switzerland, 2019; pp. 675–706. [Google Scholar]
- Ambardar, S.; Bhagat, N.; Vakhlu, J.; Gowda, M. Diversity of Rhizo-Bacteriome of Crocus sativus Grown at Various Geographical Locations and Cataloging of Putative PGPRs. Front. Sustain. Food Syst. 2021, 5, 644230. [Google Scholar] [CrossRef]
- Gupta, V.; Sharma, A.; Rai, P.; Gupta, S.; Singh, B.; Sharma, S.; Singh, S.; Hussain, R.; Razdan, V.; Kumar, D.; et al. Corm Rot of Saffron: Epidemiology and Management. Agronomy 2021, 11, 339. [Google Scholar] [CrossRef]
- Husaini, A.M.; Jiménez, A.J.L. Understanding saffron biology using omics-and bioinformatics tools: Stepping towards a better Crocus phenome. Mol. Bio. Rep. 2022, 49, 5325–5340. [Google Scholar] [CrossRef]
- Ganie, M.A.; Nusrath, A. Marketing and Trade Mechanism of Saffron (Crocus sativus). IOSR J. Econ. Financ. 2016, 7, 55–62. [Google Scholar]
- Ali, A.; Hakim, I.A. An overview of the production practices and trade mechanism of saffron in Kashmir Valley (India): Issues and challenges. Pac. Bus. Rev. Int. 2017, 10, 97–106. [Google Scholar]
- Bazoobandi, M.; Rahimi, H.; Karimi-Shahri, M.R. Saffron crop protection. In Saffron; Woodhead Publishing: Cambridge, UK, 2020; pp. 169–185. [Google Scholar]
- Khaledi, N. Evaluation of cell wall degrading enzymes of Fusarium species associated with root and corm of saffron in South Khorasan province. Saffron Agron. Technol. 2020, 8, 243–259. [Google Scholar]
- Kumar, A.; Devi, M.; Kumar, R.; Kumar, S. Introduction of high-value Crocus sativus (saffron) cultivation in non-traditional regions of India through ecological modelling. Sci. Rep. 2022, 12, 119125. [Google Scholar] [CrossRef]
- Husaini, A.M.; Hassan, B.; Ghani, M.Y.; Teixeira da Silva, J.A.; Kirmani, N.A. Saffron (Crocus sativus Kashmirianus) cultivation in Kashmir: Practices and problems. Funct. Plant Sci. Biotechnol. 2010, 4, 108–115. [Google Scholar]
- Gupta, V.; Kalha, C.S.; Razdan, V.K.; Dolly. Etiology and management of corm rot of saffron in Kishtwar district of Jammu and Kashmir, India. J. Mycol. Plant Pathol. 2011, 41, 361–366. [Google Scholar]
- Gupta, R.; Vakhlu, J. Native Bacillus amyloliquefaciens W2 as a potential biocontrol for Fusarium oxysporum R1 causing corm rot of Crocus sativus. Eur. J. Plant Pathol. 2015, 143, 123–131. [Google Scholar] [CrossRef]
- Dar, M.H.; Groach, R.; Razvi, S.M.; Singh, N. Saffron Crop (Golden Crop) in Modern Sustainable Agricultural Systems. Int. J. Res. Appl. Sci. Eng. Technol. 2017, 5, 247–259. [Google Scholar] [CrossRef]
- Jan, B.; Baba, S.A. Corm rot of saffron: Symptoms and biological management. EC Microbiol. 2018, 14, 1. [Google Scholar]
- Magotra, S.; Bhagat, N.; Ambardar, S.; Ali, T.; Hurek, B.R.; Hurek, T.; Verma, P.K.; Vakhlu, J. Field evaluation of PGP Bacillus sp. strain D5 native to Crocus sativus, in traditional and non traditional areas, and mining of PGP genes from its genome. Sci. Rep. 2021, 11, 5454. [Google Scholar] [CrossRef] [PubMed]
- Di Primo, P.; Cappelli, C.; Katan, T. Vegetative Compatibility Grouping of Fusarium oxysporum f. sp. gladioli from Saffron. Eur. J. Plant Pathol. 2002, 108, 869–875. [Google Scholar] [CrossRef]
- Gupta, V.; Kumar, K.; Fatima, K.; Razdan, V.; Sharma, B.; Mahajan, V.; Rai, P.; Sharma, A.; Gupta, V.; Hassan, M.; et al. Role of Biocontrol Agents in Management of Corm Rot of Saffron Caused by Fusarium oxysporum. Agronomy 2020, 10, 1398. [Google Scholar] [CrossRef]
- López-Díaz, C.; Rahjoo, V.; Sulyok, M.; Ghionna, V.; Martín-Vicente, A.; Capilla, J.; López-Berges, M.S. Fusaric acid contributes to virulence of Fusarium oxysporum on plant and mammalian hosts. Mol. Plant Pathol. 2018, 19, 440–453. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wu, Q.; He, W.; He, T.; Wu, Q.; Miao, Y. Combined de novo transcriptome and metabolome analysis of common bean response to Fusarium oxysporum f. sp. phaseoli infection. Int. J. of Mol. Sci. 2019, 20, 6278. [Google Scholar] [CrossRef]
- Hashem, A.; Tabassum, B.; Abd_Allah, E.F. Omics approaches in chickpea Fusarium wilt disease management. In Management of Fungal Pathogens in Pulses; Springer: Cham, Switzerland, 2020; pp. 57–72. [Google Scholar]
- Manikandan, R.; Harish, S.; Karthikeyan, G.; Raguchander, T. Comparative Proteomic Analysis of Different Isolates of Fusarium oxysporum f.sp. lycopersici to Exploit the Differentially Expressed Proteins Responsible for Virulence on Tomato Plants. Front. Microbiol. 2018, 9, 420. [Google Scholar] [CrossRef] [PubMed]
- Pegg, K.G.; Coates, L.M.; O’Neill, W.T.; Turner, D.W. The Epidemiology of Fusarium Wilt of Banana. Front. Plant Sci. 2019, 10, 1395. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.; Kim, D.W.; Park, J.S.; Khang, C.H. Live-cell fluorescence imaging to investigate the dynamics of plant cell death during infection by the rice blast fungus Magnaporthe oryzae. BMC Plant Biol. 2016, 16, 69. [Google Scholar] [CrossRef]
- Fones, H.N.; Littlejohn, G.R. From Sample to Data: Preparing, Obtaining, and Analyzing Images of Plant-Pathogen Interactions Using Confocal Microscopy. In Host-Pathogen Interactions; Humana Press: New York, NY, USA, 2018; pp. 257–262. [Google Scholar]
- Gelvin, S.B.; Schilperoort, R.A. Plant Molecular Biology Manual, 2nd ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1994. [Google Scholar]
- Nizam, S.; Singh, K.; Verma, P.K. Expression of the fluorescent proteins DsRed and EGFP to visualize early events of colonization of the chickpea blight fungus Ascochyta rabiei. Curr. Genet. 2010, 56, 391–399. [Google Scholar] [CrossRef]
- Liu, D.; Coloe, S.; Baird, R.; Pedersen, J. Rapid Mini-Preparation of Fungal DNA for PCR. J. Clin. Microbiol. 2000, 38, 471. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from Filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Jacobs, A.; Van Wyk, P.S.; Marasas, W.F.; Wingfield, B.D.; Wingfield, M.J.; Coutinho, T.A. Fusarium ananatum sp. nov. in the Gibberella fujikuroi species complex from pineapples with fruit rot in South Africa. Fungal Biol. 2010, 114, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyl, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Komada, H. Development of a selective medium for quantitative isolation of Fusarium oxysporum from natural soil. Rev. Plant Prot. Res. 1975, 8, 114–124. [Google Scholar]
- Islam, M.N.; Nizam, S.; Verma, P.K. A highly efficient Agrobacterium mediated transformation system for chickpea wilt pathogen Fusarium oxysporum f. sp ciceri using DsRed-Express to follow root colonisation. Microbiol. Res. 2012, 167, 332–338. [Google Scholar]
- Bundock, P.; Dulk-Ras, A.; Beijersbergen, A.; Hooykaas, P.J. Transkingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 1995, 14, 3206–3214. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Southern hybridization of radiolabeled probes to nucleic acids immobilized on membranes. Cold Spring Harb. Protoc. 2006, 2006, pdb-prot4044. [Google Scholar] [CrossRef]
- Collins, T.J. ImageJ for microscopy. Biotechniques 2007, 43, S25–S30. [Google Scholar] [CrossRef]
- George, D.; Mallery, P. IBM SPSS Statistics 26 Step by Step: A Simple Guide and Reference; Routledge: New York, NY, USA, 2019. [Google Scholar]
- Singh, N.; Kapoor, R. Quick and accurate detection of Fusarium oxysporum f. sp. carthami in host tissue and soil using conventional and real-time PCR assay. World J. Microbiol. Biotechnol. 2018, 34, 1–12. [Google Scholar] [CrossRef]
- Olivain, C.; Humbert, C.; Nahalkova, J.; Fatehi, J.; L’Haridon, F.; Alabouvette, C. Colonization of Tomato Root by Pathogenic and Nonpathogenic Fusarium oxysporum Strains Inoculated Together and Separately into the Soil. Appl. Environ. Microbiol. 2006, 72, 1523–1531. [Google Scholar] [CrossRef]
- Ploetz, R.C. Fusarium wilt of banana is caused by several pathogens referred to as Fusarium oxysporum f. sp cubense. Phytopathology 2006, 96, 653–656. [Google Scholar] [CrossRef]
- Dubey, S.C.; Suresh, M.; Singh, B. Evaluation of Trichoderma species against Fusarium oxysporum f. sp ciceris for integrated management of chickpea wilt. Biol. Control 2007, 40, 118–127. [Google Scholar]
- Aoki, T.; O’Donnell, K.; Geiser, D.M. Systematics of key phytopathogenic Fusarium species: Current status and future challenges. J. Gen. Plant Pathol. 2014, 80, 189–201. [Google Scholar] [CrossRef]
- Li, C.; Yang, J.; Li, W.; Sun, J.; Peng, M. Direct root penetration and rhizome vascular colonization by Fusarium oxysporum f. sp cubense are the key steps in the successful infection of Brazil Cavendish. Plant Dis. 2017, 101, 2073–2078. [Google Scholar] [PubMed]
- NirmalaDevi, D.; Venkataramana, M.; Srivastava, R.K.; Uppalapati, S.R.; Gupta, V.K.; Yli-Mattila, T.; Tsui, C.; Srinivas, C.; Niranjana, S.R.; Chandra, N.S. Molecular phylogeny, pathogenicity and toxigenicity of Fusarium oxysporum f. sp lycopersici. Sci. Rep. 2016, 6, 21367. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, C.; Devi, D.N.; Murthy, K.N.; Mohan, C.D.; Lakshmeesha, T.R.; Singh, B.; Srivastava, R.K. Fusarium oxysporum f. sp lycopersici causal agent of vascular wilt disease of tomato: Biology to diversity–A review. Saudi J. Biol. Sci. 2019, 26, 1315–1324. [Google Scholar]
- Belhi, Z.; Boulenouar, N.; Cheriti, A. The Use of Natural Products Against Fusarium oxysporum: A Review. Nat. Prod. J. 2020, 12, 27–37. [Google Scholar] [CrossRef]
- Castaño, R.; Scherm, B.; Avilés, M. Genetic Diversity of Fusarium oxysporum f. sp. dianthi in Southern Spain. J. Mycol. 2014, 2014, 582672. [Google Scholar]
- Chawla, S.; Rafie, R.A.; Likins, T.M.; Ren, S.; Ndegwa, E.; Mersha, Z. First Report of Fusarium Yellows and Rhizome Rot Caused by Fusarium oxysporum f. sp. zingiberi on Ginger in the Continental United States. Plant Dis. 2021, 105, 3289. [Google Scholar] [CrossRef]
- Liu, S.; Li, J.; Zhang, Y.; Liu, N.; Viljoen, A.; Mostert, D.; Li, C. Fusaric acid instigates the invasion of banana by Fusarium oxysporum f. sp. cubense TR 4. New Phytol. 2020, 225, 913–929. [Google Scholar] [CrossRef]
- van Dam, P.; Fokkens, L.; Schmidt, S.M.; Linmans, J.H.; Kistler, H.C.; Ma, L.-J.; Rep, M. Effector profiles distinguish formae speciales of Fusarium oxysporum. Environ. Microbiol. 2016, 18, 4087–4102. [Google Scholar] [CrossRef]
- Boba, A.; Kostyn, K.; Kozak, B.; Zalewski, I.; Szopa, J.; Kulma, A. Transcriptomic profiling of susceptible and resistant flax seedlings after Fusarium oxysporum lini infection. PLoS ONE 2021, 16, e0246052. [Google Scholar] [CrossRef]
- Bleackley, M.R.; Samuel, M.; Garcia-Ceron, D.; McKenna, J.A.; Lowe, R.G.; Pathan, M.; Anderson, M.A. Extracellular vesicles from the cotton pathogen Fusarium oxysporum f. sp. vasinfectum induce a phytotoxic response in plants. Front. Plant Sci. 2020, 10, 1610. [Google Scholar] [CrossRef]
- Zhang, M.; Su, J.; Zhang, Y.; Xu, J.; Zhang, S. Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr. Opin. Plant Biol. 2018, 45, 1–10. [Google Scholar] [CrossRef]
- Warman, N.M.; Aitken, E.A.B. The Movement of Fusarium oxysporum f.sp. cubense (Sub-Tropical Race 4) in Susceptible Cultivars of Banana. Front. Plant Sci. 2018, 9, 1748. [Google Scholar] [PubMed]
- Iacomi-Vasilescu, B.; Blanchard, D.; Guenard, M.; Molinero-Demilly, V.; Laurent, E.; Simoneau, P. Development of a PCR based diagnostic assay for detecting pathogenic Alternaria species in cruciferous seeds. Seed Sci. Technol. 2002, 30, 87–96. [Google Scholar]
- Xiao, R.F.; Zhu, Y.-J.; Li, Y.-D.; Liu, B. Studies on Vascular Infection of Fusarium oxysporum f. sp. cubense Race 4 in Banana by Field Survey and Green Fluorescent Protein Reporter. Int. J. Phytopathol. 2013, 2, 44–51. [Google Scholar]
- Pollard, A.T.; Okubara, P.A. Real-time PCR quantification of Fusarium avenaceum in soil and seeds. J. Microbiol. Methods 2018, 157, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.D.; Zellama, M.S.; Varanda, C.; Materatski, P.; Peixe, A.; Chaouachi, M.; Félix, M.D.R. Establishment of a sensitive qPCR methodology for detection of the olive-infecting viruses in Portuguese and Tunisian orchards. Front. Plant Sci. 2019, 10, 694. [Google Scholar] [CrossRef]
- Jiménez-Fernández, D.; Montes-Borrego, M.; Jiménez-Díaz, R.M.; Navas-Cortés, J.A.; Landa, B.B. In planta and soil quantification of Fusarium oxysporum f. sp. ciceris and evaluation of Fusarium wilt resistance in chickpea with a newly developed quantitative polymerase chain reaction assay. Phytopathology 2011, 101, 250–262. [Google Scholar] [PubMed]
- Matthews, M.C.; Mostert, D.; Ndayihanzamaso, P.; Rose, L.J.; Viljoen, A. Quantitative detection of economically important Fusarium oxysporum f. sp. cubense strains in Africa in plants, soil and water. PLoS ONE 2020, 15, e0236110. [Google Scholar] [CrossRef]
- Purwati, R.D.; Hidayah, N.; Sudjindro. Sudarsono Inoculation Methods and Conidial Densities of Fusarium oxysporum f.sp cubense in Abaca. HAYATI J. Biosci. 2008, 15, 1–7. [Google Scholar] [CrossRef]
- Taranto, F.; Pasqualone, A.; Mangini, G.; Tripodi, P.; Miazzi, M.M.; Pavan, S.; Montemurro, C. Polyphenol oxidases in crops: Biochemical, physiological and genetic aspects. Int. J. Mol. Sci. 2017, 18, 377. [Google Scholar] [CrossRef] [PubMed]
- van der Does, H.C.; Constantin, M.E.; Houterman, P.M.; Takken, F.L.W.; Cornelissen, B.J.C.; Haring, M.A.; Burg, H.A.V.D.; Rep, M. Fusarium oxysporum colonizes the stem of resistant tomato plants, the extent varying with the R-gene present. Eur. J. Plant Pathol. 2018, 154, 55–65. [Google Scholar] [CrossRef]
- Zvirin, T.; Herman, R.; Brotman, Y.; Denisov, Y.; Belausov, E.; Freeman, S.; Perl-Treves, R. Differential colonization and defence responses of resistant and susceptible melon lines infected by Fusarium oxysporum race 1–2. Plant Pathol. 2010, 59, 576–585. [Google Scholar] [CrossRef]
- Sampaio, A.M.; Vitale, S.; Turrà, D.; Di Pietro, A.; Rubiales, D.; van Eeuwijk, F.; Patto, M.C.V. A diversity of resistance sources to Fusarium oxysporum f. sp. pisi found within grass pea germplasm. Plant Soil 2021, 463, 19–38. [Google Scholar]
- Khan, M.R. Plant nematodes: Methodology, Morphology, Systematics, Biology and Ecology. CRC Press: Boca Raton, FL, USA, 2008. [Google Scholar]
- Mzabri, I.; Addi, M.; Berrichi, A. Traditional and Modern Uses of Saffron (Crocus Sativus). Cosmetics 2019, 6, 63. [Google Scholar] [CrossRef]
- Zhang, M.; Xu, J.H.; Liu, G.; Yao, X.F.; Li, P.F.; Yang, X.P. Characterization of the watermelon seedling infection process by Fusarium oxysporum f. sp. niveum. Plant Pathol. 2015, 64, 1076–1084. [Google Scholar] [CrossRef]
- Wu, L.; Conner, R.L.; Wang, X.; Xu, R.; Li, H. Variation in growth, colonization of maize, and metabolic parameters of GFP-and DsRed-labeled Fusarium verticillioides strains. Phytopathology 2016, 106, 890–899. [Google Scholar] [CrossRef]
- Yao, J.; Huang, P.; Chen, H.; Yu, D. Fusarium oxysporum is the pathogen responsible for stem rot of the succulent plant Echeveria ‘Perle von Nürnberg’ and observation of the infection process. Eur. J. Plant Pathol. 2021, 159, 555–568. [Google Scholar] [CrossRef]

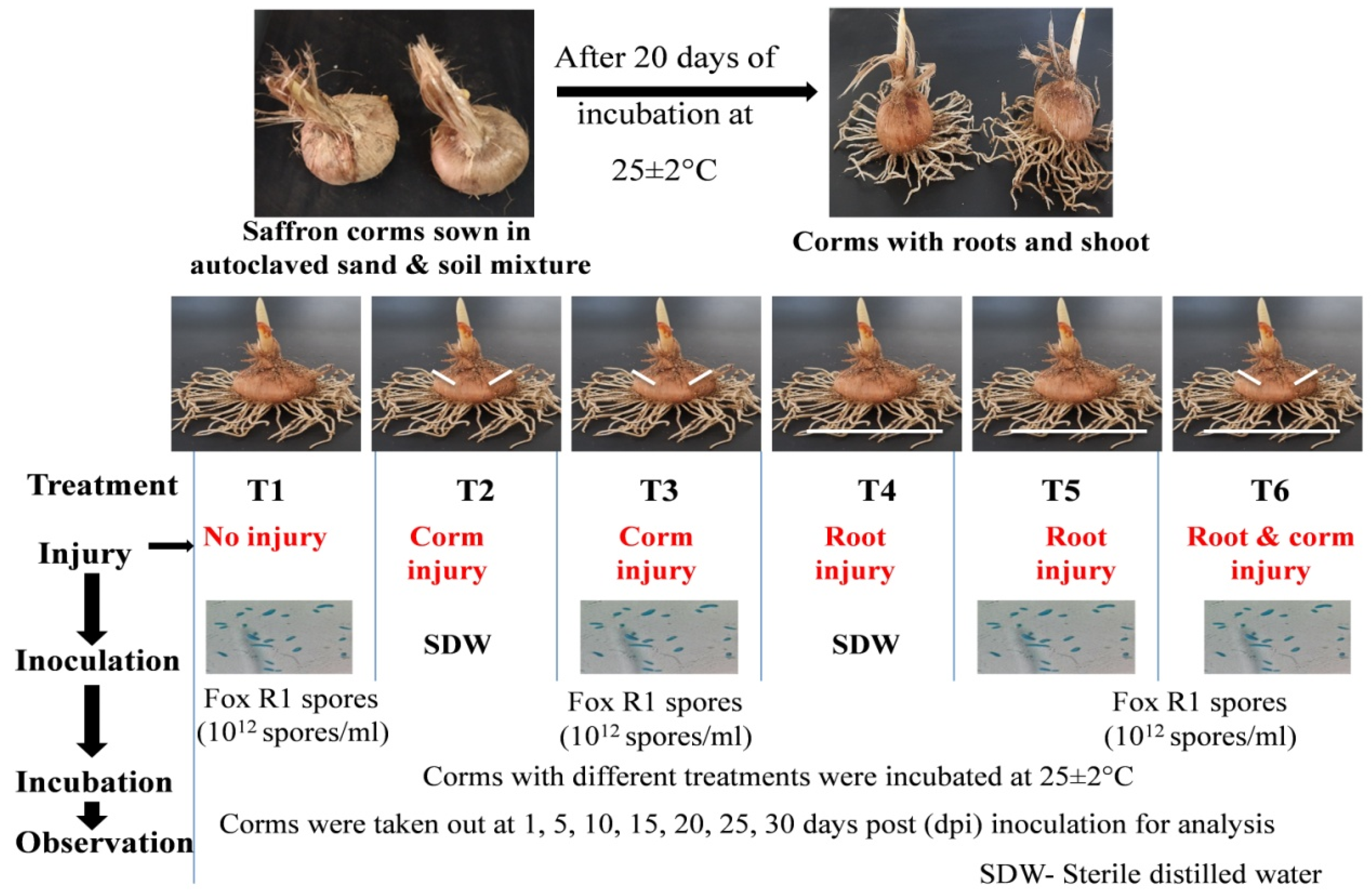


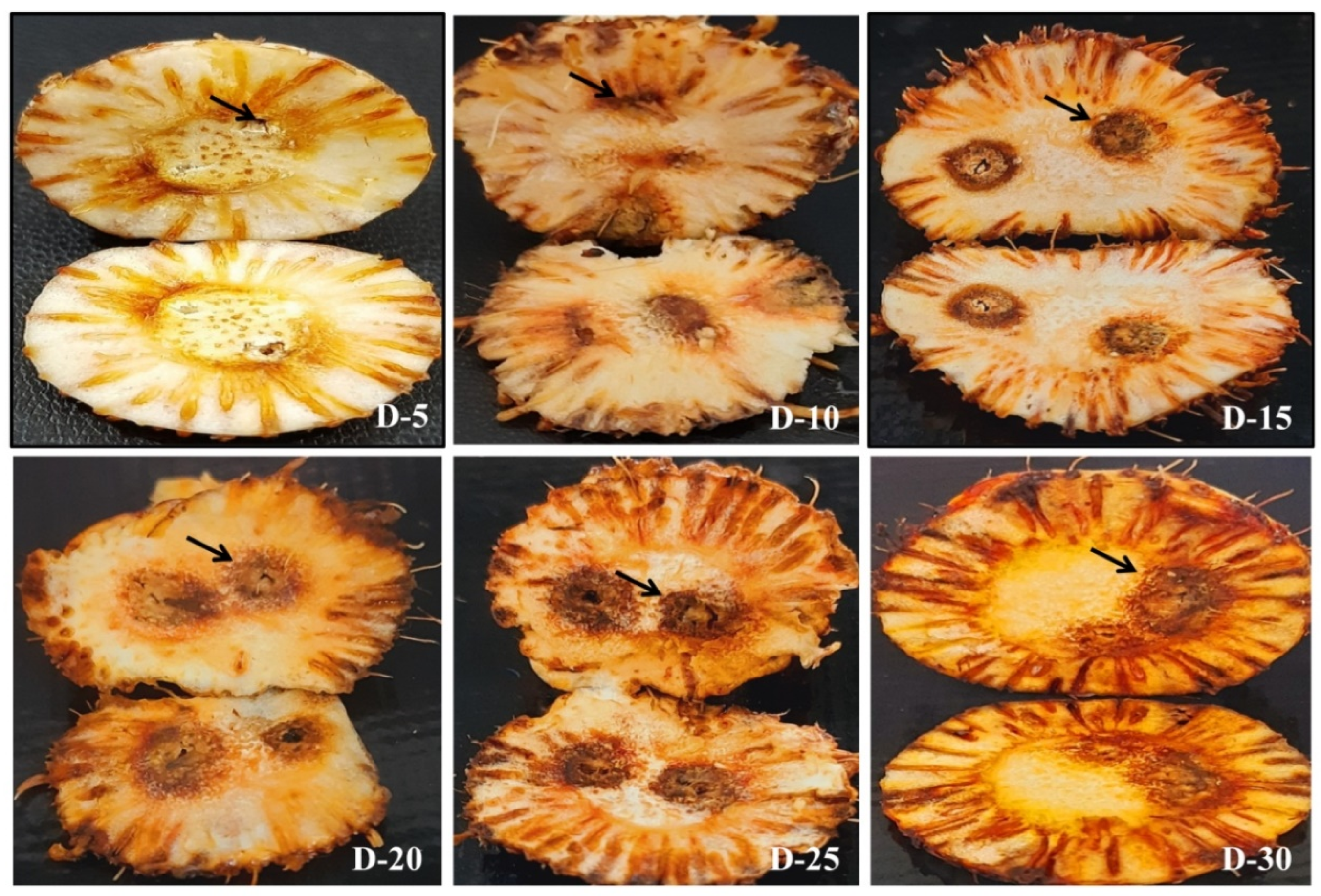
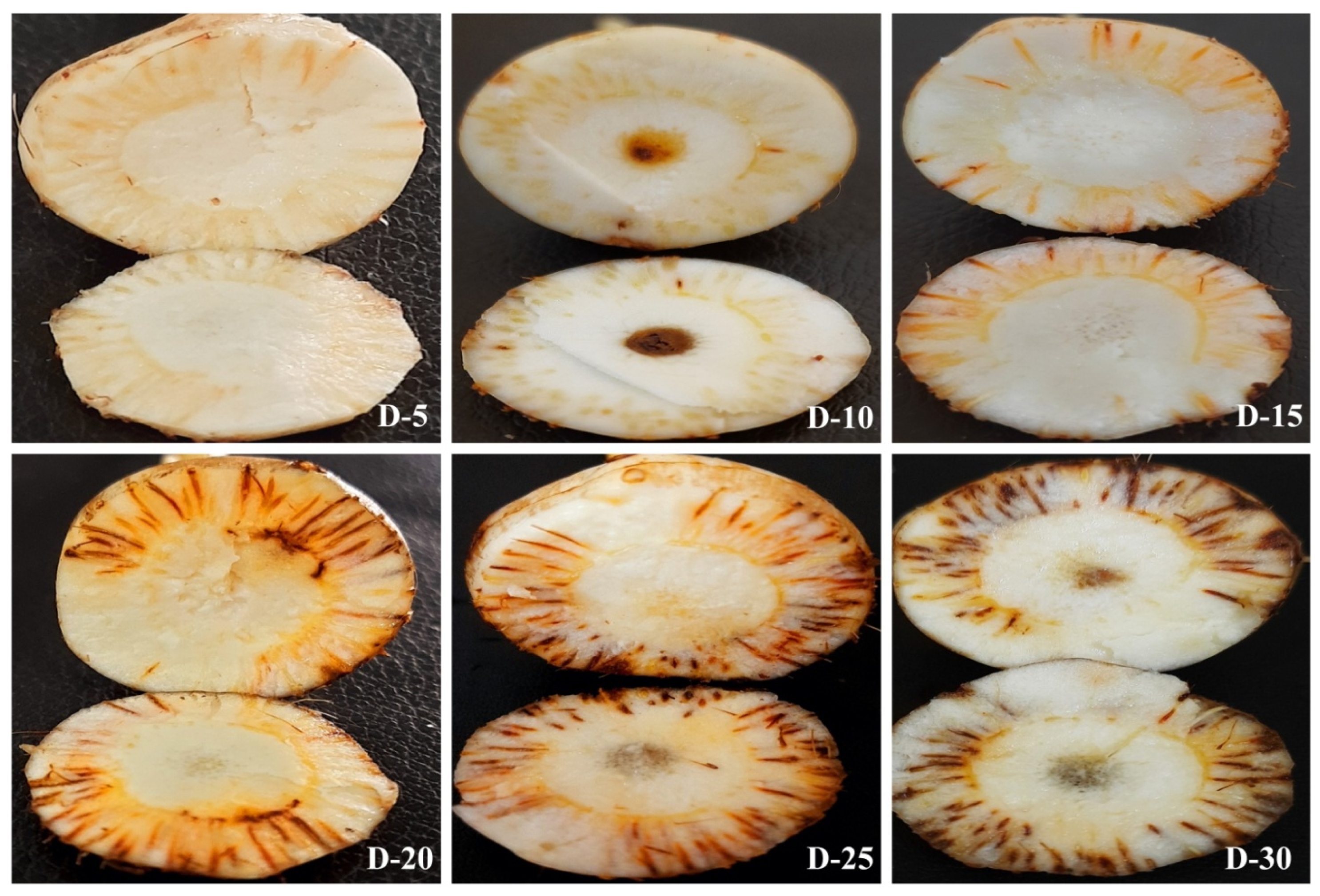
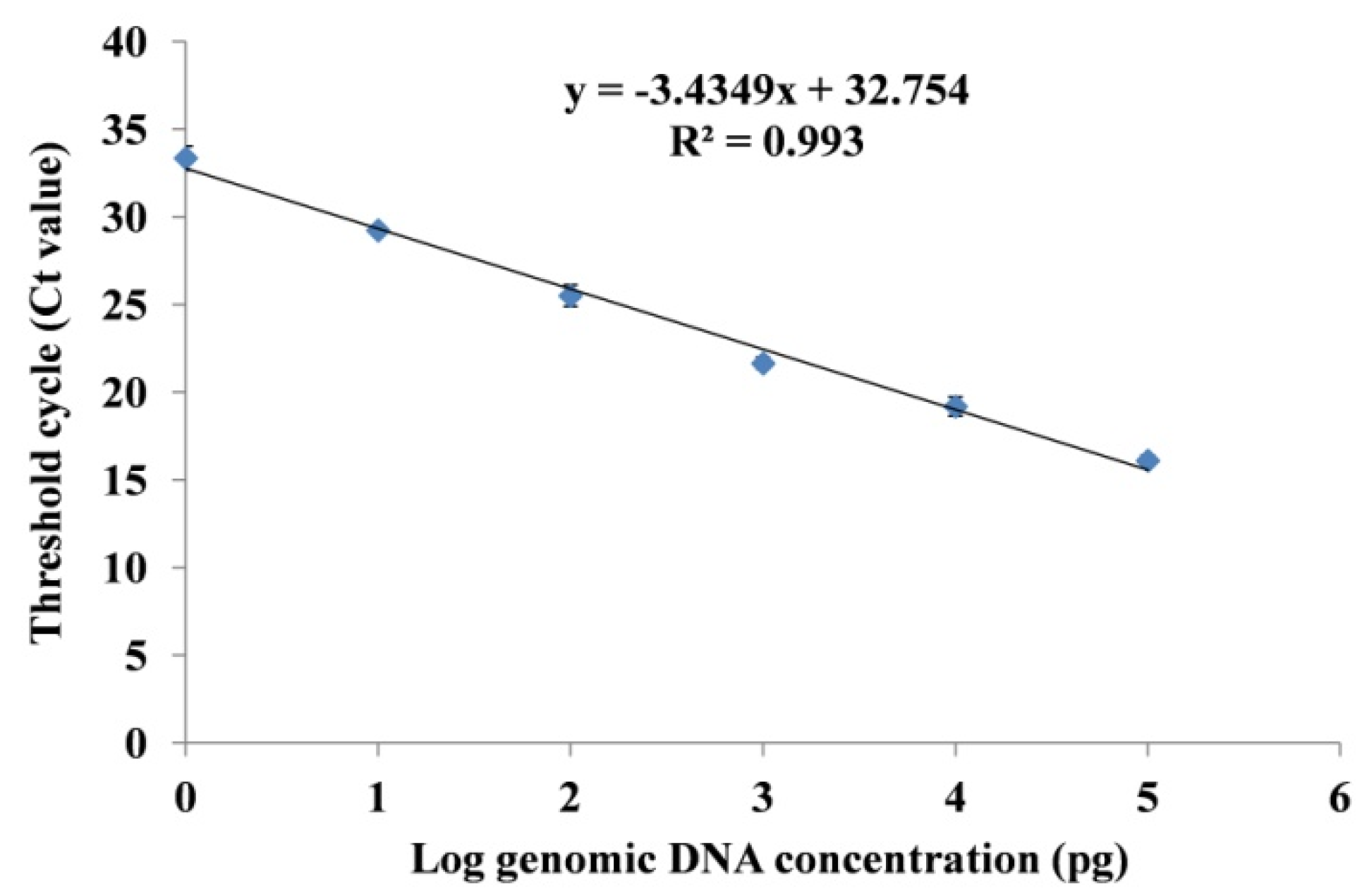





| S. No | Treatments | Description of Treatment | Site for DNA Isolation | |
|---|---|---|---|---|
| Injury | Fox R1 | |||
| 1 | T1 | No | − | Root and corm |
| 2 | T2 | Corm injury | − | Corm injury point |
| 3 | T3 | Corm injury | + | Corm basal plate and roots |
| 4 | T4 | Root injury | − | Root tips |
| 5 | T5 | Root injury | + | Roots and corm basal plate |
| 6 | T6 | Root + corm injury | + | Corm injury site and roots |
| S.No | Housekeeping Genes | Amplicon Size (bp) | GenBank Accession Number | Closest Neighbor Based on Phylogeny | Identity (%) | Accession Number |
|---|---|---|---|---|---|---|
| 1 | Actin | 300 | KJ866867 | Fusarium oxysporum strain FJDO-1 | 99.55% | MK895954 |
| 2 | Translation elongation factor 1α | 700 | KJ866868 | Fusarium oxysporum isolate A010P | 100% | MN191811 |
| 3 | β-tubulin | 350 | KJ866869 | Fusarium oxysporum f.sp. cubense isolate Foc37 | 100% | MF668108 |
| 4 | Calmudlin | 500 | KJ866870 | Fusarium oxysporum f.sp. zingiberi isolate Gf-VA-3 | 99.05% | MT802441 |
| 5 | Histone 3 | 350 | KJ866871 | Fusarium oxysporum f.sp. ciceri | 980.5% | AF346506 |
| Genus | Symptoms | Microscopy | ITS Phylogeny | Infection by Fox R1 |
|---|---|---|---|---|
| Crocus | + | Macrospores (1–3 septa), curved spores. | Fox R1 | + |
| Gladiolus | − | Aseptate spores | − | |
| Ixia | − | Single septate curved spores | - | − |
| Sparaxis | + | Macrospores (1–3 septa), curved spores. | Fox R1 | + |
| Tritonia | + | Macrospores (1–3 septa), curved spores. | Fox R1 | + |
| T3 (Injury to Corm Only + Fox R1) | T5 (Injury to Roots Only + Fox R1) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S.No. | Corm | Roots | Roots | Corm Basal Plate | |||||||||||||
| dpi | Symptoms (A) | Semi-Quantitative (B) | q-PCR (C) | Concentration (pg) (D) | Symptoms (A) | Semi-Quantitative(B) | q-PCR (C) | Concentration (pg) (D) | Symptoms (A) | Semi-Quantitative (B) | q-PCR (C) | Concentration (pg) (D) | Symptoms (A) | Semi-Quantitative (B) | q-PCR (C) | Conentration (pg) (D) | |
| 1 | 1 | + | + | +/29.08 ± 0.91 | +/11.77 ± 1.9 | - | - | - | - | + | + | 29.18 ± 0.8 | +/11.01 ± 2.5 | - | - | - | |
| 2 | 3 | + | + | +/27.13 ± 0.67 | +/43.61 ± 5.62 | - | - | - | - | + | + | 28.15 ± 0.18 | +/21.99 ± 4.8 | - | - | - | |
| 3 | 5 | + | + | +/25.26 ± 0.12 | +/153.82 ± 15.90 | - | - | +/31.06 ± 0.65 | 3.11 ± 0.9 | + | + | 26.34 ± 0.33 | +/74.12 ± 8.7 | - | - | - | |
| 4 | 10 | + | + | +/23.67 ± 0.55 | +/445.04 ± 39.7 | + | + | +/27.19 ± 0.43 | 41.89 ± 3.5 | + | + | 24.74 ± 0.6 | +/216.9 9± 19.8 | - | - | 30.99 ± 0.87 | +/3.26 ± 1.2 |
| 5 | 15 | + | + | +/21.47 ± 0.91 | +/1948.94 ± 180.7 | + | + | +/25.57 ± 0.31 | 124.29 ± 7.9 | + | + | 23.49 ± 0.12 | +/502.11 ± 34.6 | + | + | 27 ± 0.54 | +/47.593 ± 8.9 |
| 6 | 20 | + | + | +/19.69 ± 1.01 | +/6437.98 ± 192.99 | + | + | +/23.08 ± 0.50 | 661.31 ± 49.7 | + | + | 22.59 ± 0.79 | +/918.9 ± 51.7 | + | + | 24.8 ± 0.44 | +/208.425 ± 40.7 |
| 7 | 25 | + | + | +/19.32 ± 0.79 | +/8253.15 ± 204.7 | + | + | +/22.13 ± 0.59 | 1251.35 ± 104.7 | + | + | 22.09 ± 0.55 | +/1348.96 ± 1.2.9 | + | + | 21.87 ± 0.19 | +/1489.97 ± 118.7 |
| 8 | 30 | + | + | +/18.93 ± 0.34 | +/10,723.09 ± 222.1 | + | + | +/21.87 ± 0.34 | 1489.97 ± 169.7 | + | + | 21.8 ± 0.54 | +/1561.167 ± 176.4 | + | + | 20.79 ± 0.52 | +/3076.45 ± 167.3 |
| S.No. | Days Post Inoculation(dpi) | Fusarium oxysporum R1 Load (105 CFU/gm of Tissue) in Different Treatments | ||||||
|---|---|---|---|---|---|---|---|---|
| T3 (Corm Injury + FoxR1) | T6 (Corm + Root Injury +Fox R1) | T5 (Root Injury + Fox R1) | ||||||
| Sites |  | Corm basal plate | Roots | Corm tissue around injury point | Roots | Roots | Corm basal plate | |
| 1 | 1 | 0.16 ± 0.09 a | 0 | 0.4 ± 0.10 a | 0.15 ± 0.07 a | 0.14 ± 0.08 a | 0 | |
| 2 | 3 | 0.42 ± 0.11 b | 0 | 1.28 ± 0.08 b | 0.5 ± 0.08 b | 0.7 ± 0.06 b | 0 | |
| 3 | 5 | 1.50 ± 0.10 c | 0 | 4.94 ± 0.11 c | 1.42 ± 0.06 c | 1.56 ± 0.12 c | 0 | |
| 4 | 10 | 5.78 ± 0.08 d | 2.06 ± 0.08 a | 9.6 ± 0.07 d | 3.06 ± 0.11 d | 3.52 ± 0.10 d | 0 | |
| 5 | 15 | 11.84 ± 0.11e | 4.24 ± 0.11 b | 13.2 ± 0.13 e | 5.26 ± 0.05 e | 5.2 ± 0.09 e | 0.32 ± 0.13 a | |
| 6 | 20 | 15.50 ± 0.07 f | 5.72 ± 0.06 c | 17.19 ± 0.12 f | 6.72 ± 0.13 f | 6.68 ± 0.13 f | 0.82 ± 0.08 b | |
| 7 | 25 | 18.12 ± 0.13 g | 7.28 ± 0.07 d | 20.92 ± 0.08 g | 7.2 ± 0.12 g | 7.22 ± 0.07 g | 2.36 ± 0.05 c | |
| 8 | 30 | 21.32 ± 0.08 h | 7.34 ± 0.27 e | 24.9 ± 0.10 h | 7.6 ± 0.03 h | 7.61 ± 0.25 h | 6.88 ± 0.09 d | |
| 9 | p-value | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhagat, N.; Magotra, S.; Gupta, R.; Sharma, S.; Verma, S.; Verma, P.K.; Ali, T.; Shree, A.; Vakhlu, J. Invasion and Colonization of Pathogenic Fusarium oxysporum R1 in Crocus sativus L. during Corm Rot Disease Progression. J. Fungi 2022, 8, 1246. https://doi.org/10.3390/jof8121246
Bhagat N, Magotra S, Gupta R, Sharma S, Verma S, Verma PK, Ali T, Shree A, Vakhlu J. Invasion and Colonization of Pathogenic Fusarium oxysporum R1 in Crocus sativus L. during Corm Rot Disease Progression. Journal of Fungi. 2022; 8(12):1246. https://doi.org/10.3390/jof8121246
Chicago/Turabian StyleBhagat, Nancy, Shanu Magotra, Rikita Gupta, Shikha Sharma, Sandhya Verma, Praveen Kumar Verma, Tahir Ali, Ankita Shree, and Jyoti Vakhlu. 2022. "Invasion and Colonization of Pathogenic Fusarium oxysporum R1 in Crocus sativus L. during Corm Rot Disease Progression" Journal of Fungi 8, no. 12: 1246. https://doi.org/10.3390/jof8121246
APA StyleBhagat, N., Magotra, S., Gupta, R., Sharma, S., Verma, S., Verma, P. K., Ali, T., Shree, A., & Vakhlu, J. (2022). Invasion and Colonization of Pathogenic Fusarium oxysporum R1 in Crocus sativus L. during Corm Rot Disease Progression. Journal of Fungi, 8(12), 1246. https://doi.org/10.3390/jof8121246







