Stress Responses Elicited by Glucose Withdrawal in Aspergillus fumigatus
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain, Culturing Conditions
2.2. Agar Plate Assays
2.3. Detecting Growth and Metabolic Activity
2.4. Enzyme Assays
2.5. Measuring Redox Imbalance, GSH, and GSSG Contents
2.6. Reverse-Transcription Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR) Assays
2.7. High-Throughput RNA Sequencing
2.8. Detecting Allele-Specific Expression
2.9. Detecting Gene Duplications in A. fumigatus Genomes
2.10. Evaluation of Transcriptome Data
3. Results
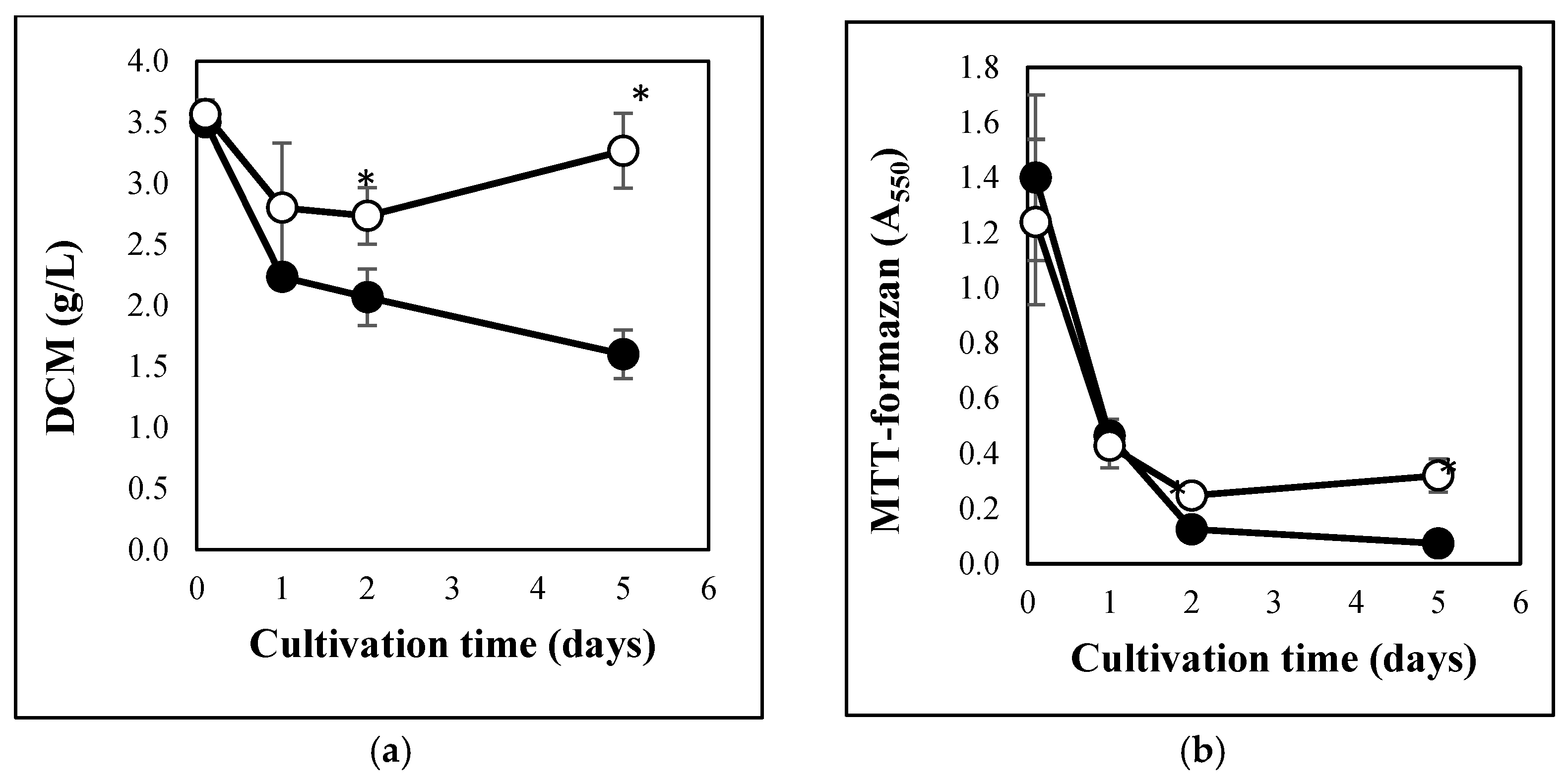
3.1. Carbon Stress-Induced Hydrolase Secretion and Increased Redox Imbalance in A. fumigatus
3.2. Glucose Withdrawal Had Substantial Consequences on the Transcriptome; Peptone Modified the Transcriptome Markedly Only in the Absence of Glucose
3.3. Autolytic Cell Wall Degradation Is Important in Carbon Stress Adaptation
3.4. GSH Depletion Was Accompanied with Upregulation of γGT, but Not DUG Pathway Genes
3.5. Antioxidative Enzymes and Secondary Metabolism Cluster Genes Had Transcriptional Patterns Characteristic of the Carbon Source
3.6. Carbon Stress Altered the Transcription of Iron Metabolism Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wong, S.S.W.; Aimanianda, V. Host soluble mediators: Defying the immunological inertness of Aspergillus fumigatus conidia. J. Fungi 2017, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Simões, D.; Caetano, L.A.; Veríssimo, C.; Viegas, C.; Sabino, R. Aspergillus collected in specific indoor settings: Their molecular identification and susceptibility pattern. Int. J. Environ. Health Rev. 2019, 31, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Verburg, K.; van Neer, J.; Duca, M.; de Cock, H. novel treatment approach for aspergilloses by targeting germination. J. Fungi 2022, 8, 758. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef] [PubMed]
- Lestrade, P.P.A.; Meis, J.F.; Melchers, W.J.G.; Verweij, P.E. Triazole resistance in Aspergillus fumigatus: Recent insights and challenges for patient management. Clin. Microbiol. Infect. 2019, 25, 799–806. [Google Scholar] [CrossRef]
- Abad, A.; Fernández-Molina, J.V.; Bikandi, J.; Ramírez, A.; Margareto, J.; Sendino, J.; Hernando, F.L.; Pontón, J.; Garaizar, J.; Rementeria, A. What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev. Iberoam. Micol. 2010, 27, 155–182. [Google Scholar] [CrossRef]
- Brown, A.J.P.; Cowen, L.E.; di Pietro, A.; Quinn, J. Stress adaptation. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Emri, T.; Forgács, K.; Pócsi, I. Biologia futura: Combinatorial stress responses in fungi. Biol. Futur. 2022, 73, 207–217. [Google Scholar] [CrossRef]
- Emri, T.; Vékony, V.; Gila, B.; Nagy, F.; Forgács, K.; Pócsi, I. Autolytic hydrolases affect sexual and asexual development of Aspergillus nidulans. Folia Microbiol. 2018, 63, 619–626. [Google Scholar] [CrossRef]
- Richie, D.L.; Fuller, K.K.; Fortwendel, J.; Miley, M.D.; McCarthy, J.W.; Feldmesser, M.; Rhodes, J.C.; Askew, D.S. Unexpected link between metal ion deficiency and autophagy in Aspergillus fumigatus. Eukaryot Cell 2007, 6, 2437–2447. [Google Scholar] [CrossRef]
- van Munster, J.; Burggraaf, A.; Pócsi, I.; Szilágyi, M.; Emri, T.; Ram, A. Post-genomic approaches to dissect carbon starvation responses in Aspergilli. In Aspergillus and Penicillium in the Post-Genomic Era; de Vries, R.P., Gelber, I., Rørdam Andersen, M., Eds.; Caister Academic Press: Norfolk, UK, 2016; pp. 89–111. [Google Scholar]
- Barratt, R.W.; Johnson, G.B.; Ogata, W.N. Wild-type and mutant stocks of Aspergillus nidulans. Genetics 1965, 52, 233–246. [Google Scholar] [CrossRef]
- Emri, T.; Molnár, Z.; Pócsi, I. The appearances of autolytic and apoptotic markers are concomitant but differently regulated in carbon-starving Aspergillus nidulans cultures. FEMS Microbiol. Lett. 2005, 251, 297–303. [Google Scholar] [CrossRef]
- Pusztahelyi, T.; Pócsi, I.; Szentirmai, A. Aging of Penicillium chrysogenum cultures under carbon starvation: II: Protease and N-acetyl-b-D-hexosaminidase production. Biotechnol. Appl. Biochem. 1997, 25, 87–93. [Google Scholar] [CrossRef]
- Fontaine, T.; Hartland, R.P.; Beauvais, A.; Diaquin, M.; Latge, J.P. Purification and characterization of an endo-1,3-beta-glucanase from Aspergillus fumigatus. Eur. J. Biochem. 1997, 243, 315–321. [Google Scholar] [CrossRef]
- Tomarelli, R.M.; Charney, J.; Harding, M.L. The Use of azoalbumin as a substrate in the colorimetric determination or peptic and tryptic activity. J. Lab. Clin. Med. 1949, 34, 428–433. [Google Scholar]
- Spitzmüller, Z.; Kwon, N.-J.; Szilágyi, M.; Keserű, J.; Tóth, V.; Yu, J.-H.; Pócsi, I.; Emri, T. γ-Glutamyl Ttanspeptidase (GgtA) of Aspergillus nidulans is not necessary for bulk degradation of glutathione. Arch. Microbiol. 2015, 197, 285–297. [Google Scholar] [CrossRef]
- Emri, T.; Molnár, Z.; Pusztahelyi, T.; Pócsi, I. Physiological and morphological changes in autolyzing Aspergillus nidulans cultures. Folia Microbiol. 2004, 49, 277–284. [Google Scholar] [CrossRef]
- Sámi, L.; Emri, T.; Pócsi, I. Autolysis and aging of Penicillium chrysogenum cultures under carbon starvation: III: Glutathione metabolism and formation of reactive oxygen species. Mycol. Res. 2001, 105, 1246–1250. [Google Scholar] [CrossRef]
- Anderson, M.E. Determination of glutathione and glutathione disulfide in biological samples. Methods Enzymol. 1985, 113, 548–555. [Google Scholar] [CrossRef]
- Emri, T.; Pócsi, I.; Szentirmai, A. Glutathione metabolism and protection against oxidative stress caused by peroxides in Penicillium chrysogenum. Free Radical. Biol. Med. 1997, 23, 809–814. [Google Scholar] [CrossRef]
- Chomczynski, P. A Reagent for the Single-Step Simultaneous Isolation of RNA, DNA and Proteins from Cell and Tissue Samples. Biotechniques 1993, 15, 532–534, 536–537. [Google Scholar] [PubMed]
- Emri, T.; Gila, B.; Antal, K.; Fekete, F.; Moon, H.; Yu, J.-H.; Pócsi, I. Atfa-independent adaptation to the toxic heavy metal cadmium in Aspergillus nidulans. Microorganisms 2021, 9, 1433. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Boeva, V.; Zinovyev, A.; Bleakley, K.; Vert, J.P.; Janoueix-Lerosey, I.; Delattre, O.; Barillot, E. Control-free calling of copy number alterations in deep-sequencing data using GC-content normalization. Bioinformatics 2011, 27, 268–269. [Google Scholar] [CrossRef] [PubMed]
- Priebe, S.; Kreisel, C.; Horn, F.; Guthke, R.; Linde, J. FungiFun2: A comprehensive online resource for systematic analysis of gene lists from fungal species. Bioinformatics 2015, 31, 445–446. [Google Scholar] [CrossRef]
- Kurucz, V.; Krüger, T.; Antal, K.; Dietl, A.M.; Haas, H.; Pócsi, I.; Kniemeyer, O.; Emri, T. Additional oxidative stress reroutes the global response of Aspergillus fumigatus to iron depletion. BMC Genomics 2018, 19, 357. [Google Scholar] [CrossRef]
- Haas, H. Iron—A key nexus in the virulence of Aspergillus fumigatus. Front. Microbiol. 2012, 3, 28. [Google Scholar] [CrossRef]
- Inglis, D.O.; Binkley, J.; Skrzypek, M.S.; Arnaud, M.B.; Cerqueira, G.C.; Shah, P.; Wymore, F.; Wortman, J.R.; Sherlock, G. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiol. 2013, 13, 91. [Google Scholar] [CrossRef]
- Lin, H.C.; Chooi, Y.H.; Dhingra, S.; Xu, W.; Calvo, A.M.; Tang, Y. The fumagillin biosynthetic gene cluster in Aspergillus fumigatus encodes a cryptic terpene cyclase involved in the formation of β-trans-bergamotene. J. Am. Chem. Soc. 2013, 135, 4616–4619. [Google Scholar] [CrossRef]
- Gila, B.C.; Moon, H.; Antal, K.; Hajdu, M.; Kovács, R.; Jónás, A.P.; Pusztahelyi, T.; Yu, J.H.; Pócsi, I.; Emri, T. The DUG pathway governs degradation of intracellular glutathione in Aspergillus nidulans. Appl. Environ. Microbiol. 2021, 87, e01321-20. [Google Scholar] [CrossRef]
- Gila, B.C.; Antal, K.; Birkó, Z.; Keserű, J.S.; Pócsi, I.; Emri, T. Strategies shaping the transcription of carbohydrate-active enzyme genes in Aspergillus nidulans. J. Fungi 2022, 8, 79. [Google Scholar] [CrossRef]
- Gasch, A.P.; Spellman, P.T.; Kao, C.M.; Carmel-Harel, O.; Eisen, M.B.; Storz, G.; Botstein, D.; Brown, P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 2000, 11, 4241–4257. [Google Scholar] [CrossRef]
- Szilágyi, M.; Kwon, N.J.; Dorogi, C.; Pócsi, I.; Yu, J.H.; Emri, T. The extracellular β-1,3-endoglucanase EngA is involved in autolysis of Aspergillus nidulans. J. Appl. Microbiol. 2010, 109, 1498–1508. [Google Scholar] [CrossRef]
- Pócsi, I.; Leiter, E.; Kwon, N.J.; Shin, K.S.; Kwon, G.S.; Pusztahelyi, T.; Emri, T.; Abuknesha, R.A.; Price, R.G.; Yu, J.H. Asexual sporulation signalling regulates autolysis of Aspergillus nidulans via modulating the chitinase ChiB production. J. Appl. Microbiol. 2009, 107, 514–523. [Google Scholar] [CrossRef]
- Emri, T.; Molnár, Z.; Szilágyi, M.; Pócsi, I. Regulation of autolysis in Aspergillus nidulans. Appl. Biochem. Biotechnol. 2008, 151, 211–220. [Google Scholar] [CrossRef]
- Nishida, H. Conservation of nucleosome positions in duplicated and orthologous gene pairs. Sci. World J. 2012, 2012, 298174. [Google Scholar] [CrossRef]
- Kaur, H.; Ganguli, D.; Bachhawat, A.K. Glutathione degradation by the alternative pathway (DUG pathway) in Saccharomyces cerevisiae is initiated by (Dug2p-Dug3p)2 complex, a novel glutamine amidotransferase (GATase) enzyme acting on glutathione. J. Biol. Chem. 2012, 287, 8920–8931. [Google Scholar] [CrossRef]
- Hagiwara, D.; Sakamoto, K.; Abe, K.; Gomi, K. Signaling pathways for stress responses and adaptation in Aspergillus species: Stress biology in the post-genomic era. Biosci. Biotechnol. Biochem. 2016, 80, 1667–1680. [Google Scholar] [CrossRef]
- Silva, P.L.; Horta, M.; Goldman, G. Genetic Interactions Between Aspergillus fumigatus Basic Leucine Zipper (bZIP) Transcription Factors AtfA, AtfB, AtfC, and AtfD. Front. Fungal Biol. 2021, 2, 632048. [Google Scholar] [CrossRef]
- Szilágyi, M.; Anton, F.; Pócsi, I.; Emri, T. Autolytic enzymes are responsible for increased melanization of carbon stressed Aspergillus nidulans cultures. J. Basic Microbiol. 2018, 58, 440–447. [Google Scholar] [CrossRef]
- Spitzmüller, Z.; Gonda, S.; Kiss-Szikszai, A.; Vasas, G.; Pócsi, I.; Emri, T. Characterization of extracellular γ-glutamyl transpeptidase from Aspergillus nidulans. Mycoscience 2016, 57, 400–403. [Google Scholar] [CrossRef]
- Bello, M.H.; Epstein, L. Clades of γ-glutamyltransferases (GGTs) in the ascomycota and heterologous expression of Colletotrichum graminicola CgGGT1, a member of the pezizomycotina-only GGT clade. J. Microbiol. 2013, 51, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.A.; Goldman, G.H. The contribution of Aspergillus fumigatus stress responses to virulence and antifungal resistance. J. Microbiol. 2016, 54, 243–253. [Google Scholar] [CrossRef] [PubMed]
- De Vries, R.P.; Riley, R.; Wiebenga, A.; Aguilar-Osorio, G.; Amillis, S.; Uchima, C.A.; Anderluh, G.; Asadollahi, M.; Askin, M.; Barry, K.; et al. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017, 18, 28. [Google Scholar] [CrossRef] [PubMed]
- Emri, T.; Sümegi-Győri, V.M.; Páll, K.; Gila, B.C.; Pócsi, I. Effect of the combinatorial iron-chelation and oxidative stress on the growth of Aspergillus species. Res. Microbiol. 2022, 173, 103969. [Google Scholar] [CrossRef]
- Emri, T.; Szarvas, V.; Orosz, E.; Antal, K.; Park, H.; Han, K.H.; Yu, J.H.; Pócsi, I. Core oxidative stress response in Aspergillus nidulans. BMC Genomics 2015, 16, 478. [Google Scholar] [CrossRef]
- Fallon, J.P.; Reeves, E.P.; Kavanagh, K. The Aspergillus fumigatus toxin fumagillin suppresses the immune response of Galleria mellonella larvae by inhibiting the action of haemocytes. Microbiology 2011, 157, 1481–1488. [Google Scholar] [CrossRef]
- Scharf, D.H.; Heinekamp, T.; Remme, N.; Hortschansky, P.; Brakhage, A.A.; Hertweck, C. Biosynthesis and function of gliotoxin in Aspergillus fumigatus. Appl. Microbiol. Biotech. 2012, 93, 467–472. [Google Scholar] [CrossRef]
- Yin, W.B.; Baccile, J.A.; Bok, J.W.; Chen, Y.; Keller, N.P.; Schroeder, F.C. A nonribosomal peptide synthetase-derived iron(III) complex from the pathogenic fungus Aspergillus fumigatus. J. Am. Chem. Soc. 2013, 135, 2064–2067. [Google Scholar] [CrossRef]
- Gallagher, L.; Owens, R.A.; Dolan, S.K.; O’Keeffe, G.; Schrettl, M.; Kavanagh, K.; Jones, G.W.; Doyle, S. The Aspergillus fumigatus protein GliK protects against oxidative stress and is essential for gliotoxin biosynthesis. Eukaryot Cell 2012, 11, 1226–1238. [Google Scholar] [CrossRef]
- Owens, R.A.; Hammel, S.; Sheridan, K.J.; Jones, G.W.; Doyle, S. A proteomic approach to investigating gene cluster expression and secondary metabolite functionality in Aspergillus fumigatus. PLoS ONE 2014, 9, e106942. [Google Scholar] [CrossRef]
- Dolan, S.K.; O’Keeffe, G.; Jones, G.W.; Doyle, S. Resistance is not futile: Gliotoxin biosynthesis, functionality and utility. Trends Microbiol. 2015, 23, 419–428. [Google Scholar] [CrossRef]
- Scharf, D.H.; Remme, N.; Habel, A.; Chankhamjon, P.; Scherlach, K.; Heinekamp, T.; Hortschansky, P.; Brakhage, A.A.; Hertweck, C. A dedicated glutathione S-transferase mediates carbon-sulfur bond formation in gliotoxin biosynthesis. J. Am. Chem. Soc. 2011, 133, 12322–12325. [Google Scholar] [CrossRef]
- Davis, C.; Carberry, S.; Schrettl, M.; Singh, I.; Stephens, J.C.; Barry, S.M.; Kavanagh, K.; Challis, G.L.; Brougham, D.; Doyle, S. The role of glutathione S-transferase GliG in gliotoxin biosynthesis in Aspergillus fumigatus. Chem. Biol. 2011, 18, 542–552. [Google Scholar] [CrossRef]
- Nairz, M.; Weiss, G. Iron in infection and immunity. Mol. Aspects Med. 2020, 75, 100864. [Google Scholar] [CrossRef]
- Misslinger, M.; Hortschansky, P.; Brakhage, A.A.; Haas, H. Fungal iron homeostasis with a focus on Aspergillus fumigatus. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118885. [Google Scholar] [CrossRef]
- Schrettl, M.; Bignell, E.; Kragl, C.; Joechl, C.; Rogers, T.; Arst, H.N., Jr.; Haynes, K.; Haas, H. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J. Exp. Med. 2004, 200, 1213–1219. [Google Scholar] [CrossRef]






| Significantly Enriched GO Terms * | ||
|---|---|---|
| Upregulated | Downregulated | |
| Carbon-starved vs. Glucose | Alpha-amino acid catabolic process; Aromatic amino acid family catabolic process; Polysaccharide catabolic process; Cellulose binding; Galacturonan metabolic process; Hemicellulose metabolic process; Pectin catabolic process; Xylan catabolic process; Carbohydrate transport; Hexose transmembrane transport; Glucose transmembrane transport; Secondary metabolic process; Phenol-containing compound biosynthetic process; DNA-binding transcription factor activity; Heme binding. | Cytosolic ribosome; Mitochondrial ribosome; Translation; Ribonucleotide biosynthetic process; Alpha-amino acid biosynthetic process; Alpha-amino acid catabolic process; Glycolysis/Gluconeogenesis; Pentose-phosphate shunt; Acetyl-CoA biosynthetic process; Citrate cycle (TCA cycle); Aerobic respiration; Sulfate assimilation; Sulfur amino acid biosynthetic process; Nitrate transport; Nitrite transport; Vitamin biosynthetic process; Ergosterol biosynthetic process; Fatty acid biosynthetic process; Fungal-type cell wall polysaccharide metabolic process; Glucan biosynthetic process; Iron-sulfur cluster binding; Heme binding; Heme biosynthetic process; Cellular response to iron ion starvation; Siderophore biosynthetic process; Cellular potassium ion homeostasis; Sodium ion transport; Sodium inorganic phosphate symporter activity; Biosynthesis of secondary metabolites; Galactose metabolism; and Serine-type peptidase activity. |
| Peptone vs. Glucose + peptone | Polysaccharide catabolic process; Carbohydrate transmembrane transporter activity DNA-binding transcription factor activity; Aromatic compound biosynthetic process; and Iron ion binding. | Cytosolic ribosome; Mitochondrial ribosome; Translation; Ribonucleotide biosynthetic process; Alpha-amino acid biosynthetic process; Alpha-amino acid catabolic process; Glycolysis/Gluconeogenesis; Pentose-phosphate shunt; Acetyl-CoA biosynthetic process; Citrate cycle (TCA cycle); Aerobic respiration; Sulfur amino acid biosynthetic process; Vitamin metabolic process; Ergosterol biosynthetic process; Fatty acid biosynthetic process; Heme binding; Heme biosynthetic process; Cellular response to iron ion starvation; Siderophore biosynthetic process; Cellular potassium ion homeostasis; Sodium ion transport; Biosynthesis of secondary metabolites; and Peroxiredoxin activity. |
| Peptone vs. Glucose | Cellular amino acid catabolic process; Aromatic amino acid family catabolic process; and Ribosome biogenesis. | Cytosolic ribosome; Translation; Ribonucleotide biosynthetic process; Alpha-amino acid biosynthetic process; Alpha-amino acid catabolic process; Glycolysis/Gluconeogenesis; Pentose-phosphate shunt; Acetyl-CoA biosynthetic process; Citrate cycle (TCA cycle); Aerobic respiration; Sulfur compound biosynthetic process; Vitamin metabolic process; Ergosterol biosynthetic process; Lipid biosynthetic process; Fungal-type cell wall organization or biogenesis; Glucan biosynthetic process; Heme binding; Cellular response to iron ion starvation; Cellular potassium ion homeostasis; Sodium ion transport; Inorganic phosphate transmembrane transporter activity; Biosynthesis of secondary metabolites; Galactose metabolism; Glycogen biosynthetic process; Serine-type peptidase activity; Peptide transport; and Peroxiredoxin activity. |
| Significantly Enriched GO Terms * | ||
|---|---|---|
| Upregulated | Downregulated | |
| Peptone vs. Carbon-starved | Ribosome biogenesis; Response to unfolded protein; N-acetylglucosamine catabolic process; and Iron ion homeostasis. | Polysaccharide catabolic process; Cell wall organization or biogenesis; Secondary metabolite biosynthetic process; Melanin metabolic process; and Heme binding. |
| Glucose + peptone vs. Glucose | Ribosome biogenesis; Translation; Gene expression; Biosynthesis of amino acids; Alpha-amino acid catabolic process; Cysteine and methionine metabolism; Ergosterol biosynthetic process; Fatty acid catabolic process; Cellular response to iron ion starvation; and Siderophore biosynthetic process. | Nitrite transmembrane transporter activity; Nitrate assimilation; Reactive nitrogen species metabolic process; Amino acid transmembrane transporter activity; Glutamate biosynthetic process; Nucleobase transmembrane transporter activity; Urea catabolic process; Maltose metabolic process; Secondary metabolite biosynthetic process; Melanin biosynthetic process; and Heme binding. |
| Gene Group a | Number of Upregulated/Downregulated Genes | ||||
|---|---|---|---|---|---|
| Peptone vs. Carbon-Starved | Glucose + Peptone vs. Glucose | Carbon-Starved vs. Glucose | Peptone vs. Glucose + Peptone | Peptone vs. Glucose | |
| Cazyme genes (566) | 39/135 b | 13/37 b | 157 b/109 | 134 b/146 b | 117 b/156 b |
| Autophagy-related genes (17) | 0/1 | 1/0 | 2/0 | 8/1 | 5/0 |
| Chitinase genes (16) | 1/6 b | 0/0 | 5/1 | 4/3 | 4/3 |
| Chitine utilization genes (6) | 6 b/0 | 0/0 | 6 b/0 | 6 b/0 | 6 b/0 |
| Glucanase genes (49) | 2/16 b | 2/3 | 15 b/14 b | 11/17 b | 11/19 b |
| Secreted peptidase genes (36) | 2/11 b | 1/2 | 8/14 b | 8/17 b | 4/19 b |
| Antioxidative enzyme genes (34) | 5/5 | 2/1 | 9/8 | 7/12 b | 7/8 |
| Catalases, peroxidases, SODs (15) | 2/3 | 1/1 | 6 b/2 | 4/4 | 4/2 |
| Thioredoxin, glutaredoxin, glutathione systems (19) | 3/2 | 1/0 | 3/6 | 3/8 b | 3/6 |
| Heme binding protein genes (121) | 6/38 b | 6/18 b | 43 b/36 b | 36 b/38 b | 32 b/45 b |
| Heme biosynthesis genes (11) | 2/0 | 0/0 | 0/6 b | 1/6 b | 1/4 |
| Fe-S cluster protein genes (43) | 7 b/4 | 1/2 | 5/18 b | 8/16 b | 6/16 b |
| Fe-S cluster assembly genes (15) | 1/0 | 0/0 | 1/3 | 2/4 | 3/3 |
| Fe acquisition genes (30) | 9 b/4 | 8b/0 | 4/16 b | 5/16 b | 4/16 b |
| Siderophore metabolism genes (14) | 6 b/1 | 7 b/0 | 0/11 b | 0/11 b | 0/10 b |
| RIA genes (3) | 0/1 | 1/0 | 0/3 b | 0/3 b | 0/3 b |
| other iron transporter genes (8) | 2/2 | 0/0 | 4/0 | 4/0 | 4/1 |
| Comparison | Number of Upregulated/Downregulated | |
|---|---|---|
| Clusters a | Genes b | |
| Peptone vs. Carbon-starved | 2/10 | 27/99 |
| Glucose + peptone vs. Glucose | 3/8 | 19/51 |
| Carbon-starved vs. Glucose | 9/8 | 81/86 |
| Peptone vs. Glucose + peptone | 4/8 | 57/104 |
| Peptone vs. Glucose | 4/10 | 50/105 |
| Cluster a | Number of Upregulated and Downregulated Genes | ||||
|---|---|---|---|---|---|
| Peptone vs. Carbon-Starved | Glucose + Peptone vs. Glucose | Carbon-Starved vs. Glucose | Peptone vs. Glucose + Peptone | Peptone vs. Glucose | |
| DHN-melanin cluster (10) | 1/7 b | 0/4 b | 5 b/1 | 3/2 | 2/5 b |
| Endocrocin cluster (9) | 2/6 b | 1/4 b | 7 b/0 | 5 b/0 | 5 b/0 |
| Fumagillin cluster (15) | 0/13 b | 0/11 b | 0/10 b | 0/13 b | 0/13 b |
| Fumigaclavine C (fga) cluster (11) | 0/11 b | 0/2 | 6 b/0 | 0/3 | 0/4 |
| Fumipyrrole cluster (7) | 2/1 | 0/0 | 0/7 b | 0/7 b | 0/7 b |
| Fumiquizoline cluster (5) | 0/5 b | 0/3 b | 5 b/0 | 2/1 | 0/1 |
| Fumitremorgin B (ftm) cluster (9) | 0/8 b | 0/1 | 0/8 b | 0/8 b | 0/8 b |
| Gliotoxin (gli) cluster (12) | 3 b/0 | 0/7 b | 0/10 b | 2/6 b | 1/9 b |
| Hexadehydro-astechrome cluster (8) | 0/7 b | 0/7 b | 0/7 b | 2/3 | 0/7 b |
| Pseurotin A cluster (4) | 0/4 b | 0/4 b | 0/4 b | 0/4 b | 0/4 b |
| Siderophore cluster (18) | 6 b/0 | 9 b/2 | 2/9 b | 3/10 b | 1/9 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Emri, T.; Antal, K.; Gila, B.; Jónás, A.P.; Pócsi, I. Stress Responses Elicited by Glucose Withdrawal in Aspergillus fumigatus. J. Fungi 2022, 8, 1226. https://doi.org/10.3390/jof8111226
Emri T, Antal K, Gila B, Jónás AP, Pócsi I. Stress Responses Elicited by Glucose Withdrawal in Aspergillus fumigatus. Journal of Fungi. 2022; 8(11):1226. https://doi.org/10.3390/jof8111226
Chicago/Turabian StyleEmri, Tamás, Károly Antal, Barnabás Gila, Andrea P. Jónás, and István Pócsi. 2022. "Stress Responses Elicited by Glucose Withdrawal in Aspergillus fumigatus" Journal of Fungi 8, no. 11: 1226. https://doi.org/10.3390/jof8111226
APA StyleEmri, T., Antal, K., Gila, B., Jónás, A. P., & Pócsi, I. (2022). Stress Responses Elicited by Glucose Withdrawal in Aspergillus fumigatus. Journal of Fungi, 8(11), 1226. https://doi.org/10.3390/jof8111226






