The Mitochondrial Alternative Oxidase in Ustilago maydis Is Not Involved in Response to Oxidative Stress Induced by Paraquat
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Strains and Growth Conditions
2.3. Cell Viability
2.4. Oxygen Consumption
2.5. Flow Cytometry
2.6. Production of Hydrogen Peroxide by U. maydis Cells
2.7. Detection of Intracellular Reactive Oxygen Species
2.8. Confocal Microscopy
2.9. Preparation of Cell-Free Extracts
2.10. Glutathione Reductase
2.11. Glutathione Peroxidase
2.12. Superoxide Dismutase
2.13. Catalase-Peroxidase
2.14. Glutathione and Glutathione Disulfide Determination
2.15. Statistical Analysis
3. Results
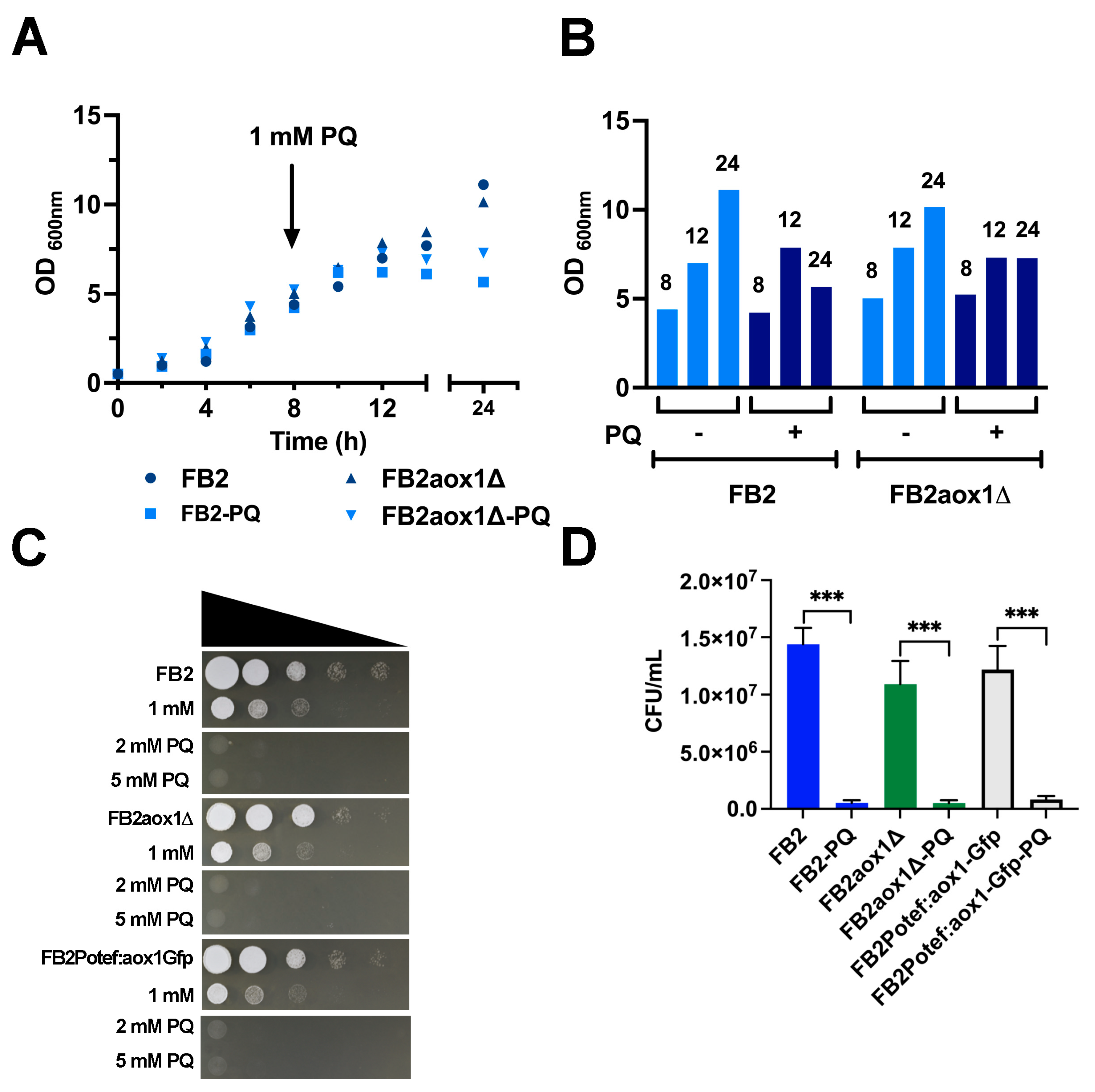
3.1. Effect of Paraquat on U. maydis Growth
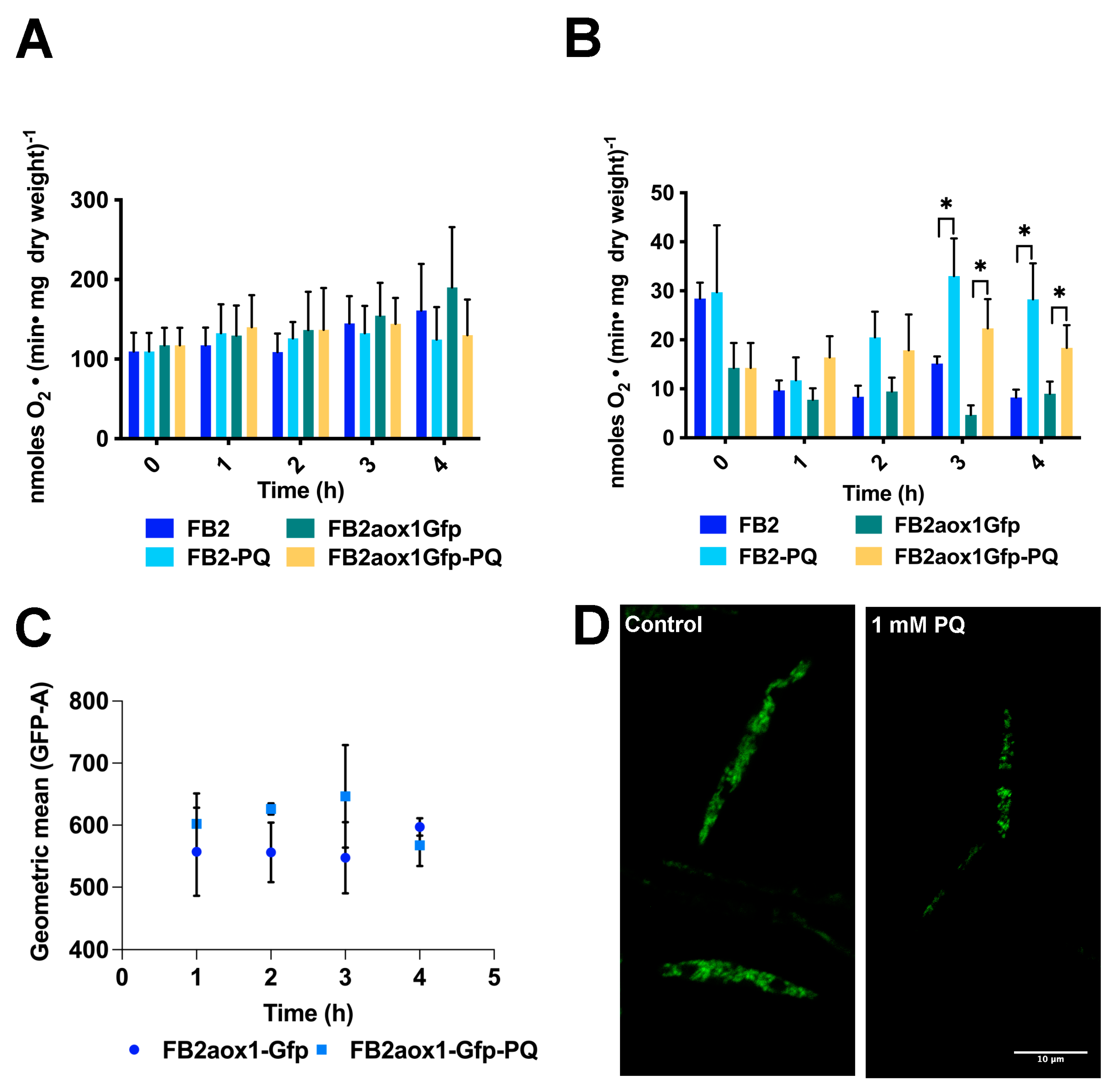
3.2. Paraquat and the Mitochondrial Respiratory Activity
3.3. Paraquat and the Aox1 Activity
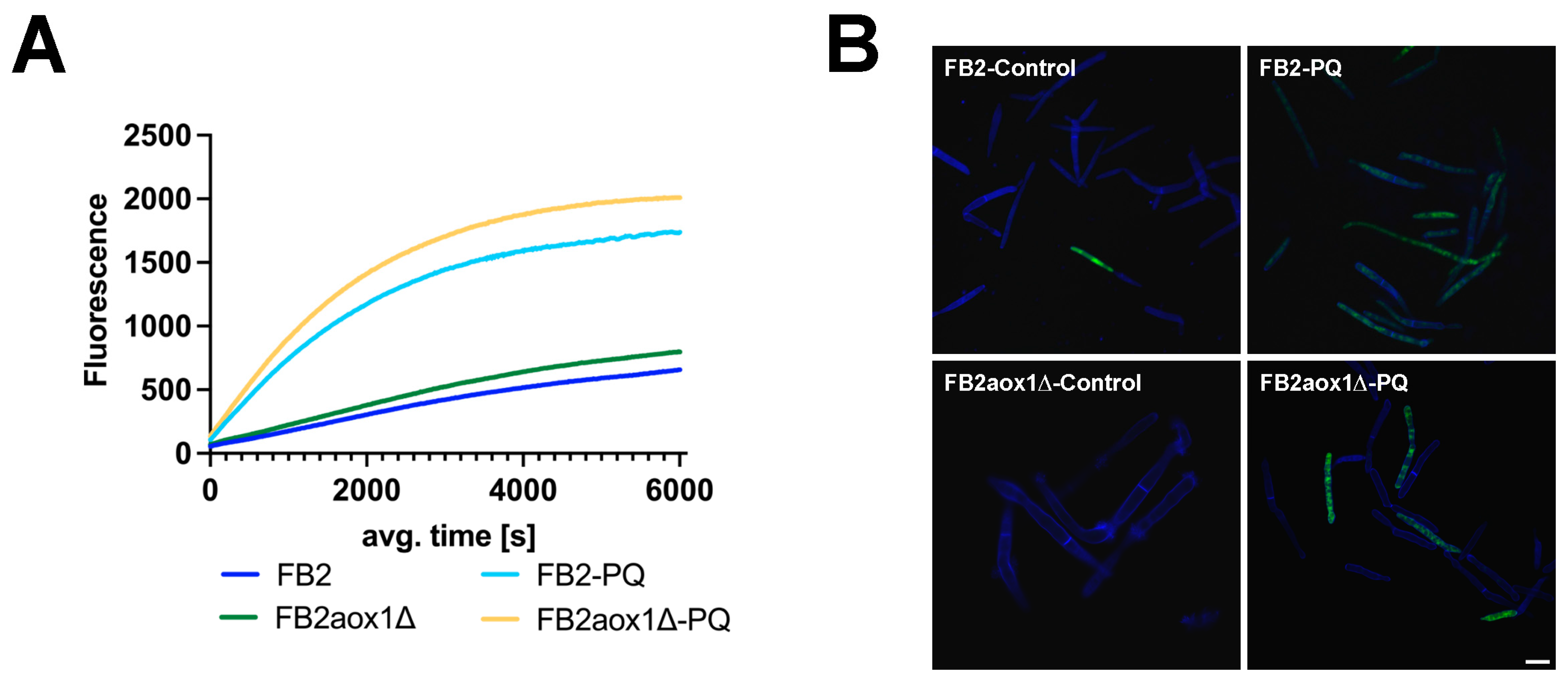
3.4. Production of H2O2 in Cells Treated with Paraquat
3.5. Effect of Paraquat on the Activity of Antioxidant Enzymes
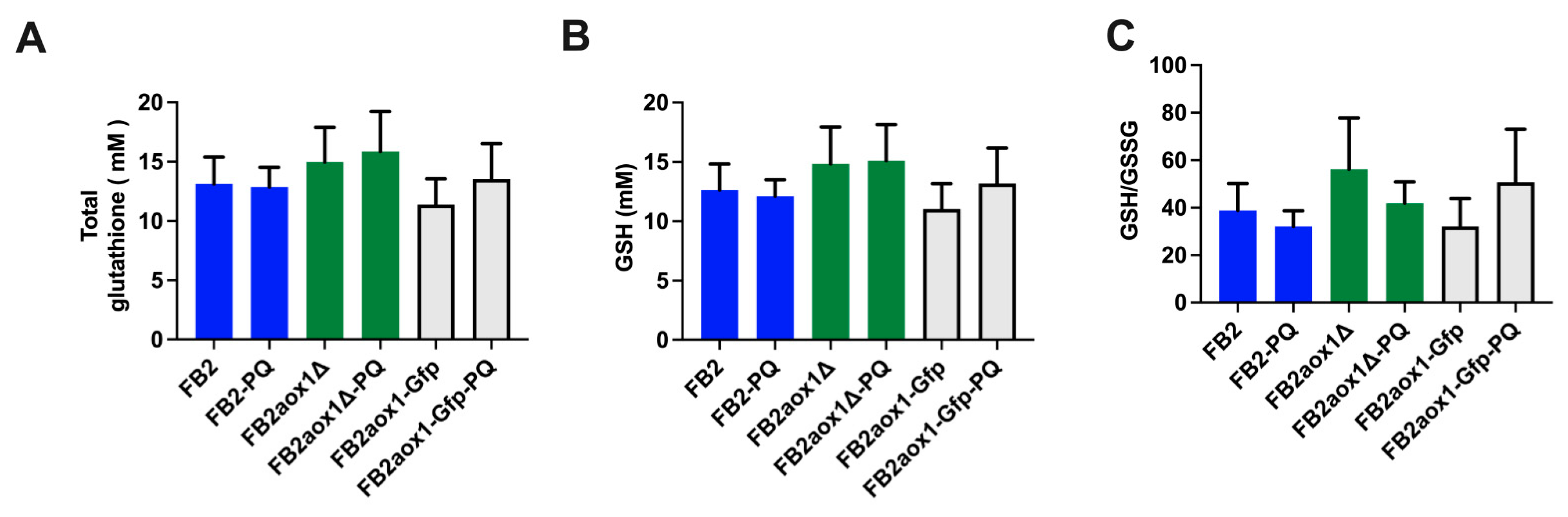
3.6. Glutathione Content in Cells Incubated in the Presence of Paraquat
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Steinberg, G.; Perez-Martin, J. Ustilago maydis, A New Fungal Model System for Cell Biology. Trends Cell Biol. 2008, 18, 61–67. [Google Scholar] [CrossRef]
- Brefort, T.; Doehlemann, G.; Mendoza-Mendoza, A.; Reissmann, S.; Djamei, A.; Kahmann, R. Ustilago maydis as a Pathogen. Annu. Rev. Phytopathol. 2009, 47, 423–445. [Google Scholar] [CrossRef]
- Ferris, A.C.; Walbot, V. Understanding Ustilago maydis Infection of Multiple Maize Organs. J. Fungi 2021, 7, 8. [Google Scholar] [CrossRef]
- Juárez, O.; Guerra, G.; Martínez, F.; Pardo, J.P. The Mitochondrial Respiratory Chain of Ustilago maydis. Biochim. Biophys. Acta 2004, 1658, 244–251. [Google Scholar] [CrossRef][Green Version]
- Juarez, O.; Guerra, G.; Velazquez, I.; Flores-Herrera, O.; Rivera-Perez, R.E.; Pardo, J.P. The Physiologic Role of Alternative Oxidase in Ustilago maydis. FEBS J. 2006, 273, 4603–4615. [Google Scholar] [CrossRef]
- Sierra-Campos, E.; Velázquez, I.; Matuz-Mares, D.; Villavicencio-Queijeiro, A.; Pardo, J.P. Functional Properties of the Ustilago maydis Alternative Oxidase under Oxidative Stress Conditions. Mitochondrion 2009, 9, 96–102. [Google Scholar] [CrossRef]
- Cárdenas-Monroy, C.A.; Pohlmann, T.; Piñón-Zárate, G.; Matus-Ortega, G.; Guerra, G.; Feldbrügge, M.; Pardo, J.P. The Mitochondrial Alternative Oxidase Aox1 Is Needed to Cope with Respiratory Stress but Dispensable for Pathogenic Development in Ustilago maydis. PLoS ONE 2017, 12, e0173389. [Google Scholar] [CrossRef]
- Saavedra, E.; Ramos-Casillas, L.E.; Marín-Hernández, A.; Moreno-Sánchez, R.; Guerra-Sánchez, G. Glycolysis in Ustilago maydis. FEMS Yeast Res. 2008, 8, 1313–1323. [Google Scholar] [CrossRef]
- Lee, J.; Hilgers, F.; Loeschke, A.; Jaeger, K.-E.; Feldbrügge, M. Ustilago maydis Serves as a Novel Production Host for the Synthesis of Plant and Fungal Sesquiterpenoids. Front. Microbiol. 2020, 11, 1655. [Google Scholar] [CrossRef]
- Aguilar, L.R.; Pardo, J.P.; Lomeli, M.M.; Bocardo, O.I.L.; Juarez Oropeza, M.A.; Sanchez, G.G. Lipid Droplets Accumulation and Other Biochemical Changes Induced in the Fungal Pathogen Ustilago maydis under Nitrogen-Starvation. Arch. Microbiol. 2017, 199, 1195–1209. [Google Scholar] [CrossRef]
- Matuz-Mares, D.; Matus-Ortega, G.; Cárdenas-Monroy, C.; Romero-Aguilar, L.; Villalobos-Rocha, J.C.; Vázquez-Meza, H.; Guerra-Sánchez, G.; Peña-Díaz, A.; Pardo, J.P. Expression of Alternative NADH Dehydrogenases (NDH-2) in the Phytopathogenic Fungus Ustilago maydis. FEBS Open Bio 2018, 8, 1267–1279. [Google Scholar] [CrossRef]
- Zhu, Y.; Lu, J.; Wang, J.; Chen, F.; Leng, F.; Li, H. Regulation of Thermogenesis in Plants: The Interaction of Alternative Oxidase and Plant Uncoupling Mitochondrial Protein. J. Integr. Plant Biol. 2011, 53, 7–13. [Google Scholar] [CrossRef]
- Liu, M.; Guo, X. A Novel and Stress Adaptive Alternative Oxidase Derived from Alternative Splicing of Duplicated Exon in Oyster Crassostrea Virginica. Sci. Rep. 2017, 7, 10785. [Google Scholar] [CrossRef]
- Chaudhuri, M.; Ott, R.D.; Hill, G.C. Trypanosome Alternative Oxidase: From Molecule to Function. Trends Parasitol. 2006, 22, 484–491. [Google Scholar] [CrossRef]
- Smith, C.A.; Melino, V.J.; Sweetman, C.; Soole, K.L. Manipulation of Alternative Oxidase Can Influence Salt Tolerance in Arabidopsis Thaliana. Physiol. Plant. 2009, 137, 459–472. [Google Scholar] [CrossRef]
- Vishwakarma, A.; Tetali, S.D.; Selinski, J.; Scheibe, R.; Padmasree, K. Importance of the Alternative Oxidase (AOX) Pathway in Regulating Cellular Redox and ROS Homeostasis to Optimize Photosynthesis during Restriction of the Cytochrome Oxidase Pathway in Arabidopsis Thaliana. Ann. Bot. 2015, 116, 555–569. [Google Scholar] [CrossRef]
- Saha, B.; Borovskii, G.; Panda, S.K. Alternative Oxidase and Plant Stress Tolerance. Plant Signal. Behav. 2016, 11, e1256530. [Google Scholar] [CrossRef]
- Avila-Adame, C.; Köller, W. Disruption of the Alternative Oxidase Gene in Magnaporthe Grisea and Its Impact on Host Infection. Mol. Plant-Microbe Interact. 2002, 15, 493–500. [Google Scholar] [CrossRef][Green Version]
- Marin-Felix, Y.; Guarro, J.; Cano-Lira, J.F.; García, D.; Miller, A.N.; Stchigel, A.M. Melanospora (Sordariomycetes, Ascomycota) and Its Relatives. MycoKeys 2018, 44, 81–122. [Google Scholar] [CrossRef]
- Mendoza, H.; Culver, C.D.; Lamb, E.A.; Schroeder, L.A.; Khanal, S.; Müller, C.; Schirawski, J.; Perlin, M.H. Identification and Functional Characterization of a Putative Alternative Oxidase (Aox) in Sporisorium Reilianum f. Sp. Zeae. J. Fungi 2022, 8, 148. [Google Scholar] [CrossRef]
- Mitsopoulos, P.; Suntres, Z.E. Cytotoxicity and Gene Array Analysis of Alveolar Epithelial A549 Cells Exposed to Paraquat. Chem. Biol. Interact. 2010, 188, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Bora, S.; Vardhan, G.S.H.; Deka, N.; Khataniar, L.; Gogoi, D.; Baruah, A. Paraquat Exposure over Generation Affects Lifespan and Reproduction through Mitochondrial Disruption in C. Elegans. Toxicology 2021, 447, 152632. [Google Scholar] [CrossRef] [PubMed]
- Brunharo, C.A.C.G.; Hanson, B.D. Vacuolar Sequestration of Paraquat Is Involved in the Resistance Mechanism in Lolium perenne L. Spp. Multiflorum. Front. Plant Sci. 2017, 8, 1485. [Google Scholar] [CrossRef]
- Cochemé, H.M.; Murphy, M.P. Complex I Is the Major Site of Mitochondrial Superoxide Production by Paraquat. J. Biol. Chem. 2008, 283, 1786–1798. [Google Scholar] [CrossRef]
- Castello, P.R.; Drechsel, D.A.; Patel, M. Mitochondria Are a Major Source of Paraquat-Induced Reactive Oxygen Species Production in the Brain. J. Biol. Chem. 2007, 282, 14186–14193. [Google Scholar] [CrossRef] [PubMed]
- Reczek, C.R.; Birsoy, K.; Kong, H.; Martínez-Reyes, I.; Wang, T.; Gao, P.; Sabatini, D.M.; Chandel, N.S. A CRISPR Screen Identifies a Pathway Required for Paraquat-Induced Cell Death. Nat. Chem. Biol. 2017, 13, 1274–1279. [Google Scholar] [CrossRef]
- de Oliveira, M.V.; de Freitas Oliveira, A.C.; Shida, C.S.; de Oliveira, R.C.; Nunes, L.R. Gene Expression Modulation by Paraquat-Induced Oxidative Stress Conditions in Paracoccidioides Brasiliensis. Fungal Genet. Biol. 2013, 60, 101–109. [Google Scholar] [CrossRef]
- Martins, V.P.; Dinamarco, T.M.; Soriani, F.M.; Tudella, V.G.; Oliveira, S.C.; Goldman, G.H.; Curti, C.; Uyemura, S.A. Involvement of an Alternative Oxidase in Oxidative Stress and Mycelium-to-Yeast Differentiation in Paracoccidioides Brasiliensis. Eukaryot Cell 2011, 10, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Kretschmer, M.; Lambie, S.; Croll, D.; Kronstad, J.W. Acetate Provokes Mitochondrial Stress and Cell Death in Ustilago maydis. Mol. Microbiol. 2018, 107, 488–507. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Measurement of Glutathione Reductase Activity—Mannervik—1999—Current Protocols in Toxicology—Wiley Online Library. Available online: https://currentprotocols.onlinelibrary.wiley.com/doi/full/10.1002/0471140856.tx0702s00 (accessed on 4 October 2022).
- Chapter 15—Antioxidant Properties of Wheat Bran against Oxidative Stress|Elsevier Enhanced Reader. Available online: https://reader.elsevier.com/reader/sd/pii/B9780124017160000155?token=F1058F34E11B5310325CADE2C5C810E4446DE7FFD38C67A639F66627287117F136A4C5FBF8235DF49168ED0331823631&originRegion=us-east-1&originCreation=20221004213325 (accessed on 4 October 2022).
- Crouch, R.K.; Gandy, S.E.; Kimsey, G.; Galbraith, R.A.; Galbraith, G.M.P.; Buse, M.G. The Inhibition of Islet Superoxide Dismutase by Diabetogenic Drugs. Diabetes 1981, 30, 235–241. [Google Scholar] [CrossRef]
- Rahman, I.; Kode, A.; Biswas, S.K. Assay for Quantitative Determination of Glutathione and Glutathione Disulfide Levels Using Enzymatic Recycling Method. Nat. Protoc. 2006, 1, 3159–3165. [Google Scholar] [CrossRef] [PubMed]
- Akerboom, T.P.; Sies, H. Assay of Glutathione, Glutathione Disulfide, and Glutathione Mixed Disulfides in Biological Samples. Methods Enzymol. 1981, 77, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.P.; Heck, D.E.; Mishin, V.; Smith, P.J.S.; Hong, J.-Y.; Thiruchelvam, M.; Cory-Slechta, D.A.; Laskin, D.L.; Laskin, J.D. Paraquat Increases Cyanide-Insensitive Respiration in Murine Lung Epithelial Cells by Activating an NAD(P)H: Paraquat Oxidoreductase: IDENTIFICATION OF THE ENZYME AS THIOREDOXIN REDUCTASE. J. Biol. Chem. 2007, 282, 7939–7949. [Google Scholar] [CrossRef] [PubMed]
- Hassan, H.M.; Fridovich, I. Paraquat and Escherichia coli. Mechanism of Production of Extracellular Superoxide Radical. J. Biol. Chem. 1979, 254, 10846–10852. [Google Scholar] [CrossRef]
- Matsubara, M.; Yamagami, K.; Kitazawa, Y.; Kawamoto, K.; Tanaka, T. Paraquat Causes S-Phase Arrest of Rat Liver and Lung Cells in Vivo. Arch. Toxicol. 1996, 70, 514–518. [Google Scholar] [CrossRef] [PubMed]
- Tiên Nguyên-nhu, N.; Knoops, B. Mitochondrial and Cytosolic Expression of Human Peroxiredoxin 5 in Saccharomyces Cerevisiae Protect Yeast Cells from Oxidative Stress Induced by Paraquat. FEBS Lett. 2003, 544, 148–152. [Google Scholar] [CrossRef]
- Li, Y.; Zhong, X.; Ye, J.; Guo, H.; Long, Y. Proteome of Saccharomyces Cerevisiae under Paraquat Stress Regulated by Therapeutic Concentration of Copper Ions. Ecotoxicol. Environ. Saf. 2021, 217, 112245. [Google Scholar] [CrossRef]
- Lopes, M.; Mota, M.; Belo, I. Comparison of Yarrowia Lipolytica and Pichia Pastoris Cellular Response to Different Agents of Oxidative Stress. Appl. Biochem. Biotechnol. 2013, 170, 448–458. [Google Scholar] [CrossRef]
- Garrido-Bazán, V.; Pardo, J.P.; Aguirre, J. DnmA and FisA Mediate Mitochondria and Peroxisome Fission, and Regulate Mitochondrial Function, ROS Production and Development in Aspergillus Nidulans. Front. Microbiol. 2020, 11, 837. [Google Scholar] [CrossRef]
- Lin, C.-H.; Yang, S.L.; Chung, K.-R. Cellular Responses Required for Oxidative Stress Tolerance, Colonization, and Lesion Formation by the Necrotrophic Fungus Alternaria Alternata in Citrus. Curr. Microbiol. 2011, 62, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Wiemer, M.; Osiewacz, H.D. Effect of Paraquat-Induced Oxidative Stress on Gene Expression and Aging of the Filamentous Ascomycete Podospora Anserina. Microb. Cell 2014, 1, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Forman, H.J.; Zhang, H.; Rinna, A. Glutathione: Overview of Its Protective Roles, Measurement, and Biosynthesis. Mol. Aspects Med. 2009, 30, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sipari, N.; Lihavainen, J.; Shapiguzov, A.; Kangasjärvi, J.; Keinänen, M. Primary Metabolite Responses to Oxidative Stress in Early-Senescing and Paraquat Resistant Arabidopsis Thaliana Rcd1 (Radical-Induced Cell Death1). Front. Plant Sci. 2020, 11, 194. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Wu, J.; Jamieson, P.A.; Zhang, C. Alternative Oxidase Is Involved in the Pathogenicity, Development, and Oxygen Stress Response of Botrytis Cinerea. Phytopathology 2019, 109, 1679–1688. [Google Scholar] [CrossRef]
- Wang, J.; Rajakulendran, N.; Amirsadeghi, S.; Vanlerberghe, G.C. Impact of Mitochondrial Alternative Oxidase Expression on the Response of Nicotiana Tabacum to Cold Temperature. Physiol. Plant. 2011, 142, 339–351. [Google Scholar] [CrossRef]
- Ali, S.; Jain, S.K.; Abdulla, M.; Athar, M. Paraquat Induced DNA Damage by Reactive Oxygen Species. IUBMB Life 1996, 39, 63–67. [Google Scholar] [CrossRef]
- Xu, T.; Yao, F.; Liang, W.-S.; Li, Y.-H.; Li, D.-R.; Wang, H.; Wang, Z.-Y. Involvement of Alternative Oxidase in the Regulation of Growth, Development, and Resistance to Oxidative Stress of Sclerotinia Sclerotiorum. J. Microbiol. 2012, 50, 594–602. [Google Scholar] [CrossRef]
- Zhang, X.; Yin, H.; Chen, S.; He, J.; Guo, S. Changes in Antioxidant Enzyme Activity and Transcript Levels of Related Genes in Limonium sinense Kuntze Seedlings under NaCl Stress. J. Chem. 2014, 2014, e749047. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romero-Aguilar, L.; Vázquez-Meza, H.; Guerra-Sánchez, G.; Luqueño-Bocardo, O.I.; Pardo, J.P. The Mitochondrial Alternative Oxidase in Ustilago maydis Is Not Involved in Response to Oxidative Stress Induced by Paraquat. J. Fungi 2022, 8, 1221. https://doi.org/10.3390/jof8111221
Romero-Aguilar L, Vázquez-Meza H, Guerra-Sánchez G, Luqueño-Bocardo OI, Pardo JP. The Mitochondrial Alternative Oxidase in Ustilago maydis Is Not Involved in Response to Oxidative Stress Induced by Paraquat. Journal of Fungi. 2022; 8(11):1221. https://doi.org/10.3390/jof8111221
Chicago/Turabian StyleRomero-Aguilar, Lucero, Héctor Vázquez-Meza, Guadalupe Guerra-Sánchez, Oscar Ivan Luqueño-Bocardo, and Juan Pablo Pardo. 2022. "The Mitochondrial Alternative Oxidase in Ustilago maydis Is Not Involved in Response to Oxidative Stress Induced by Paraquat" Journal of Fungi 8, no. 11: 1221. https://doi.org/10.3390/jof8111221
APA StyleRomero-Aguilar, L., Vázquez-Meza, H., Guerra-Sánchez, G., Luqueño-Bocardo, O. I., & Pardo, J. P. (2022). The Mitochondrial Alternative Oxidase in Ustilago maydis Is Not Involved in Response to Oxidative Stress Induced by Paraquat. Journal of Fungi, 8(11), 1221. https://doi.org/10.3390/jof8111221




