Abstract
Ectomycorrhiza (ECM) function has been well studied; however, there is little detailed information regarding the establishment of ECM symbioses. We investigated the morphological and transcriptional changes that occur during the establishment of the Pinus massoniana–Suillus bovinus ECM. S. bovinus promoted the growth of P. massoniana via the release of volatile organic compounds and exudates during the pre-symbiotic stage. Exudate-induced effects showed host plant specificity. At seven days post-inoculation (dpi), the mycelium started to penetrate P. massoniana roots. At 28 dpi, the Hartig net and mantle formed. At the pre-symbiotic stage, most differentially expressed genes in P. massoniana roots were mapped to the biosynthesis of secondary metabolites, signal transduction, and carbohydrate metabolism. At the symbiotic stage, S. bovinus colonization induced the reprogramming of pathways involved in genetic information processing in P. massoniana, particularly at the Hartig net and mantle formation stage. Phenylpropanoid biosynthesis was present at all stages and was regulated via S. bovinus colonization. Enzyme inhibitor tests suggested that hydroxycinnamoyl-CoA shikimate/quinate transferase is involved in the development of the Hartig net. Our findings outline the mechanism involved in the P. massoniana–S. bovinus ECM. Further studies are needed to clarify the role of phenylpropanoid biosynthesis in ECM formation.
1. Introduction
An ectomycorrhiza (ECM) is a mutualistic association formed by ECM fungi and tree roots [1,2]. In this symbiotic relationship, ECM fungi improve host growth and fitness by promoting nutrient absorption and enhancing resistance to biotic stresses (such as pests and diseases) and abiotic stresses (such as drought and heavy metals) [3,4]. In exchange, fungi rely on the carbohydrates provided by their plant partners for vegetative growth and fruit body differentiation [5,6,7]. In addition, many ECM fungi form edible fruiting bodies that are of high economic value [8,9].
The development of ECM is a dynamic process, requiring elaborately regulated interactions between plant roots and compatible fungi. First, the host plant and fungus recognize each other by releasing and receiving signals, which induce lateral root formation, fungal spore germination, and mycelial branching. These changes increase the chances of ECM fungal hyphae encountering plant roots [10,11,12,13,14,15,16,17,18]. Next, the mycelium attaches to the root and starts to colonize. The mycelium stretches along the root surface and the intercellular space, and eventually differentiates to form a mantle and Hartig net [1,19]. In general, most ECM associations can be characterized by both mantle and Hartig net structures, and the formation of Hartig net is defined as signs of functional ECM establishment, which is considered functioning in nutrient exchanges between the two partners [20,21]. However, in some ECM associations formed between ascomycetes and broadleaf trees, the mantle may be poorly developed or essentially non-existent [1].
ECM formation is accompanied by changes in gene expression [16,19,22,23,24], involving cell growth and differentiation, signaling, defense, energy production, and other functional genes [21,22,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40]. The expression patterns of these genes follow a complex series of sequential steps [24,27]. Small secretory proteins induced by mycorrhizae, such as MiSSP7 [41,42,43], MiSSP7.6 [44], MiSSP8 [45], and MiSSP10b [46], help fungi evade host defenses and play an important role during the early stages of ECM formation. Genes associated with symbiosis-induced malate synthase [47], arginine methyltransferase [33], endoglucanase (LbGH5-CBM1) [48], polygalacturonase (LbGH28A) [49], and pectin methylesterases [21] are involved during the late stages of ECM development. A phosphorus transporter (HcPT2) [50] and an ammonium transporter (AMT2.2) [51,52] play important roles in maintaining the function of the ECM when functional ECM structures—the mantle and Hartig net—are formed. These findings have greatly contributed to our understanding of the molecular mechanism of ECM symbiosis. However, these previous findings have been limited to a few specific ECM combinations, such as Populus-Laccaria bicolor [21,41], Betula-Paxillus involutus [25,27], Eucalyptus-Pisolithus tinctorius/Pisolithus microcarpus [21,26], and Pinus pinaster-Hebeloma cylindrosporum [32]. Given the huge numbers of fungi and tree species that can form ECM and the complexity of these relationships, the establishment process of different ECM combinations is likely to be specific [53]. Therefore, it is necessary to expand our studies to other ECM combinations to gain a comprehensive understanding of this important symbiotic association. Another problem is that there are little data on the molecular regulation of the ECM symbiotic process because previous studies have mainly focused on specific stages (usually the functional stage) of the ECM formation process.
Pinus massoniana is one of the main timbers and a pioneer afforestation tree species in China; however, the survival and breed of this unique native tree species are highly dependent on ECM fungi [54,55,56,57]. Our group previously reported that Suillus bovinus is the dominant ECM fungus in P. massoniana forests. In addition, S. bovinus produces edible fruit bodies with a high economic value [58]. As well as other species in the genus of Suillus, S. bovinus present a high degree of host specificity towards conifers, and its distribution coincides with the natural distribution of Pinaceae in the Northern Hemisphere [59,60]. Suillus species are also recognized as the earliest colonizers of pine seedlings which play vital roles in conifer invasions [61,62]. Although we have a good understanding of the function of the P. massoniana ECM, the mechanisms involved in the formation of the ECM remain unclear, particularly at the transcriptional level. In this study, we investigated the morphological and transcriptional characteristics of P. massoniana roots when inoculated with S. bovinus. We clarified the stages of the formation process of this ECM association by morphological profiling and then performed transcriptional profiling. Both transcriptional and physiological data suggest that the phenylpropanoid biosynthesis pathway may play important roles in ECM formation.
2. Materials and Methods
2.1. Plant and Fungal Materials
Seeds of P. massoniana (collected from the P. massoniana national base at Maanshan Forest Farm, Duyun City, Guizhou Province, China) were cleaned with 0.01% Tween 20, and then sterilized, first with 0.5% KMnO4 solution for 2 h, and finally with 0.01% Tween 20 and 0.5% carbendazim for 1 h, and then with antibiotics—200 mg/L streptomycin and 100 mg/L gentamicin for 20 min. At the end of each of these steps, the seeds were cleaned with sterile water three times for 5 min each time. The sterilized seeds were then placed in wet vermiculite and incubated in 25 °C climate chambers with 14 h of light (150 μmol m−2s−1) and 10 h of darkness (light 0 μmol m−2s−1) per day. Thirty-day-old P. massoniana seedlings were used in the ECM formation trials.
Seeds of wild-type Arabidopsis thaliana Columbia-0 (kindly provided by Prof. Fuhua Fan) were sterilized with 75% alcohol for 1 min and with 5% NaClO solution for 10 min. At the end of each of these steps, the seeds were cleaned with sterile water three times for 5 min each time. Seeds were then transferred to Murashige and Skoog medium and incubated in 25 °C climate chambers with 14 h of light per day (150 μmol m−2s−1) and 10 h of darkness.
The strain S. bovinus LL-1 was isolated from a fruiting body collected in a P. massoniana forest [52]. The mycelium of S. bovinus was subcultured and maintained on modified Melin-Norkran’s (MMN) [63] medium at 25 °C in the dark. The composition of MMN medium is 25 mg/L NaCl; 250 mg/L (NH4)2HPO4; 500 mg/L KH2PO4; 5 mg/L FeCl3; 50 mg/L CaCl2; 150 mg/L MgSO4·7H2O; 100 mg/L VB1; 10 g/L glucose; 1.00 g/L casamino acids; 5.00 g/L malt; and 10 g/L agar.
2.2. In Vitro Mycorrhizal Formation between P. massoniana and S. bovinus
To investigate the morphological features of the pre-symbiotic phase (before physical contact has been made between the host and the fungus), we carried out two trials to test the effects of volatile organic compounds (VOCs) and exudates released by S. bovinus on P. massoniana growth.
(a) VOC effects: 30 mL of basal medium (DCR) (composed of 400 mg/L NH4NO3; 556 mg/L Ca(NO3)2·4H2O; 370 mg/L MgSO4·7H2O; 85 mg/L CaCl2·2H2O; 170 mg/L KH2PO4; 6.2 mg/L H3BO3; 22.3 mg/L MnSO4·H2O; 8.6 mg/L ZnSO4·7H2O; 0.25 mg/L CuSO4·5H2O; 0.83 mg/L KI; 0.025 mg/L CoCl2·6H2O; 0.025 mg/L LiCl; 0.25 mg/L NaMoO4·2H2O; 27.8 mg/L FeSO4·7H2O; 37.3 mg/L EDTA-2Na; 1.0 mg/L VB1; 0.5 mg/L VB6; 0.5 mg/L nicotinic acid; 2.0 mg/L glycine; 200 mg/L myo-inositol; 10 g/L sucrose; and 10 g/L agar) [64] was poured into 13 cm × 13 cm Petri dishes. Once the medium had solidified, the medium in one half of each Petri dish was cut out with a scalpel, and then 15 mL of MMN medium was poured into the empty half. Once the MMN medium had solidified, a 2 cm × 13 cm strip of medium at the junction of the two media was cut with a scalpel and removed to leave a 2 cm gap between the two 5 cm × 13 cm blocks of media in each dish. The MMN medium was inoculated with an S. bovinus plug (1 cm in diameter) and a P. massoniana seedling was transplanted onto the DCR medium (VOC treatment). Plates that were not inoculated with S. bovinus were considered to be controls (NVOC treatment).
(b) Exudate effects: a P. massoniana seedling was transplanted onto DCR medium (30 mL) in 13 cm × 13 cm Petri dishes. A cellophane membrane was placed over the roots and then covered with a thin layer of MMN medium. The MNM medium was then inoculated with an S. bovinus plug (1 cm in diameter) (Exud treatment). Plates that were not inoculated with S. bovinus were considered to be controls (NExud treatment).
To verify whether the effects of S. bovinus VOCs and exudates are host-specific, we also set up plates to observe the effects of S. bovinus VOCs and exudates on the root growth of the non-host A. thaliana using the same experimental procedures as those described above.
To investigate the morphological features of the symbiotic phase, a P. massoniana seedling was transplanted onto DCR medium (30 mL) in 13 cm × 13 cm Petri dishes and an S. bovinus plug (1 cm in diameter) was placed in direct contact with the P. massoniana taproot (M treatment). P. massoniana taproots that were not inoculated with an S. bovinus plug were considered to be controls (NM treatment). There were 20 replicates of each treatment. The bottom of each Petri dish was wrapped with tinfoil to cover the root growth area before placing the Petri dish vertically in a climate chamber at 25 °C with 14 h of light (light 120 μmol m−2s−1) and 10 h of darkness (light 0 μmol m−2s−1) per day.
2.3. Morphological Observations
Morphological observations at the pre-symbiosis phase were recorded by scanning Petri dishes (Epson Perfection V330 Photo) every 7 days. Root length and branch measurements were obtained using the ImageJ SmartRoot plug-in. However, because it was difficult to observe the number of Arabidopsis root branches in the Petri dish, the number of root branches was only recorded on the day of harvest at 28 days post-inoculation (dpi).
To characterize the morphology at the symbiosis stage, five seedlings were randomly selected for observation every 7 days from inoculation with S. bovinus until ECM formation. First, the contact between hyphae and roots was examined under a stereomicroscope (M205FA, Leica Microsystems, Wetzlar, Germany). Second, transversal cross-sections of at least 20 independent root segments were cleared (5% KOH solution, 90 °C for 2 h) to make them transparent, acidified (1% HCl solution w/v, 10 min at room temperature), and then stained with 0.03% chlorazol black (90 °C for 20 min). The stained sections were then mounted in glycerol and observed under a light microscope (DM3000, Leica Microsystems, Wetzlar, Germany).
2.4. RNA Extraction, Sequencing and Analysis
Given that the exudate-induced effects were host-specific, whereas the VOC-induced effects were not, plants treated with exudates were selected as materials for transcription analysis at the pre-symbiotic stage. Transcriptome profiling was conducted on P. massoniana roots treated with S. bovinus exudates for 14 days and on P. massoniana roots 7 dpi (inoculated M7, uninoculated NM7) and 28 dpi (inoculated M28, uninoculated NM28) with S. bovinus. For each treatment, four biological replicates were collected. The samples were frozen in liquid nitrogen and stored at −80 °C until RNA extraction.
Total RNA was extracted using an RNAprep Pure Plant Kit (TIANGEN, Beijing, China). Samples were first subjected to quality control using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and then sent to Novogene (Beijing, China) for sequencing on the Illumina HiSeqTM2000 platform.
Raw data obtained by sequencing included a small number of reads with a sequencing adapter or that were of low sequencing quality. To ensure the quality and reliability of data analysis, the original data were filtered as follows to remove: (1) reads containing an adapter; (2) reads containing N (N indicates that base information cannot be determined); (3) low-quality reads (a Qphred score of ≤20 bases for more than 50% of the total read length).
2.5. Determination of Differentially Expressed Genes and Enrichment Analysis
Differentially expressed genes (DEGs) were analyzed with DESeq2 [65], and genes with an absolute log2-fold change value of ≥1 and an adjusted p-value of <0.05 were deemed to be differentially expressed. Gene Ontology (GO) enrichment analysis was performed based on the GOseq method [66], and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was performed using KOBAS (2.0) [67].
RNA-seq data for all samples are available at the Sequence Read Archive of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/sra, accessed on 24 October 2022) under accession number PRJNA886481.
2.6. Effect of Acibenzolar Acid on Mycorrhizal Development
Hydroxycinnamoyl-CoA shikimate/quinate transferase (HCT) plays an important role in lignin synthesis; however, acibenzolar acid has an inhibitory effect on this enzyme [68]. We analyzed the effect of acibenzolar acid on HCT activity and mycorrhizal development in P. massoniana.
Acibenzolar acid (CAS: 35272-27-6; Dr. Ehrensorfer GmbH, Augsburg, Germany) dissolved in ethanol and filtered through a 0.22 μm diameter aperture was added to the DCR medium at concentrations of 0, 100, 300, or 500 μM. After four weeks of colonization, mycorrhizal development was examined according to the method described above. The ImageJ platform was used to measure the Hartig net depth and area (the area of fungal hyphae between cells). Five micrographs were measured for each treatment and three measurements were recorded per micrograph.
The HCT activity of P. massoniana roots was analyzed using an HCT ELISA detection kit (JingMei, JiangSu, China; www.jsjmsw.com, accessed on 17 October 2022) according to the manufacturer’s instructions.
2.7. Statistical Analysis
Apart from the DEG analysis data, data were analyzed using SPSS 25.0 software (IBM® SPSS® Statistics). The effects of VOCs and exudates on root growth (root length and number of root branches) were analyzed using one-way analysis of variance (ANOVA). A Student’s t-test was used to determine significant differences between means. ANOVA of P. massoniana HCT activity under different treatments, the Hartig net depth, and area were analyzed by performing a Duncan’s test. The principal component analysis (PCA) of the DEGs was performed using the OmicShare tools (https://www.omicshare.com/tools, accessed on 29 October 2022).
3. Results
3.1. Pre-Symbiotic Interactions between P. massoniana and S. bovinus
VOCs and exudates released by S. bovinus significantly promoted the root length and number of root branches of P. massoniana from the 14th day of treatment compared with control treatments (Figure 1). In addition, VOCs and exudates significantly increased the biomass of pine seedlings by 29.37% and 15.07%, respectively, compared with the NVOC and NExud control treatments (Figure 1a,c).


Figure 1.
Effects of volatile organic compounds (VOCs) and exudates released by Suillus bovinus on the growth of Pinus massoniana. (a) Number of root branches, root length, and fresh weight of P. massoniana seedlings treated with VOCs and without VOCs (NVOC), n = 15; (b) P. massoniana seedlings subjected to VOC (28 d) and NVOC (28 d) treatments, scale bars = 2 cm; (c) root branches, root length, and fresh weight of P. massoniana seedlings subjected to the exudate (Exud) (28 d) and non-exudate (NExud) (28 d) treatments, n = 20; (d) P. massoniana seedlings subjected to Exud (28 d) and NExud (28 d) treatments, scale bars = 2 and 3 cm. ** p < 0.01; * p < 0.05. Bars represent mean values ± the SE. Different letters above bars indicate significant differences between treatments.
However, S. bovinus VOCs and exudates had the opposite effects on A. thaliana growth. Although S. bovinus VOCs promoted A. thaliana root branching, they inhibited the elongation of the taproot (Figure 2a,b), and S. bovinus exudates inhibited both the shoot and root growth of A. thaliana (Figure 2c,d).

Figure 2.
Effects of VOCs and exudates released by S. bovinus on the growth of Arabidopsis thaliana. The effect of VOCs on: (a) A. thaliana root branch number, n = 15; (b) A. thaliana plants, scale bars = 2 cm. Effects of S. bovinus exudates on: (c) A. thaliana root branch number, n = 20; (d) A. thaliana plants, scale bars = 2 and 3 cm. Bars represent mean values ± the SE. Different letters above bars indicate significant differences between treatments.
3.2. Symbiotic Interactions between P. massoniana Roots and S. bovinus over Time
At 7 dpi, the mycelium of S. bovinus had proliferated on the surface of P. massoniana roots and started to invade the intercellular space (the invasion stage) (Figure 3a,b). By 14 dpi, more hyphae had aggregated around the root surface and grown into inter-radical spaces (Figure 3c,d). By 21 dpi, hyphae had covered the root surface to form a mantle-like structure (Figure 3e), and hyphae had penetrated the root intercellularly to form a Hartig net structure (a Hartig net was considered to have formed when hyphae had invaded the root system and completely wrapped one to two layers of cortical cells, Figure 3f). A functional mycorrhiza had established by 28 dpi, at which point the Hartig net and mantle (a mantle was considered to have formed when more than four layers of hyphae were tightly wrapped around a root; Figure 3i) were fully developed, and the root tips showed dichotomous branching and were swollen in shape (mantle and Hartig net formation stage) (Figure 3g,h).
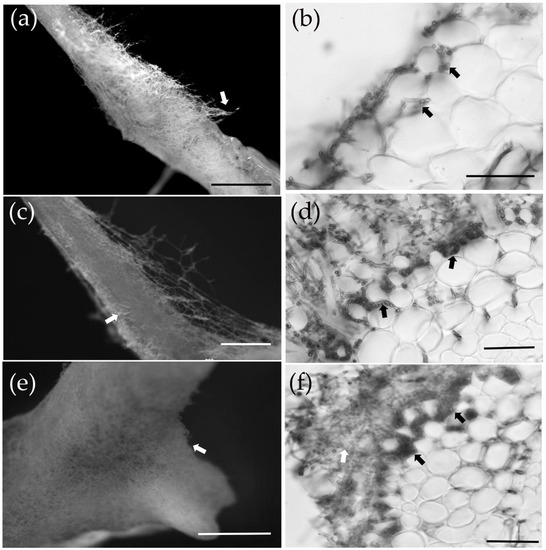
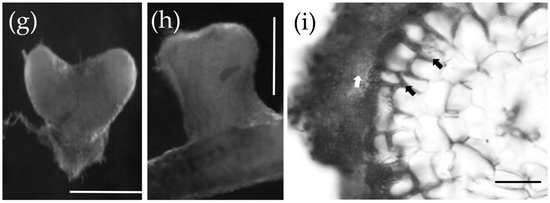
Figure 3.
The ectomycorrhizal (ECM) formation process between P. massoniana and S. bovinus. (a,b) At 7 days post-inoculation (dpi): scale bars = 5 mm and 500 µm, respectively. The white arrow indicates hyphae wrapped around the root system; black arrows indicate the intercellular mycelium. (c,d) At 14 dpi: scale bars = 1 mm and 100 µm, respectively. The white arrow indicates hyphae wrapped around the root system; black arrows indicate the developing Hartig net. (e,f) At 21 dpi: scale bars = 1 mm and 50 µm, respectively. The white arrow in (e) indicates hyphae wrapped around the root system. The white arrow in (f) indicates the developing mantle and black arrows indicate the Hartig net. (g–i) At 28 dpi: scale bars = 1 mm, 500 μm, and 50 μm, respectively. (g) Dichotomous branching hypha, (h) swollen hypha. (i) Cross-section of the root: the white arrow indicates the mantle and black arrows indicate the Hartig net.
3.3. Root Transcriptional Analysis during Symbiotic Interactions between P. massoniana and S. bovinus
3.3.1. Quality Analysis
Transcriptome sequencing analysis of all P. massoniana root samples resulted in mean raw reads of 40,419,484 and 35,330,493 for the Exud and NExud treatments, respectively; 70,190,257 and 68,570,981 for the M7 and NM7 treatments, respectively; and 41,427,706 and 35,802,254 for the M28 and NM28 treatments, respectively. By removing reads with adapters and low-quality reads from raw reads, mean clean reads of 38,892,360 and 34,386,741 were obtained for the Exud and NExud treatments, 68,939,003 and 67,174,335 for M7 and NM7 treatments, respectively, and 40,267,781 and 34,747,055 for the M28 and NM28 treatments, respectively. The Q20 and Q30 of all samples were above 97% and 92%, respectively, and the GC content was also at a normal level (Table S1).
3.3.2. Analysis of DEGs
In total, across the three colonization time points there were 35,297 DEGs due to S. bovinus colonization. The lowest number of DEGs was detected during the pre-symbiotic phase, when the plant and fungus were separated physically by a cellophane membrane. There were nearly equal numbers of upregulated and downregulated genes at this time point (Figure 4a). The largest number of DEGs was found at the invasion stage (7 dpi), followed by the Hartig net and mantle formation stage (28 dpi). In addition, 11 DEGs were co-expressed at the pre-symbiotic stage and the invasion stage, while at the symbiosis stage, 34 DEGs were co-expressed at the invasion and the Hartig net and mantle formation stage (Figure 4b). PCA showed that DEGs in samples from inoculated and non-inoculated treatments at the three stages of symbiosis were significantly separated (Figure 4c).

Figure 4.
Number of differentially expressed genes (DEGs). (a) Pie charts showing the number of significantly upregulated and downregulated DEGs at each timepoint (p < 0.05); (b) Venn diagram showing the number of DEGs that are common to more than one timepoint and the number of genes that are unique to individual timepoints; (c) principle component analysis (PCA) of DEGs detected in the Exud vs. NExud treatment, 7 dpi with S. bovinus (M7) vs. the uninoculated (NM7) treatment, and 28 dpi with S. bovinus (M28) vs. the uninoculated (NM28) treatment.
3.3.3. Enrichment Analysis of DEGs
GO was used to functionally classify DEGs during P. massoniana–S. bovinus ECM formation. Here, we focused on the enrichment of DEGs in the biological process category. We found 693 DEGs at the pre-symbiotic stage, 2493 at 7 dpi, and 6636 at 28 dpi (Figure 5a). At the pre-symbiotic stage, the top two terms were “cellular protein modification process” and “carbohydrate metabolic process”; at 7 dpi, the top two terms were “oxidation–reduction process” and “multi-organism process”; and at 28 dpi, the top two terms were “biosynthetic process” and “transport”. In addition, we found that DEGs associated with cell walls were significantly enriched during all three stages.
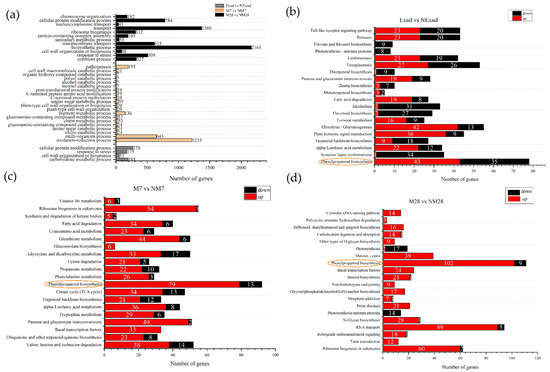
Figure 5.
Enrichment of DEGs. (a) Enriched Gene Ontology biological processes. (b–d) Kyoto Encyclopedia of Genes and Genome enrichment for Exud vs. NExud, M7 vs. NM7, and M28 vs. NM28. The numbers shown on the red and black bars indicate the number of upregulated and downregulated DEGs, respectively. The phenylpropanoid biosynthesis pathway (circled) was common to all three symbiosis stages.
These DEGs were further analyzed using KEGG to fully explore their functions. At the pre-symbiotic stage, most DEGs were mapped to the biosynthesis of other secondary metabolites, carbohydrate and lipid metabolism, signal transduction, and the immune system, e.g., phenylpropanoid biosynthesis (78; ko00940), glycolysis/gluconeogenesis (55; ko00010), plant hormone signal transduction (45; ko04075), the Toll-like receptor signaling pathway (43; ko04620), and alpha-linolenic acid metabolism (34; ko00592), and the number of upregulated and downregulated DEGs was similar (Figure 5b). At 7 days of symbiosis, a number of DEGs were mapped to the biosynthesis of secondary metabolites, genetic information processing, amino acids, and carbohydrate metabolism, e.g., phenylpropanoid biosynthesis (92), ribosome biogenesis in eukaryotes (55; ko03008), valine, leucine, and isoleucine degradation (52; ko00280), pentose and glucuronate interconversions (51; ko00040) and glyoxylate and dicarboxylate metabolism (50; ko00630), and most of these DEGs were upregulated (Figure 5c). At 28 days of symbiosis, functional ECM had formed. Most DEGs were involved in the biosynthesis of other secondary metabolites, genetic information processing, and glycan biosynthesis and metabolism, e.g., phenylpropanoid biosynthesis (111), RNA transport (94; ko03013), ribosome biogenesis in eukaryotes (62), N-glycan biosynthesis (29; ko00510) and basal transcription factors (24; ko03022), and most of these DEGs were upregulated (Figure 5d). Phenylpropanoid biosynthesis was a common pathway in ECM development processes in P. massoniana and S. bovinus (Figure 5b–d). Furthermore, the pathway with the most DEGs at each stage was the phenylpropanoid biosynthesis pathway. The number of DEGs associated with the phenylpropanoid biosynthesis pathway increased over time and their expression patterns differed at different stages of symbiosis. The phenylpropanoid biosynthesis pathway exhibited a mixture of both positive and negative gene regulation during the pre-symbiotic stage. In the invasion and functional stages, most genes involved were generally upregulated. Moreover, as the ECM formation process progressed, the number of upregulated DEGs in this pathway gradually increased, while the number of downregulated DEGs decreased.
3.3.4. Genes Involved in the Phenylpropanoid Biosynthesis Pathway throughout Colonization
In this study, phenylpropanoid biosynthesis was found to be a common pathway and to have the largest number of DEGs that were shared at different stages of the symbiotic process during the formation of the ECM between P. massoniana and S. bovinus. We focused our attention on the lignin synthesis process of this pathway. The genes encoding phenylalanine ammonia-lyase (PAL) and cinnamyl alcohol dehydrogenase (CAD), which are involved in lignin synthesis, were significantly expressed during all three phases of ECM formation. We also found that genes involved in PAL synthesis had similar expression patterns to those of genes encoding CAD during the pre-symbiotic and mantle and Hartig net formation stages. Prior to physical contact, PAL genes (Cluster-21925.46012, Cluster-21925.78638, Cluster-21925.44510, Cluster-21925.71279) and the CAD gene (Cluster-21925.8529) were downregulated as a result of the S. bovinus exudate treatment (Figure 6a). However, at the Hartig net and mantle formation stage, PAL (Cluster-21925.21162) and CAD genes (Cluster-9078.0) were upregulated (Figure 6c). At the invasion stage, the expression patterns of the genes encoding PAL were mixed and the CAD gene was upregulated (Cluster-20437.23132, Cluster-20437.24246, Cluster-20437.24240, Cluster-20437.24241) (Figure 6b). Furthermore, the genes encoding HCT were not significantly expressed during the pre-symbiotic stage; however, at the symbiotic stage, they were generally upregulated; coumarate 3-hydroxylase (C3H) also showed the same expression pattern (Figure 6, Table S2).
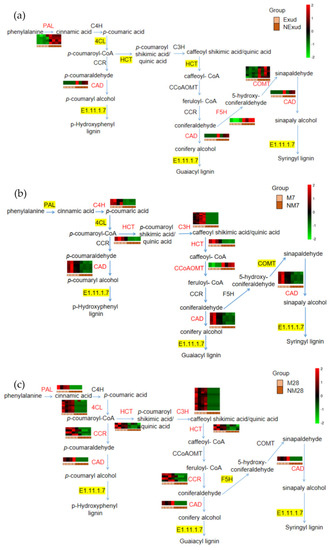
Figure 6.
Phenylpropanoid biosynthesis pathway with heatmaps showing fold changes in the expression of genes encoding key enzymes in this pathway. (a) Exud vs. NExud; (b) M7 vs. NM7; (c) M28 vs. NM28. Key enzymes encoded by DEGs that were annotated in metabolic pathways are shown in red. An enzyme highlighted in yellow indicates that both upregulated and downregulated genes encode the enzyme. An enzyme that is not highlighted in yellow indicates that no genes encoding the enzyme were annotated. Enzymes: PAL, phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate:CoA ligase; HCT, hydroxycinnamoyl-CoA shikimate/quinate transferase; C3H, coumarate 3-hydroxylase; CCoAOMT, caffeoyl-CoA-O-methyltransferase; CCR, cinnamoyl CoA reductase; F5H, ferulate 5-hydroxylase; COMT, caffeic acid O-methyltransferase; CAD, cinnamyl alcohol dehydrogenase.
3.4. Effect of Inhibiting HCT Activity on ECM Morphogenesis
HCT enzyme activity was significantly inhibited by the presence of 300 μM acibenzolar acid (Figure S1), and the development of Hartig nets was further promoted. Following the 300 μM acibenzolar acid treatment, hyphae penetrated the intercellular spaces of the third layer of cortical cells (Figure 7c,d), layers 1–2 at 100 μM (Figure 7b), and layers 2–3 at 500 μM (Figure 7e,f). In contrast, in the absence of acibenzolar acid, mycelia only encased the first layer of cortical cells, and most of the mycelia spread intercellularly between epidermal cells to form a labyrinthine structure (Figure 7a). A significantly deeper Hartig net developed following the 300 μM treatment (70.4 ± 4.43 μm; p < 0.05), compared with the 500 μM (26.33 ± 1.86 μm), 100 μM (25.49 ± 1.16 μm) or 0 μM (23.08 ± 3.12 μm) acibenzolar acid treatments (Figure 8a). The presence of acibenzolar acid also affected the Hartig net area intercellular spaces (500 μM > 100 μM > 300 μM > 0 μM); however, these data were not statistically significant (Figure 8b).

Figure 7.
Effects of acibenzolar acid treatment on ECM development. Transverse cross-sections of a P. massoniana lateral root colonized by S. bovinus: (a) without the addition of acibenzolar acid; (b) in the presence of 100 μM acibenzolar acid; (c,d) 300 μM acibenzolar acid; and (e,f) 500 μM acibenzolar acid. Scale bars = 50 µm. Black arrows and ellipses indicate Hartig nets, white arrows indicate the mantle. Abbreviations: CC, cortical cells; EC, epidermal cells.

Figure 8.
Effects of acibenzolar acid treatment on Hartig net development. (a) Hartig net depth; (b) Hartig net area. Bars represent means ± the SE, n = 5; different letters above bars indicate significant differences between treatments at p < 0.05.
4. Discussion
An ECM association between P. massoniana and S. bovinus is typical in forests of south China. In this study, we focused on the morphological and transcriptional changes that occurred over time during the formation of this ECM symbiotic association. Our findings showed that during the formation of the ECM between P. massoniana and S. bovinus, the symbiotic morphogenesis was coordinated with root transcriptional adjustments.
The establishment of a mycorrhizal association between P. massoniana and S. bovinus can be divided into two stages: the pre-symbiotic stage (signal recognition before physical contact) and the symbiosis stage. Studies have shown that before physical contact, mycorrhizal fungi can affect host root development by releasing signaling molecules (including VOCs and exudates) to promote, for example, root elongation and branching [13,14,15,69,70,71], so as to increase the contact opportunities between fungi and host roots [1]. We also found that both VOCs and exudates released by S. bovinus stimulated P. massoniana growth and root branching. However, the exudate-induced effects showed host plant specificity, whereas VOC-induced effects did not. Previous studies have also reported that ECM fungal volatiles can promote A. thaliana root growth [13,15,69,70,71]. Compared with exudates, VOCs have long-distance diffusion features. We speculate that ECM fungi stimulate plant root growth from a long distance, and when ECM fungi and plant roots come within a certain distance of each other, exudate-induced effects further help ECM fungi to discriminate potential host plants from non-host plants.
Fungi and hosts enter the symbiotic stage after successful mutual recognition. The mycelium gathered around the root surface develops into a mantle, and the intraradical mycelium invades the intercellular space among cortical cells to form a Hartig net [1,72,73]. In this study, we found that at 7 dpi, mycelium began to invade the intercellular space among root cortical cells, and by 28 dpi, the mantle and Hartig net had developed and matured. The time required for ECM formation differs in different studies, ranging from four days to two weeks, or up to one month [26,74,75,76,77,78]. These differences may be due to the species of ECM fungus involved and the host specificity. Differences in the time required for ECM formation have also been observed between strains, even when they are forming an ECM with the same host. Yu (2007) [78] observed that different strains of Cortinarius sp. and Picea koraiensis formed ECM at different rates, ranging from 21 days to more than one month. Moreover, the experimental system and culture environment may also affect the time needed for mycorrhizal formation. Opinions about the development sequence of the Hartig net and mantle also vary. Some studies have reported that the Hartig net forms before the mantle [53,79,80,81], while others have suggested the opposite [26,69,74,82,83]. These different conclusions may be due to the specificities of the ECM formation process between distinct ECM fungal species and host plant combinations (particularly gymnosperms and angiosperms). In this study, the Hartig net and mantle developed synchronously during the ECM formation between P. massoniana and S. bovinus; however, the Hartig net formed earlier than the mantle.
ECM formation is a dynamic process, and all morphological changes involve coordinated changes in gene expression [22,25,28,29,30,36]. Our data showed that most DEGs were present at the invasion stage (7 dpi), indicating that P. massoniana seedlings underwent a greater range of transcriptional reprogramming at the beginning of the symbiotic stage in response to S. bovinus infection. Moreover, there were no common DEGs at the three stages of symbiosis, suggesting that there may be a unique set of molecular mechanisms supporting the formation of a symbiotic association between P. massoniana and S. bovinus. Using GO enrichment analysis, we focused on those biological processes that were most significantly enriched and found that some DEGs at each stage were significantly enriched in categories relating to cell structure. Given that the invasion of ECM fungi can cause a significant loosening of plant root cells [84,85], these DEGs may play an important role in ECM symbiosis. KEGG enrichment was used to further analyze the function of DEGs. During the pre-symbiotic stage, the main task of the host and fungus is to recognize each other by releasing signals [16,17,18,19], which include many secondary metabolites and plant hormones [11,12,13,15,76,84]. In this study, we found that during this stage a large number of DEGs were associated with the biosynthesis of secondary metabolites and signal transduction. Successful colonization by fungi often induces the breakdown of host carbohydrate to meet the autogenous growth needs [1]. Such a reprogramming of carbohydrate metabolism induced by S. bovinus may be initiated prior to physical contact because a large proportion of DEGs were associated with carbohydrate metabolism during the pre-symbiotic stage. These metabolic pathway changes lay the foundation for successful colonization and symbiosis later on. At the infection phase (7 dpi), in addition to DEGs involved in the biosynthesis of secondary metabolites and carbohydrate metabolism, a large number of DEGs were associated with genetic information processing and amino acid metabolism, and most of these DEGs were upregulated. Amino acids are important sinks for carbon assimilation [86], particularly aliphatic amino acids such as valine, leucine, and isoleucine. Thus, genes involved in the degradation of these amino acids may provide carbon sources for S. bovinus. Once a functional ECM was established (28 dpi), the DEGs were mainly associated with RNA transport, ribosome biogenesis in eukaryotes, basal transcription factors, and the cytosolic DNA-sensing pathway, which may further facilitate substance exchanges between the two symbionts [76].
Phenylpropanoid biosynthesis was a common pathway during the development of this ECM symbiosis. Related DEGs showed a mixture of both up-and downregulated expression patterns at the pre-symbiotic stage; however, related DEGs were upregulated at 7 dpi and 28 dpi. Different studies have reported different findings regarding the expression patterns of DEGs involved in this pathway. During the early stages of ECM colonization, Weiss et al. [87] and Plett et al. [88] reported that phenylpropanoids were increased; however, Hill et al. [77] suggested that this pathway was downregulated during the invasion phase and upregulated during the functional symbiosis phase. Some studies have shown that the colonization of ECM could induce phenylpropanoid metabolism in hosts and hinder the colonization process [89,90]. For example, certain metabolites produced by this pathway can limit hyphal penetration and the formation of the Hartig net [90]. However, Behr et al. [91] found that transgenic lines with downregulated genes in the phenylpropanoid pathway all showed lower colonization rates compared with the wild type, indicating that metabolite production in this pathway favors mycorrhizal formation. The reason for these two different conclusions may be that colonization by different ECM fungi results in different end products being produced by the host phenylpropane biosynthesis pathway.
HCT is a rate-limiting enzyme of phenylpropanoid biosynthesis and is important for the products synthesized by the phenylpropane biosynthesis pathway [92]. According to our data, at both 7 and 28 dpi, the colonization of S. bovinus significantly induced the expression of HCT genes in P. massoniana roots. To gain more information about the function of HCT, we investigated the effects of the activity of HCT on ECM formation between S. bovinus and P. massoniana by using a specific enzyme inhibitor: acibenzolar acid [68]. Acibenzolar acid has only weak inhibitory effects on HCT activity, and a significant inhibitory effect could only be found with 300 μM acibenzolar acid. The use of acibenzolar acid had little influence on the symbiotic process but deepened the degree of mycelial infection. Under the 300 μM treatment, the depth of the Hartig net increased significantly, which may be because acibenzolar acid enlarges the intercellular spaces among cortical cells, which facilitates the infection of mycelium.
5. Conclusions
We investigated the characteristics of morphological and transcriptional changes during the establishment of the ECM symbiosis between P. massoniana and S. bovinus. ECM formation between P. massoniana roots and S. bovinus can be divided into two stages: the pre-symbiotic stage and the symbiotic stage (Figure 9). During the pre-symbiotic stage, VOCs and/or exudates released by S. bovinus induced host root growth. At 7 dpi, the mycelia invaded the intercellular space of the root cortex, and at 28 dpi, the functional ECM established with the mantle and Hartig net fully developed. Meanwhile, the biosynthesis of secondary metabolites, signal transduction, genetic information processing, and carbohydrate and lipid metabolism in P. massoniana roots changed in response to colonization by S. bovinus. The phenylpropanoid biosynthesis pathway was common to all three stages during the development of this ECM symbiosis, and the activity of a key enzyme-HCT-was related to the formation of the Hartig net. These findings highlight the need for a comprehensive investigation of the roles of the phenylpropanoid biosynthesis pathway in ECM formation, and perhaps also in ECM functions.
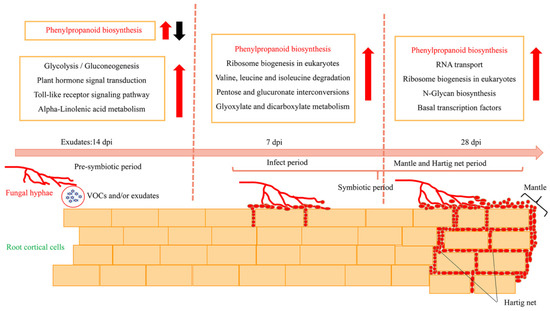
Figure 9.
Schematic diagram showing morphological and transcriptional changes in a P. massoniana root during the formation of an ECM symbiosis with S. bovinus. Red arrows indicate upregulation; black arrow indicates downregulation. The phenylpropanoid biosynthesis pathway is common to all three ECM stages.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8111162/s1. Figure S1: Effect of acibenzolar acid on the HCT enzyme activity of P. massoniana; Table S1: Quality analysis of transcriptome data; Table S2: HCT-and C3H-related genes at three stages of symbiosis.
Author Contributions
Conceptualization, X.S. and G.D.; methodology, W.F. and X.S.; formal analysis, W.F.; writing—original draft preparation, W.F.; writing—review and editing, W.F.; X.S. and G.D.; funding acquisition, X.S. and G.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (31971572 and 31500090) (from X.S.), the Science and Technology Project of Guizhou Province, China [2018]5261 (from X.S.), and the Cultivation Project of Guizhou University [2020]47 (from G.D.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original data presented in this study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis, 3rd ed.; Academic Press: London, UK, 2008; pp. 192–268. [Google Scholar]
- Becquer, A.; Guerrero-Galán, C.; Eibensteiner, J.L.; Houdinet, G.; Bücking, H.; Zimmermann, S.D.; Garcia, K. The ectomycorrhizal contribution to tree nutrition. In Advances in Botanical Research; Cánovas, F.M., Ed.; Academic Press: London, UK, 2019; Volume 89, pp. 77–126. [Google Scholar]
- Nehls, U.; Plassard, C. Nitrogen and phosphate metabolism in ectomycorrhizas. New Phytol. 2018, 220, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Zak, D.R.; Pellitier, P.T.; Argiroff, W.; Castillo, B.; James, T.Y.; Nave, L.E.; Averill, C.; Beidler, K.V.; Bhatnagar, J.; Blesh, J.; et al. Exploring the role of ectomycorrhizal fungi in soil carbon dynamics. New Phytol. 2019, 223, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Nehls, U.; Grunze, N.; Willmann, M.; Reich, M.; Kuster, H. Sugar for my honey: Carbohydrate partitioning in ectomycorrhizal symbiosis. Phytochemistry 2007, 68, 82–91. [Google Scholar] [CrossRef]
- Casieri, L.; Ait Lahmidi, N.; Doidy, J.; Veneault-Fourrey, C.; Migeon, A.; Bonneau, L.; Courty, P.E.; Garcia, K.; Charbonnier, M.; Delteil, A.; et al. Biotrophic transportome in mutualistic plant–fungal interactions. Mycorrhiza 2013, 23, 597–625. [Google Scholar] [CrossRef]
- Lindahl, B.D.; Tunlid, A. Ectomycorrhizal fungi–potential organic matter decomposers, yet not saprotrophs. New Phytol. 2015, 205, 1443–1447. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yu, F.Q.; Zhang, C.; Liu, C.; Yang, M.; Li, S. Edible ectomycorrhizal fungi and their cultivation in China. In Mushrooms, Humans and Nature in a Changing World; Pérez-Moreno, J., Guerin-Laguette, A., Flores Arzú, R., Yu, F.Q., Eds.; Springer: Cham, Germany, 2020; pp. 31–60. [Google Scholar]
- Zhao, Y.Z.; Sun, X.G.; Feng, J.W.; Guo, Q.Q. Diversity of edible mycorrhizal fungi in Pinus massoniana forests of central Guizhou, southwest China. Mycosystema 2021, 40, 108–123. (In Chinese) [Google Scholar]
- Horan, D.P.; Chilvers, G.A. Chemotropism-the key to ectomycorrhizal formation? New Phytol. 1990, 116, 297–301. [Google Scholar] [CrossRef]
- Lagrange, H.; Jay-Allemand, C.; Lapeyrie, F. Rutin, the phenolglycoside from eucalyptus root exudates, stimulates Pisolithus hyphal growth at picomolar concentrations. New Phytol. 2001, 149, 349–355. [Google Scholar] [CrossRef]
- Kikuchi, K.; Matsushita, N.; Suzuki, K.; Hogetsu, T. Flavonoids induce germination of basidiospores of the ectomycorrhizal fungus Suillus bovinus. Mycorrhiza 2007, 17, 563–570. [Google Scholar] [CrossRef]
- Splivallo, R.; Fischer, U.; Göbel, C.; Feussner, I.; Karlovsky, P. Truffles regulate plant root morphogenesis via the production of auxin and ethylene. Plant Physiol. 2009, 150, 2018–2029. [Google Scholar] [CrossRef]
- Krause, K.; Henke, C.; Asiimwe, T.; Ulbricht, A.; Klemmer, S.; Schachtschabel, D.; Boland, W.; Kothe, E. Biosynthesis and secretion of indole-3-acetic acid and its morphological effects on Tricholoma vaccinum–spruce ectomycorrhiza. App. Environ. Microb. 2015, 81, 7003–7011. [Google Scholar] [CrossRef] [PubMed]
- Ditengou, F.A.; Müller, A.; Rosenkranz, M.; Felten, J.; Lasok, H.; van Doorn, M.M.; Legué, V.; Palme, K.; Schnitzler, J.P.; Polle, A. Volatile signalling by sesquiterpenes from ectomycorrhizal fungi reprogrammes root architecture. Nat. Commun. 2015, 6, 6729. [Google Scholar] [CrossRef] [PubMed]
- Garcia, K.; Delaux, P.M.; Cope, K.R.; Ané, J.M. Molecular signals required for the establishment and maintenance of ectomycorrhizal symbioses. New Phytol. 2015, 208, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Daguerre, Y.; Plett, J.M.; Veneault-Fourrey, C. Signaling pathways driving the development of ectomycorrhizal symbiosis. In Molecular Mycorrhizal Symbiosis; Martin, F., Ed.; John Wiley & Sons, Inc.: Hoboken, UK, 2016; pp. 141–157. [Google Scholar]
- Feng, W.Y.; Sun, X.G.; Ding, G.J. Advance of ectomycorrhizal signal recognition mechanism in pre-symbiosis. Plant Physiol. J. 2021, 57, 749–758. (In Chinese) [Google Scholar]
- Martin, F.; Duplessis, S.; Ditengou, F.; Lagrange, H.; Voiblet, C.; Lapeyrie, F. Developmental cross talking in the ectomycorrhizal symbiosis: Signals and communication genes. New Phytol. 2001, 151, 145–154. [Google Scholar] [CrossRef]
- Plett, J.M.; Tisserant, E.; Brun, A.; Morin, E.; Grigoriev, I.V.; Kuo, A.; Martin, F.; Kohler, A. The mutualist Laccaria bicolor expresses a core gene regulon during the coloniza-tion of diverse host plants and a variable regulon to counteract host-specific defenses. Mol. Plant Microbe Interact. 2015, 28, 261–273. [Google Scholar] [CrossRef]
- Chowdhury, J.; Kemppainen, M.; Delhomme, N.; Shutava, I.; Zhou, J.J.; Takahashi, J.; Pardo, A.G.; Lundberg-Felten, J. Laccaria bicolor pectin methylesterases are involved in ectomycorrhiza development with Populus tremula × Populus tremuloides. New Phytol. 2022, 236, 639–655. [Google Scholar] [CrossRef]
- Voiblet, C.; Duplessis, S.; Encelot, N.; Martin, F. Identification of symbiosis-regulated genes in Eucalyptus globulus–Pisolithus tinctorius ectomycorrhiza by differential hybridization of arrayed cDNAs. Plant J. 2001, 25, 181–191. [Google Scholar] [CrossRef]
- Hogekamp, C.; Küster, H. A roadmap of cell-type specific gene expression during sequential stages of the arbuscular mycorrhiza symbiosis. BMC Genom. 2013, 14, 306. [Google Scholar] [CrossRef]
- Larsen, P.E.; Sreedasyam, A.; Trivedi, G.; Desai, S.; Dai, Y.; Cseke, L.J.; Collart, F.R. Multi-omics approach identifies molecular mechanisms of plant-fungus mycorrhizal interaction. Front. Plant Sci. 2016, 6, 1061. [Google Scholar] [CrossRef]
- Johansson, T.; Le Quéré, A.; Ahren, D.; Söderström, B.; Erlandsson, R.; Lundeberg, J.; Uhlén, M.; Tunlid, A. Transcriptional responses of Paxillus involutus and Betula pendula during formation of ectomycorrhizal root tissue. Mol. Plant Microbe. Interact. 2004, 17, 202–215. [Google Scholar] [CrossRef] [PubMed]
- Duplessis, S.; Courty, P.E.; Tagu, D.; Martin, F. Transcript patterns associated with ectomycorrhiza development in Eucalyptus globulus and Pisolithus microcarpus. New Phytol. 2005, 165, 599–611. [Google Scholar] [CrossRef]
- Le Quéré, A.; Wright, D.P.; Söderström, B.; Tunlid, A.; Johansson, T. Global patterns of gene regulation associated with the development of ectomycorrhiza between birch (Betula pendula Roth.) and Paxillus involutus (Batsch) Fr. Mol. Plant Microbe. Interact. 2005, 18, 659–673. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Da Silva Coelho, I.; de Queiroz, M.V.; Costa, M.D.; Kasuya, M.C.M.; de Araújo, E.F. Identification of differentially expressed genes of the fungus Hydnangium sp. during the pre-symbiotic phase of the ectomycorrhizal association with Eucalyptus grandis. Mycorrhiza 2010, 20, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Flores-Monterroso, A.; Canales, J.; de la Torre, F.; Ávila, C.; Cánovas, F.M. Identification of genes differentially expressed in ectomycorrhizal roots during the Pinus pinaster–Laccaria bicolor interaction. Planta 2013, 237, 1637–1650. [Google Scholar] [CrossRef] [PubMed]
- Sebastiana, M.; Vieira, B.; Lino-Neto, T.; Monteiro, F.; Figueiredo, A.; Sousa, L.; Salomé Pais, M.; Tavares, R.; Paulo, O.S. Oak root response to ectomycorrhizal symbiosis establishment: RNA-Seq derived transcript identification and expression profiling. PLoS ONE 2014, 9, e98376. [Google Scholar]
- Li, H.; Peng, H.; Wang, L.; Wei, H.; Li, N.; Jing, Q. Identification of fungal genes involved in the preinfection events between ectomycorrhizal association (Pisolithus tinctorius and Pinus massoniana). Mycol. Prog. 2014, 13, 123–130. [Google Scholar] [CrossRef]
- Doré, J.; Perraud, M.; Dieryckx, C.; Kohler, A.; Morin, E.; Henrissat, B.; Lindquist, E.; Zimmermann, S.D.; Girard, V.; Kuo, A.; et al. Comparative genomics, proteomics and transcriptomics give new insight into the exoproteome of the basidiomycete Hebeloma cylindrosporum and its involvement in ectomycorrhizal symbiosis. New Phytol. 2015, 208, 1169–1187. [Google Scholar] [CrossRef]
- Plett, K.L.; Raposo, A.E.; Anderson, I.C.; Piller, S.C.; Plett, J.M. Protein arginine methyltransferase expression affects ectomycorrhizal symbiosis and the regulation of hormone signaling pathways. Mol. Plant Microbe Interact. 2019, 32, 1291–1302. [Google Scholar] [CrossRef]
- Bouffaud, M.L.; Herrmann, S.; Tarkka, M.T.; Bönn, M.; Feldhahn, L.; Buscot, F. Oak displays common local but specific distant gene regulation responses to different mycorrhizal fungi. BMC Genom. 2020, 21, 399. [Google Scholar] [CrossRef]
- Tang, N.; Lebreton, A.; Xu, W.; Dai, Y.; Yu, F.; Martin, F.M. Transcriptome profiling reveals differential gene expression of secreted proteases and highly specific gene repertoires involved in Lactarius–Pinus symbioses. Front. Plant Sci. 2021, 12, 1775. [Google Scholar] [CrossRef] [PubMed]
- Doré, J.; Kohler, A.; Dubost, A.; Hundley, H.; Singan, V.; Peng, Y.; Kuo, A.; Grigoriev, I.V.; Martin, F.; Marmeisse, R.; et al. The ectomycorrhizal basidiomycete Hebeloma cylindrosporum undergoes early waves of transcriptional reprogramming prior to symbiotic structures differentiation. Environ. Microbiol. 2017, 19, 1338–1354. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.L.; Chen, Y.; Vilgalys, R. Metatranscriptomic study of common and host-specific patterns of gene expression between pines and their symbiotic ectomycorrhizal fungi in the genus Suillus. PLoS Genet. 2016, 12, e1006348. [Google Scholar] [CrossRef]
- Heller, G.; Adomas, A.; Li, G.; Osborne, J.; Zyl, L.V.; Sederoff, R.; Finlay, R.D.; Stenlid, J.; Asiegbu, F.O. Transcriptional analysis of Pinus sylvestris roots challenged with the ectomycorrhizal fungus Laccaria bicolor. BMC Plant Biol. 2008, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Veneault-Fourrey, C.; Commun, C.; Kohler, A.; Morin, E.; Balestrini, R.; Plett, J.; Danchin, E.; Coutinho, P.; Wiebenga, A.; de Vries, R.P.; et al. Genomic and transcriptomic analysis of Laccaria bicolor CAZome reveals insights into polysaccharides remodelling during symbiosis establishment. Fungal Genet Biol. 2014, 72, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Sebastiana, M.; Figueiredo, A.; Acioli, B.; Sousa, L.; Pessoa, F.; Baldé, A.; Pais, M.S. Identification of plant genes involved on the initial contact between ectomycorrhizal symbionts (Castanea sativa–European chestnut and Pisolithus tinctorius). Eur. J. Soil Biol. 2009, 45, 275–282. [Google Scholar] [CrossRef]
- Plett, J.M.; Kemppainen, M.; Kale, S.D.; Kohler, A.; Legué, V.; Brun, A.; Tyler, B.M.; Pardo, A.G.; Martin, F. A secreted effector protein of Laccaria bicolor is required for symbiosis development. Curr. Biol. 2011, 21, 1197–1203. [Google Scholar] [CrossRef]
- Plett, J.M.; Daguerre, Y.; Wittulsky, S.; Vayssieres, A.; Deveau, A.; Melton, S.J.; Brun, A.; Veneault-Fourrey, C.; Martin, F. Effector MiSSP7 of the mutualistic fungus Laccaria bicolor stabilizes the Populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proc. Natl. Acad. Sci. USA 2014, 111, 8299–8304. [Google Scholar] [CrossRef]
- Daguerre, Y.; Basso, V.; Hartmann-Wittulski, S.; Schellenberger, R.; Meyer, L.; Bailly, J.; Kohler, A.; Plett, J.M.; Martin, F.; Veneault-Fourrey, C. The mutualism effector MiSSP7 of Laccaria bicolor alters the interactions between the Poplar JAZ6 protein and its associated proteins. Sci. Rep. 2020, 10, 20362. [Google Scholar] [CrossRef]
- Kang, H.; Chen, X.; Kemppainen, M.; Pardo, A.G.; Veneault-Fourrey, C.; Kohler, A.; Kohler, A.; Martin, F.M. The small secreted effector protein MiSSP7.6 of Laccaria bicolor is required for the establishment of ectomycorrhizal symbiosis. Environ. Microbiol. 2020, 22, 1435–1446. [Google Scholar] [CrossRef]
- Pellegrin, C.; Daguerre, Y.; Ruytinx, J.; Guinet, F.; Kemppainen, M.; Frey, N.F.D.; Puech-Pagès, V.; Hecker, A.; Pardo, A.; Martin, F.; et al. Laccaria bicolor MiSSP8 is a small-secreted protein decisive for the establishment of the ectomycorrhizal symbiosis. Environ. Microbiol. 2019, 21, 3765–3779. [Google Scholar] [CrossRef] [PubMed]
- Plett, J.M.; Plett, K.L.; Wong-Bajracharya, J.; Freitas Pereira, M.; Costa, M.D.; Kohler, A.; Martin, F.; Anderson, I.C. Mycorrhizal effector PaMiSSP10b alters polyamine biosynthesis in Eucalyptus root cells and promotes root colonization. New Phytol. 2020, 228, 712–727. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Kim, S.J.; Podila, G.K. Differential expression of a malate synthase gene during the preinfection stage of symbiosis in the ectomycorrhizal fungus Laccaria bicolor. New Phytol. 2002, 154, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Anasontzis, G.E.; Labourel, A.; Champion, C.; Haon, M.; Kemppainen, M.; Commun, C.; Deveau, A.; Pardo, A.; Veneault-Fourrey, C.; et al. The ectomycorrhizal basidiomycete Laccaria bicolor releases a secreted β-1,4 endoglucanase that plays a key role in symbiosis development. New Phytol. 2018, 220, 1309–1321. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Labourel, A.; Haon, M.; Kemppainen, M.; Machado, E.D.S.; Brouilly, N.; Veneault-Fourrey, C.; Kohler, A.; Rosso, M.N.; Pardo, A.; et al. The ectomycorrhizal basidiomycete Laccaria bicolor releases a GH28 polygalacturonase that plays a key role in symbiosis establishment. New Phytol. 2022, 233, 2534–2547. [Google Scholar] [CrossRef]
- Becquer, A.; Garcia, K.; Amenc, L.; Rivard, C.; Doré, J.; Trives-Segura, C.; Szponarski, W.; Russet, S.; Baeza, Y.; Lassalle-Kaiser, B.; et al. The Hebeloma cylindrosporum HcPT2 Pi transporter plays a key role in ectomycorrhizal symbiosis. New Phytol. 2018, 220, 1185–1199. [Google Scholar] [CrossRef]
- Lucic, E.; Fourrey, C.; Kohler, A.; Martin, F.; Chalot, M.; Brun-Jacob, A. A gene repertoire for nitrogen transporters in Laccaria bicolor. New Phytol. 2008, 180, 343–364. [Google Scholar] [CrossRef]
- Martin, F.; Aerts, A.; Ahrén, D.; Brun, A.; Danchin, E.G.J.; Duchaussoy, F.; Gibon, J.; Kohler, A.; Lindquist, E.; Pereda, V.; et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature 2008, 452, 88–92. [Google Scholar] [CrossRef]
- Tranvan, H.; Habricot, Y.; Jeannette, E.; Gay, G.; Sotta, B. Dynamics of symbiotic establishment between an IAA-overproducing mutant of the ectomycorrhizal fungus Hebeloma cylindrosporum and Pinus pinaster. Tree Physiol. 2000, 20, 123–129. [Google Scholar] [CrossRef][Green Version]
- Chen, L.Q. Studies on symbiotic mycorrhiza fungi with Masson pine. For. Res. 1989, 2, 357–362. (In Chinese) [Google Scholar]
- Zhou, Z.X. Chinese Masson Pine; China Forestry Press: Beijing, China, 2001. [Google Scholar]
- Ding, G.; Zhou, Z.; Wang, Z. Cultivation and Utilization of Pulpwood Stand for Pinus massoniana; China Forestry Publishing House: Beijing, China, 2006. [Google Scholar]
- Wu, X.Q.; Sun, M.Q. Mycorrhizal formation between seven ectomycorrhizal fungi and seedlings of three pines species. Acta Ecol. Sin. 2006, 26, 4186–4191. (In Chinese) [Google Scholar]
- Sun, X.G.; Feng, W.Y.; Li, M.; Shi, J.; Ding, G.J. Phenology and cultivation of Suillus bovinus, an edible mycorrhizal fungus, in a Pinus massoniana plantation. Can. J. For. Res. 2019, 48, 960–968. [Google Scholar] [CrossRef]
- Ruiz-Díez, B.; Rincón, A.M.; De Felipe, M.R.; Fernández-Pascual, M. Molecular characterization and evaluation of mycorrhizal capacity of Suillus isolates from Central Spain for the selection of fungal inoculants. Mycorrhiza 2006, 16, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Murata, H.; Yamada, A.; Yokota, S.; Maruyama, T.; Shimokawa, T.; Neda, H. Innate traits of Pinaceae-specific ectomycorrhizal symbiont Suillus luteus that differentially associates with arbuscular mycorrhizal broad-leaved trees in vitro. Mycoscience 2015, 56, 606–611. [Google Scholar] [CrossRef]
- Peay, K.G.; Schubert, M.G.; Nguyen, N.H.; Bruns, T.D. Measuring ectomycorrhizal fungal dispersal: Macroecological patterns driven by microscopic propagules. Mol. Ecol. 2012, 21, 4122–4136. [Google Scholar] [CrossRef]
- Rudawska, M.; Wilgan, R.; Janowski, D.; Iwański, M.; Leski, T. Shifts in taxonomical and functional structure of ectomycor-rhizal fungal community of Scots pine (Pinus sylvestris L.) underpinned by partner tree ageing. Pedobiologia 2018, 71, 20–30. [Google Scholar] [CrossRef]
- Marx, D.H. The influence of ectotrophic fungi on the resistance of pine root to pathogenic infections. I. Antagonism of mycorrhizal fungi to root pathogenic fungi and soil bacteria. Phytopathology 1969, 59, 153–163. [Google Scholar]
- Gupta, P.K.; Durzan, D.J. Shoot multiplication from mature trees of Douglas-fir (Pseudotsuga menziesii) and sugar pine (Pinus lambertiana). Plant Cell Rep. 1985, 4, 177–179. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Negrel, J.; Klinguer, A.; Adrian, M. In vitro inhibition of shikimate hydroxycinnamoyltransferase by acibenzolar acid, the first metabolite of the plant defence inducer acibenzolar-S-methyl. Plant Physiol. Biochem. 2021, 163, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Felten, J.; Kohler, A.; Morin, E.; Bhalerao, R.P.; Palme, K.; Martin, F.; Ditengou, F.A.; Legué, V. The ectomycorrhizal fungus Laccaria bicolor stimulates lateral root formation in poplar and Arabidopsis through auxin transport and signaling. Plant Physiol. 2009, 151, 1991–2005. [Google Scholar] [CrossRef]
- Felten, J.; Legué, V.; Anicet Ditengou, F. Lateral root stimulation in the early interaction between Arabidopsis thaliana and the ectomycorrhizal fungus Laccaria bicolor: Is fungal auxin the trigger? Plant Signal Behav. 2010, 5, 864–867. [Google Scholar] [CrossRef]
- Sitrit, Y.; Roth-Bejerano, N.; Kagan-Zur, V.; Turgeman, T. Pre-symbiotic interactions between the desert truffle Terfezia boudieri and its host plant Helianthemum sessiliflorum. In Desert Truffles; Kagan-Zur, V., Roth-Bejerano, N., Sitrit, Y., Morte, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 81–92. [Google Scholar]
- Balestrini, R.; Kottke, I. Structure and development of ectomycorrhizal roots. In Molecular Mycorrhizal Symbiosis; Martin, F., Ed.; John Wiley & Sons, Inc.: New York, NY, USA, 2016; pp. 47–61. [Google Scholar]
- Martin, F.; Kohler, A.; Murat, C.; Veneault-Fourrey, C.; Hibbett, D.S. Unearthing the roots of ectomycorrhizal symbioses. Nat. Rev. Microbiol. 2016, 14, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Horan, D.P.; Chilvers, G.A.; Lapeyrie, F.F. Time sequence of the infection process eucalypt ectomycorrhizas. New Phytol. 1988, 109, 451–458. [Google Scholar] [CrossRef]
- Burgess, T.; Dell, B.; Malajczuk, N. In vitro synthesis of Pisolithus-Eucalyptus ectomycorrhizae: Synchronization of lateral tip emergence and ectomycorrhizal development. Mycorrhiza 1996, 6, 189–196. [Google Scholar] [CrossRef]
- Plett, J.M.; Khachane, A.; Ouassou, M.; Sundberg, B.; Kohler, A.; Martin, F. Ethylene and jasmonic acid act as negative modulators during mutualistic symbiosis between Laccaria bicolor and Populus roots. New Phytol. 2014, 202, 270–286. [Google Scholar] [CrossRef]
- Hill, R.A.; Wong-Bajracharya, J.; Anwar, S.; Coles, D.; Wang, M.; Lipzen, A.; Ng, V.; Grigoriev, I.V.; Martin, F.; Anderson, I.C.; et al. Abscisic acid supports colonization of Eucalyptus grandis roots by the mutualistic ectomycorrhizal fungus Pisolithus microcarpus. New Phytol. 2022, 233, 966–982. [Google Scholar] [CrossRef]
- Yu, J.Y. Infecting Micromechanism of Ectomycorrhizal Fungi to Picea koraiensis. Master’s Thesis, Northeast Forestry University, Harbin, China, 2007. [Google Scholar]
- Nylund, J.; Unestam, T. Structure and physiology of ectomycorrhizae. I. The process of mycorrhiza formation in Norway spruce in vitro. New Phytol. 1982, 91, 63–79. [Google Scholar] [CrossRef]
- Melville, L.H.; Massicotte, H.B.; Peterson, R.L. Ontogeny of early stages of ectomycorrhizae synthesized between Dryas integrifolia and Hebeloma cylindrosporum. Bot. Gaz. 1987, 148, 332–341. [Google Scholar] [CrossRef]
- Brunner, I. Comparative studies on ectomycorrhizae synthesized with various in vitro techniques using Picea abies and two Hebeloma species. Trees 1991, 5, 90–94. [Google Scholar] [CrossRef]
- Fusconi, A. The development of the fungal sheath on Cistus incanus short roots. Can. J. Bot. 1983, 61, 2546–2553. [Google Scholar] [CrossRef]
- Grellier, B.; Letouze, R.; Strullu, D.G. Micropropagation of birch and mycorrhizal formation in vitro. New Phytol. 1984, 97, 591–599. [Google Scholar] [CrossRef]
- Gay, G.; Normand, L.; Marmeisse, R.; Sotta, B.; Debaud, J.C. Auxin overproducer mutants of Hebeloma cylindrosporum Romagnesi have increased mycorrhizal activity. New Phytol. 1994, 128, 645–657. [Google Scholar] [CrossRef]
- Balestrini, R.; Hahn, M.G.; Bonfante, P. Location of cell-wall components in ectomycorrhizae of Corylus avellana and Tuber magnatum. Protoplasma 1996, 191, 55–69. [Google Scholar] [CrossRef]
- Martin, F.; Ramstedt, M.; Söderhäll, K.; Canet, D. Carbohydrate and amino acid metabolism in the ectomycorrhizal ascomycete Sphaerosporella brunnea during glucose utilization: A 13C NMR study. Plant Physiol. 1988, 86, 935–940. [Google Scholar] [CrossRef]
- Weiss, M.; Mikolajewski, S.; Peipp, H.; Schmitt, U.; Schmidt, J.; Wray, V.; Strack, D. Tissue-specific and development-dependent accumulation of phenylpropanoids in larch mycorrhizas. Plant Physiol. 1997, 114, 15–27. [Google Scholar] [CrossRef]
- Plett, K.L.; Buckley, S.; Plett, J.M.; Anderson, I.C.; Lundberg-Felten, J.; Jämtgård, S. Novel microdialysis technique reveals a dramatic shift in metabolite secretion during the early stages of the interaction between the ectomycorrhizal fungus Pisolithus microcarpus and its host Eucalyptus grandis. Microorganisms 2021, 9, 1817. [Google Scholar] [CrossRef]
- Weiss, M.; Schmidt, J.; Neumann, D.; Wray, V.; Christ, R.; Strack, D. Phenylpropanoids in mycorrhizas of the Pinaceae. Planta 1999, 208, 491–502. [Google Scholar] [CrossRef]
- Feugey, L.; Strullu, D.G.; Poupard, P.; Simoneau, P. Induced defence responses limit Hartig net formation in ectomycorrhizal birch roots. New Phytol. 1999, 144, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Behr, M.; Baldacci-Cresp, F.; Kohler, A.; Morreel, K.; Goeminne, G.; Van Acker, R.; Veneault-Fourrey, C.; Mol, A.; Pilate, G.; Boerjan, W.; et al. Alterations in the phenylpropanoid pathway affect poplar ability for ectomycorrhizal colonisation and susceptibility to root-knot nematodes. Mycorrhiza 2020, 30, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, L.; Besseau, S.; Geoffroy, P.; Ritzenthaler, C.; Meyer, D.; Lapierre, C.; Pollet, B.; Legrand, M. Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 2004, 16, 1446–1465. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).