Metabolic Profiling and Metabolite Correlation Network Analysis Reveal That Fusarium solani Induces Differential Metabolic Responses in Lotus japonicus and Lotus tenuis against Severe Phosphate Starvation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material, Inoculation, Growth Conditions and Sampling
2.2. In Vitro Cultivation of Fusarium solani 142L52B and Sampling of Mycelia and Secreted Compounds
2.3. Metabolite Extraction and Profiling of Plant Tissues, Mycelia and Fungal Supernatant
2.4. Statistical Analysis of Metabolite Data
2.5. Network Analysis of the Metabolite Data
3. Results
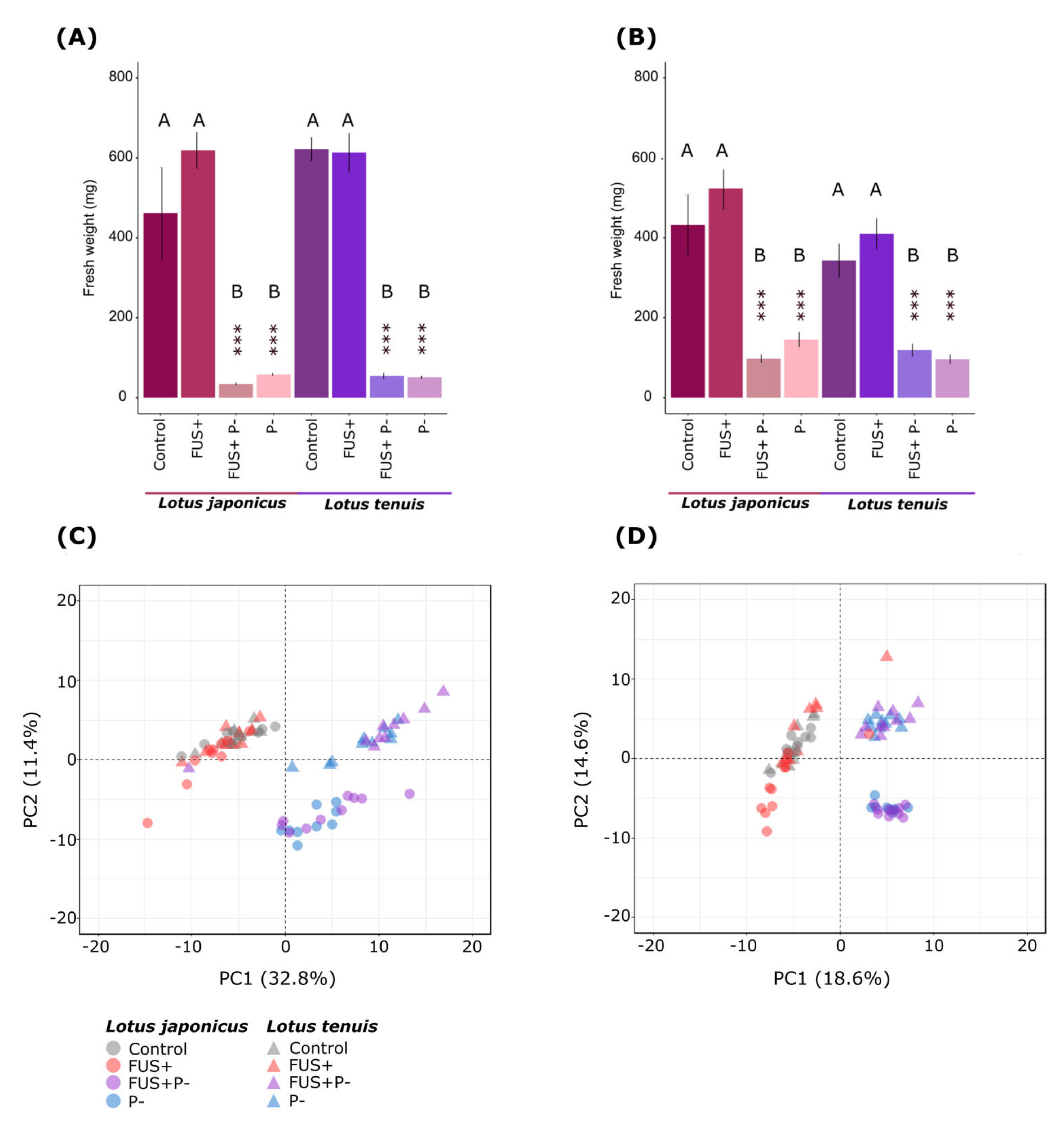
3.1. Interaction between the Fusarium solani Endophyte and Lotus spp. under Extreme Pi Starvation
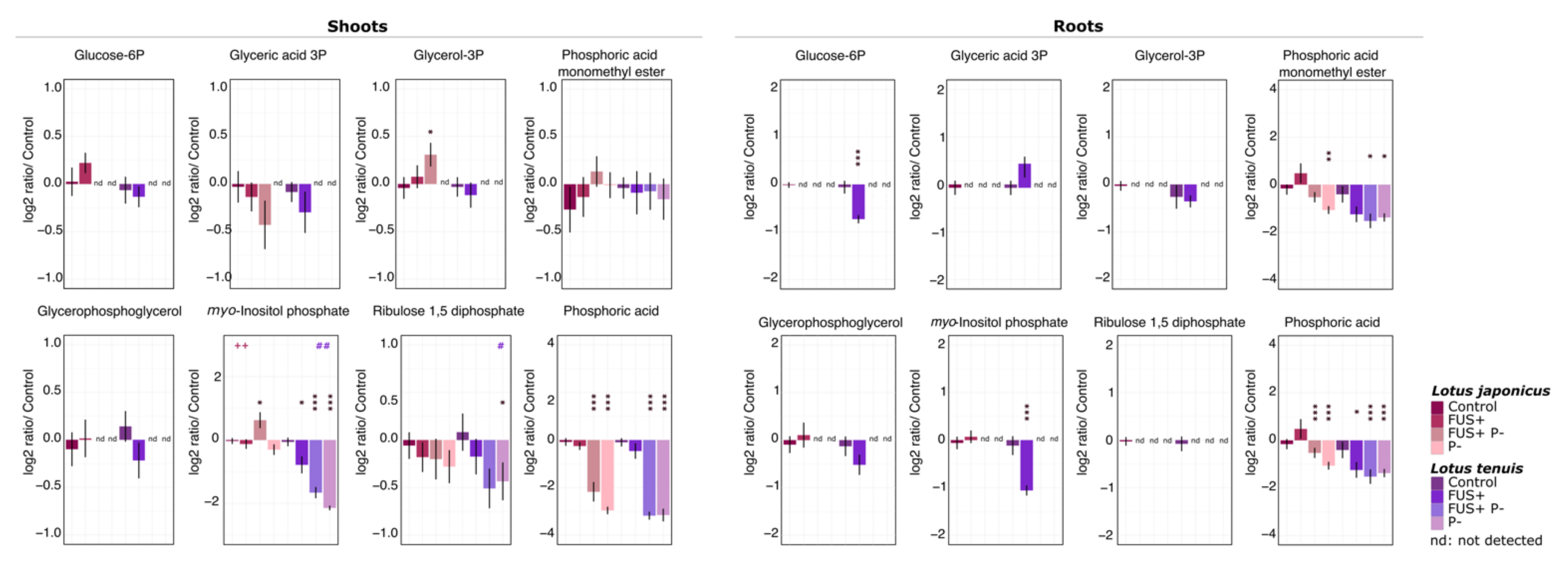
3.2. Phosphorylated Compounds
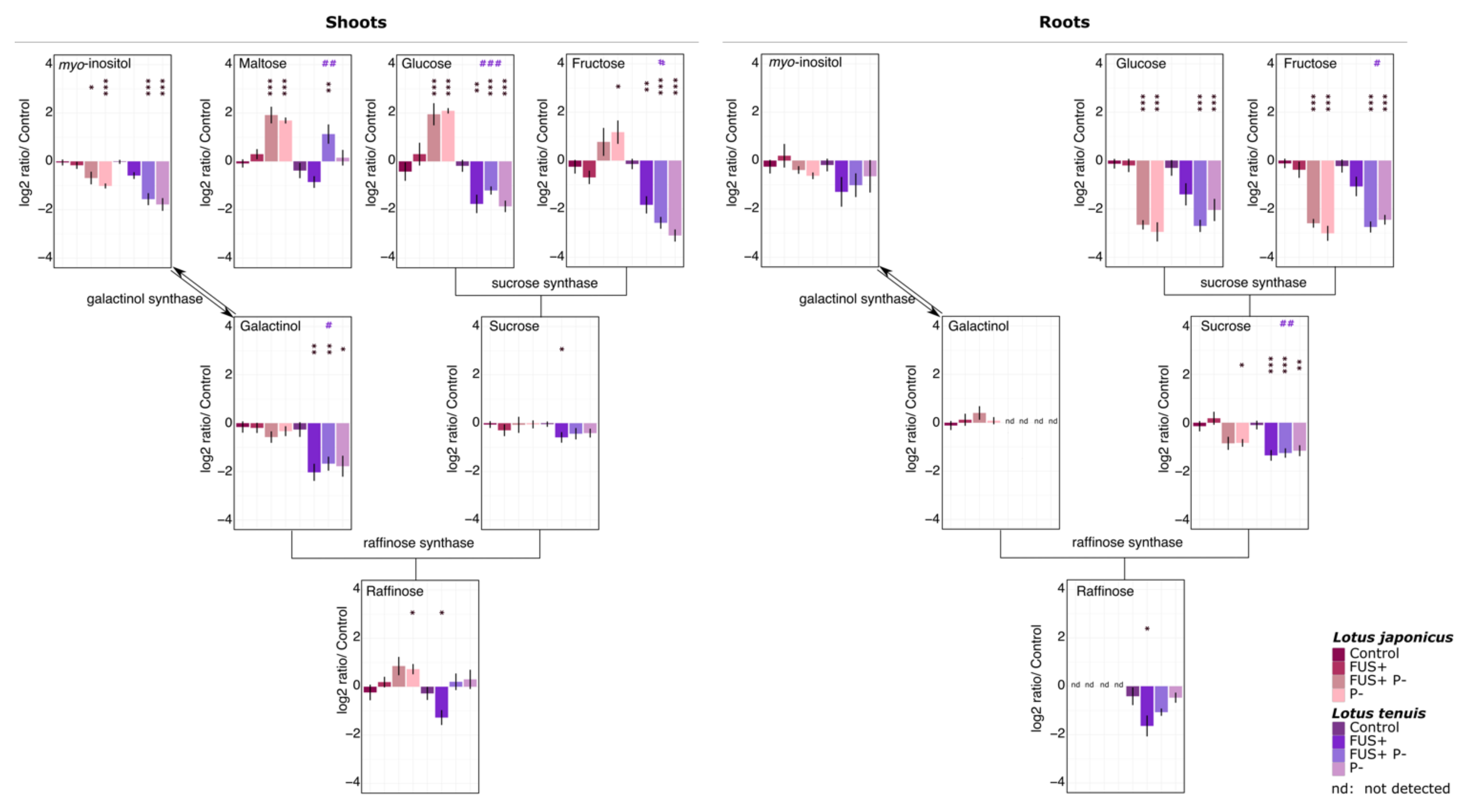
3.3. Metabolites Representing Potential Carbon Sources
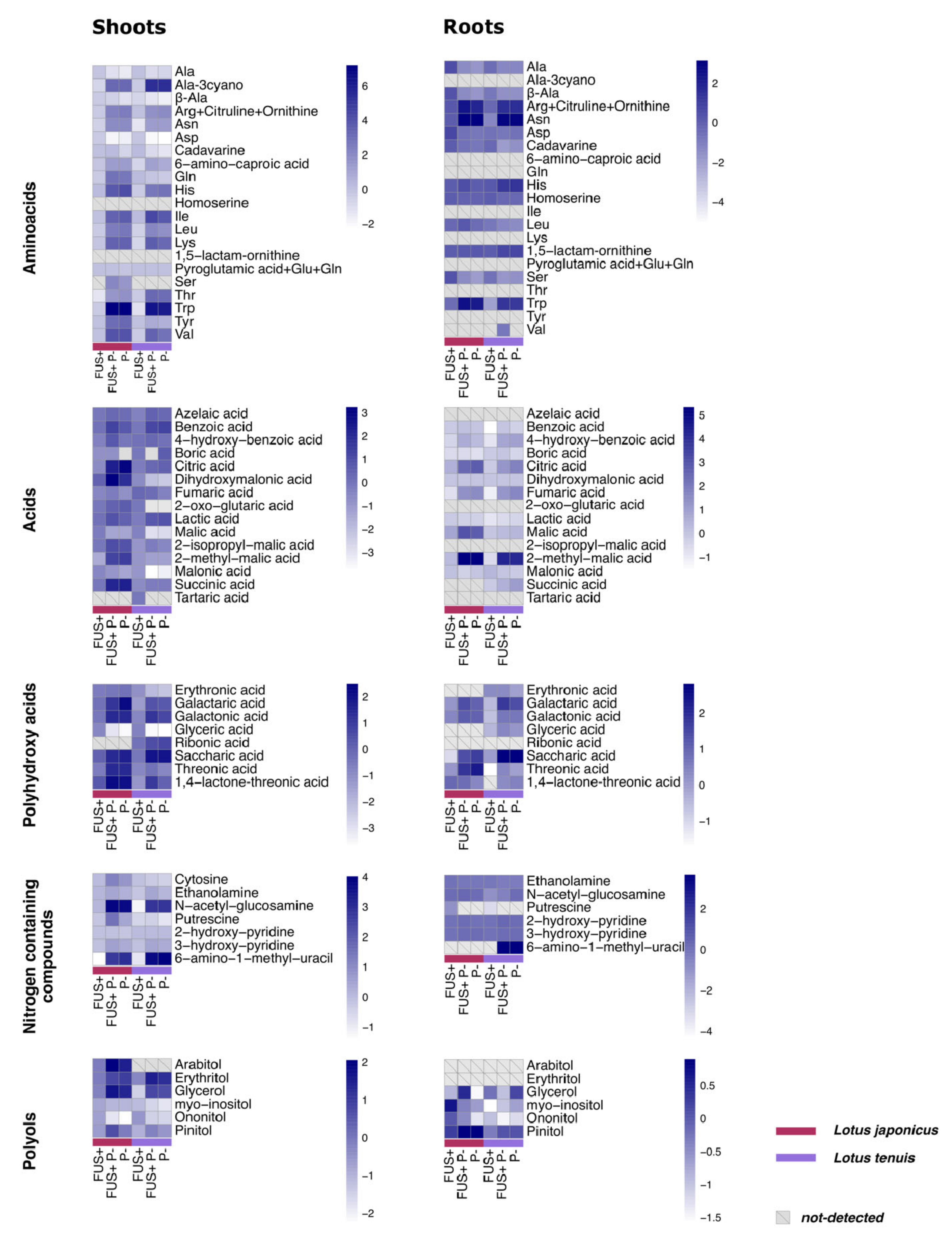
3.4. Organic Acids and Polyhydroxy Acids
3.5. Amino Acids and Other Nitrogen-Containing Compounds
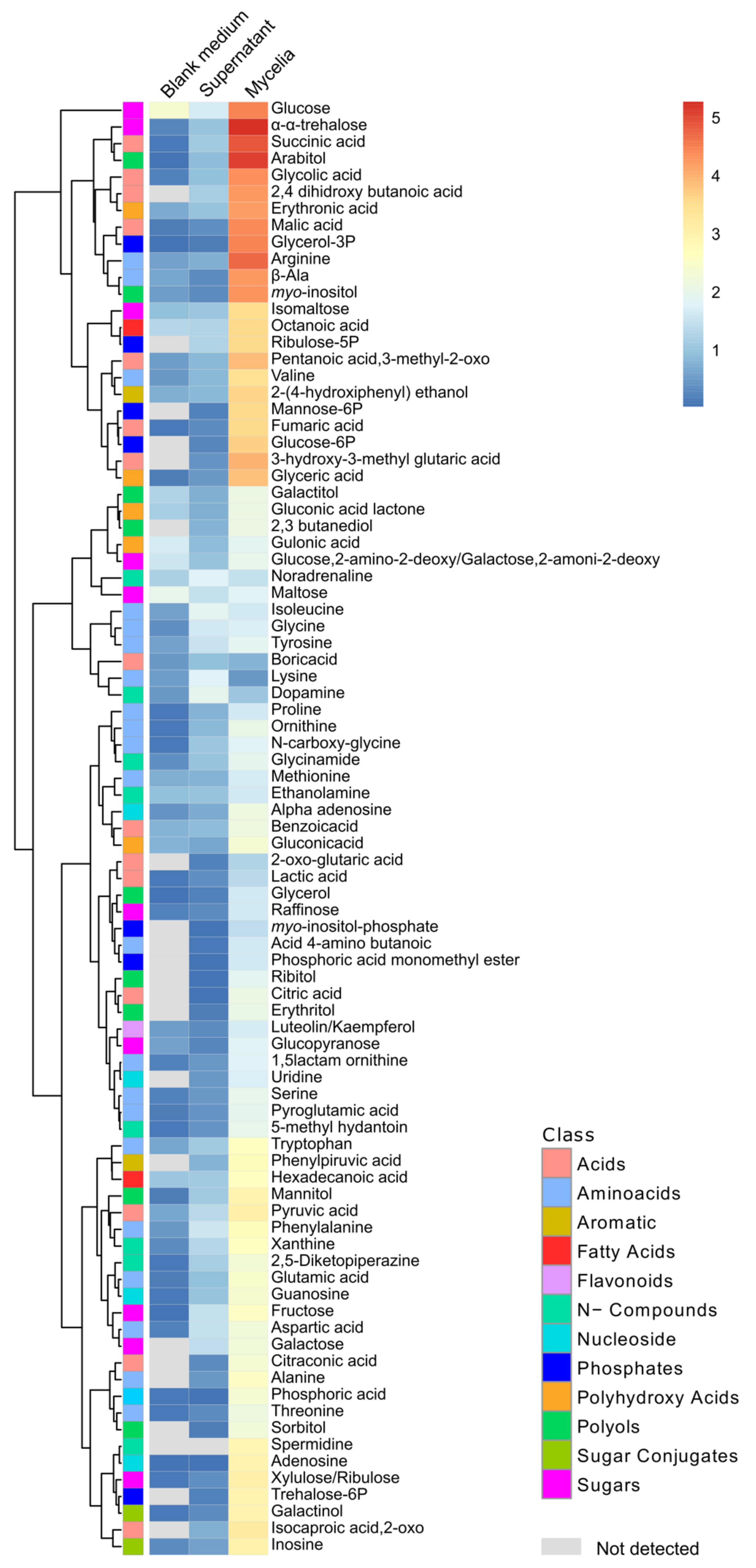
3.6. Metabolites of Mycelia and Secreted into the Medium by Fusarium solani
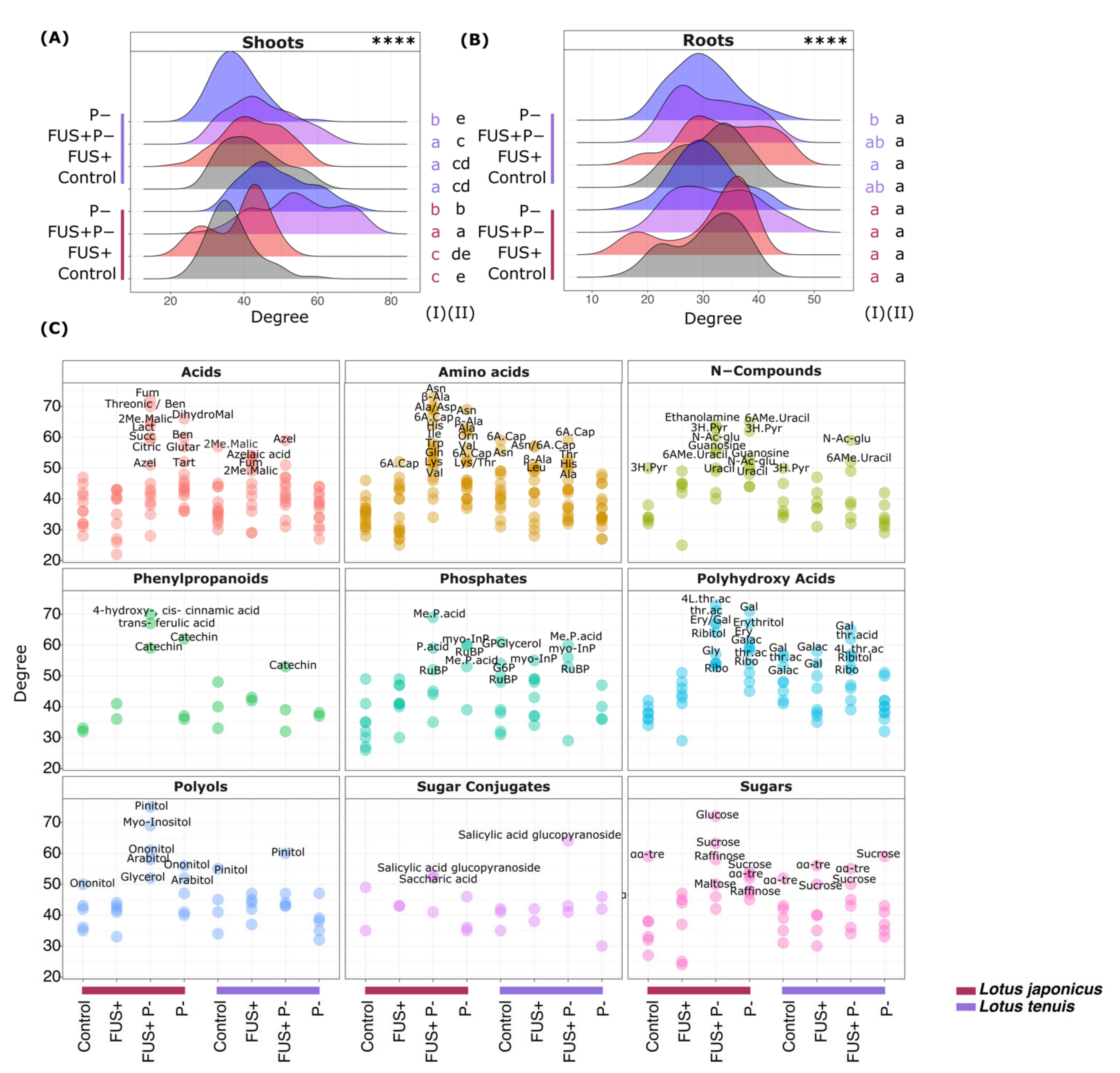
3.7. Correlation Network Analysis
4. Discussion
4.1. Pi Starvation Masks the Effect of the Endophyte Fusarium solani in Both Lotus Species
4.2. Modulation of Carbohydrate Availability by Lotus spp.-Fusarium solani Interactions under Pi Starvation Varies According to the Lotus Specie
4.3. Nitrogen-Related Compounds Responded Differentially to Pi Starvation
4.4. Organic Acids Might Play an Important Role in the Response to Pi Starvation
4.5. Role of Polyols in the Pi Starvation Response
4.6. Phosphorylated Compounds Are Accumulated Instead of Excreted by Fusarium solani
4.7. Correlation Network Analysis Remarks
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Escaray, F.J.; Menendez, A.B.; Gárriz, A.; Pieckenstain, F.L.; Estrella, M.J.; Castagno, L.N.; Carrasco, P.; Sanjuán, J.; Ruiz, O.A. Ecological and agronomic importance of the plant genus Lotus. Its application in grassland sustainability and the amelioration of constrained and contaminated soils. Plant Sci. 2012, 182, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Antonelli, C.J.; Calzadila, P.I.; Escaray, F.J.; Babuin, M.F.; Campestre, M.P.; Rocco, R.; Bordenave, C.; Perea Garcia, A.; Nieva, A.S.; Llames, M.E.; et al. Lotus sp.: Biotechnological Strategies to Improve the Bioeconomy of Lowlands in the Salado River Basin (Argentina). AGROFOR Int. J. 2016, 1, 43–53. [Google Scholar] [CrossRef] [Green Version]
- Nieva, A.S.; Ruiz, O.A. Lotus spp.: A foreigner that came to stay forever: Economic and environmental changes caused by its naturalization in the Salado River Basin (Argentina). In Saline and Alkaline Soils in Latin America: Natural Resources, Management and Productive Alternatives; Taleisnik, E., Lavado, R.S., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 431–446. [Google Scholar] [CrossRef]
- Handberg, K.; Stougaard, J. Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J. 1992, 2, 487–496. [Google Scholar] [CrossRef]
- Antonelli, C.J.; Calzadilla, P.I.; Vilas, J.M.; Campestre, M.P.; Escaray, F.J.; Ruiz, O.A. Physiological and anatomical traits associated with tolerance to long-term partial submergence stress in the Lotus genus: Responses of forage species, a model and an interspecific hybrid. J. Agron. Crop Sci. 2019, 205, 65–76. [Google Scholar] [CrossRef] [Green Version]
- Babuin, M.F.; Campestre, M.P.; Rocco, R.; Bordenave, C.D.; Escaray, F.J.; Antonelli, C.; Calzadilla, P.; Gárriz, A.; Serna, E.; Carrasco, P.; et al. Response to long-term NaHCO3-derived alkalinity in model Lotus japonicus ecotypes Gifu B-129 and Miyakojima MG-20: Transcriptomic profiling and physiological characterization. PLoS ONE 2014, 9, e97106. [Google Scholar] [CrossRef]
- Campestre, M.P.; Antonelli, C.; Calzadilla, P.I.; Maiale, S.J.; Rodríguez, A.A.; Ruiz, O.A. The alkaline tolerance in Lotus japonicus is associated with mechanisms of iron acquisition and modification of the architectural pattern of the root. J. Plant Physiol. 2016, 206, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Paz, R.C.; Rocco, R.A.; Reinoso, H.; Menéndez, A.B.; Pieckenstain, F.L.; Ruiz, O.A. Comparative study of alkaline, saline, and mixed saline–alkaline stresses with regard to their effects on growth, nutrient accumulation, and root morphology of Lotus tenuis. J. Plant Growth Regul. 2012, 31, 448–459. [Google Scholar] [CrossRef]
- Sanchez, D.H.; Cuevas, J.C.; Chiesa, M.A.; Ruiz, O.A. Free spermidine and spermine content in Lotus glaber under long-term salt stress. Plant Sci. 2005, 168, 541–546. [Google Scholar] [CrossRef]
- Calzadilla, P.I.; Signorelli, S.; Escaray, F.J.; Menéndez, A.B.; Monza, J.; Ruiz, O.A.; Maiale, S.J. Photosynthetic responses mediate the adaptation of two Lotus japonicus ecotypes to low temperature. Plant Sci. 2016, 250, 59–68. [Google Scholar] [CrossRef]
- Bordenave, C.D.; Rocco, R.; Maiale, S.J.; Campestre, M.P.; Ruiz, O.A.; Rodríguez, A.A.; Menéndez, A.B. Chlorophyll a fluorescence analysis reveals divergent photosystem II responses to saline, alkaline and saline–alkaline stresses in the two Lotus japonicus model ecotypes MG20 and Gifu-129. Acta Physiol. Plant. 2019, 41, 167. [Google Scholar] [CrossRef]
- Druille, M.; Cabello, M.N.; Omacini, M.; Golluscio, R.A. Glyphosate reduces spore viability and root colonization of arbuscular mycorrhizal fungi. Appl. Soil Ecol. 2013, 64, 99–103. [Google Scholar] [CrossRef]
- Nieva, A.S.; Bailleres, M.A.; Llames, M.E.; Taboada, M.A.; Ruiz, O.A.; Menéndez, A. Promotion of Lotus tenuis in the flooding Pampa (Argentina) increases the soil fungal diversity. Fungal Ecol. 2018, 33, 80–91. [Google Scholar] [CrossRef]
- Nieva, A.S.; Vilas, J.M.; Gárriz, A.; Maiale, S.J.; Menéndez, A.B.; Erban, A.; Kopka, J.; Ruiz, O.A. The fungal endophyte Fusarium solani provokes differential effects on the fitness of two Lotus species. Plant Physiol. Biochem. 2019, 144, 100–109. [Google Scholar] [CrossRef]
- Saikkonen, K.; Wäli, P.; Helander, M.; Faeth, S.H. Evolution of endophyte–plant symbioses. Trends Plant Sci. 2004, 9, 275–280. [Google Scholar] [CrossRef]
- Schulz, B.; Römmert, A.-K.; Dammann, U.; Aust, H.-J.; Strack, D. The endophyte-host interaction: A balanced antagonism? Mycol. Res. 1999, 103, 1275–1283. [Google Scholar] [CrossRef]
- Vance, C.P.; Uhde-Stone, C.; Allan, D.L. Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003, 157, 423–447. [Google Scholar] [CrossRef] [Green Version]
- Bolan, N.S. A critical review on the role of mycorrhizal fungi in the uptake of phosphorus by plants. Plant Soil 1991, 134, 189–207. [Google Scholar] [CrossRef]
- George, E.; Marschner, H.; Jakobsen, I. Role of arbuscular mycorrhizal fungi in uptake of phosphorus and nitrogen from soil. Crit. Rev. Biotechnol. 1995, 15, 257–270. [Google Scholar] [CrossRef]
- Mendoza, R. Phosphorus nutrition and mycorrhizal growth response of broadleaf and narrowleaf birdsfoot trefoils. J. Plant Nutr. 2001, 24, 203–214. [Google Scholar] [CrossRef]
- Mendoza, R.; Bailleres, M.; García, I.; Ruiz, O. Phosphorus fertilization of a grass-legume mixture: Effect on plant growth, nutrients acquisition and symbiotic associations with soil microorganisms. J. Plant Nutr. 2016, 39, 691–701. [Google Scholar] [CrossRef]
- Malinowski, D.; Brauer, D.; Belesky, D. The endophyte Neotyphodium coenophialum affects root morphology of tall fescue grown under phosphorus deficiency. J. Agron. Crop Sci. 1999, 183, 53–60. [Google Scholar] [CrossRef]
- Saito, K.; Kuga-Uetake, Y.; Saito, M.; Peterson, R.L. Vacuolar localization of phosphorus in hyphae of Phialocephala fortinii, a dark septate fungal root endophyte. Can. J. Microbiol. 2006, 52, 643–650. [Google Scholar] [CrossRef]
- Berger, S.; Sinha, A.K.; Roitsch, T. Plant physiology meets phytopathology: Plant primary metabolism and plant–pathogen interactions. J. Exp. Bot. 2007, 58, 4019–4026. [Google Scholar] [CrossRef]
- Vargas, W.A.; Mandawe, J.C.; Kenerley, C.M. Plant-derived sucrose is a key element in the symbiotic association between Trichoderma virens and maize plants. Plant Physiol. 2009, 151, 792–808. [Google Scholar] [CrossRef] [Green Version]
- Van der Nest, M.A.; Steenkamp, E.T.; McTaggart, A.R.; Trollip, C.; Godlonton, T.; Sauerman, E.; Roodt, D.; Naidoo, K.; Coetzee, M.P.A.; Wilken, P.M.; et al. Saprophytic and pathogenic fungi in the Ceratocystidaceae differ in their ability to metabolize plant-derived sucrose. BMC Evol. Biol. 2015, 15, 273. [Google Scholar] [CrossRef] [Green Version]
- Paul, M.J.; Stitt, M. Effects of nitrogen and phosphorus deficiencies on levels of carbohydrates, respiratory enzymes and metabolites in seedlings of tobacco and their response to exogenous sucrose. Plant Cell Environ. 1993, 16, 1047–1057. [Google Scholar] [CrossRef]
- Ciereszko, I.; Johansson, H.; Hurry, V.; Kleczkowski, L.A. Phosphate status affects the gene expression, protein content and enzymatic activity of UDP-glucose pyrophosphorylase in wild-type and pho mutants of Arabidopsis. Planta 2001, 212, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Müller, R.; Morant, M.; Jarmer, H.; Nilsson, L.; Nielsen, T.H. Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol. 2006, 143, 156–171. [Google Scholar] [CrossRef] [Green Version]
- Steuer, R.; Kurths, J.; Fiehn, O.; Weckwerth, W. Interpreting correlations in metabolomic networks. Biochem. Soc. Trans. 2003, 31, 1476–1478. [Google Scholar] [CrossRef] [PubMed]
- Camacho, D.; de la Fuente, A.; Mendes, P. The origin of correlations in metabolomics data. Metabolomics 2005, 1, 53–63. [Google Scholar] [CrossRef]
- Rosato, A.; Tenori, L.; Cascante, M.; De Atauri Carulla, P.R.; Martins dos Santos, V.A.P.; Saccenti, E. From correlation to causation: Analysis of metabolomics data using systems biology approaches. Metabolomics 2018, 14, 37. [Google Scholar] [CrossRef] [Green Version]
- Odds, F.C. Sabouraud(‘s) agar. J. Med. Vet. Mycol. 1991, 29, 355–359. [Google Scholar] [CrossRef]
- Evans, C.G.T.; Herbert, D.; Tempest, D.W. Chapter XIII The Continuous Cultivation of Micro-organisms: 2. Construction of a Chemostat. In Methods Microbiol; Norris, J.R., Ribbons, D.W., Eds.; Academic Press: Cambridge, MA, USA, 1970; Volume 2, pp. 277–327. [Google Scholar] [CrossRef]
- Sanchez, D.H.; Schwabe, F.; Erban, A.; Udvardi, M.K.; Kopka, J. Comparative metabolomics of drought acclimation in model and forage legumes. Plant Cell Environ. 2012, 35, 136–149. [Google Scholar] [CrossRef] [PubMed]
- Erban, A.; Martinez-Seidel, F.; Rajarathinam, Y.; Dethloff, F.; Orf, I.; Fehrle, I.; Alpers, J.; Beine-Golovchuk, O.; Kopka, J. Multiplexed profiling and data processing methods to identify temperature-regulated primary metabolites using gas chromatography coupled to mass spectrometry. In Plant Cold Acclimation: Methods and Protocols; Hincha, D.K., Zuther, E., Eds.; Springer: New York, NY, USA, 2020; pp. 203–239. [Google Scholar] [CrossRef]
- Luedemann, A.; Strassburg, K.; Erban, A.; Kopka, J. TagFinder for the quantitative analysis of gas chromatography—mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics 2008, 24, 732–737. [Google Scholar] [CrossRef] [PubMed]
- Hummel, J.; Strehmel, N.; Selbig, J.; Walther, D.; Kopka, J. Decision tree supported substructure prediction of metabolites from GC-MS profiles. Metabolomics 2010, 6, 322–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hastie, T.; Tibshirani, R.; Narasimhan, B.; Chu, G. Impute: Imputation for Microarray Data. Available online: https://bioconductor.org/packages/release/bioc/html/impute.html (accessed on 10 September 2021).
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. Available online: https://CRAN.R-project.org/package=factoextra (accessed on 10 September 2021).
- Faith, J.J.; Hayete, B.; Thaden, J.T.; Mogno, I.; Wierzbowski, J.; Cottarel, G.; Kasif, S.; Collins, J.J.; Gardner, T.S. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007, 5, e8. [Google Scholar] [CrossRef]
- Saccenti, E.; Suarez-Diez, M.; Luchinat, C.; Santucci, C.; Tenori, L. Probabilistic networks of blood metabolites in healthy subjects as indicators of latent cardiovascular risk. J. Proteome Res. 2015, 14, 1101–1111. [Google Scholar] [CrossRef]
- Sales, G.; Romualdi, C. Parmigene—A parallel R package for mutual information estimation and gene network reconstruction. Bioinformatics 2011, 27, 1876–1877. [Google Scholar] [CrossRef] [PubMed]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. InterJournal Complex Syst. 2006, 16995, 1–9. [Google Scholar]
- Toubiana, D.; Fernie, A.R.; Nikoloski, Z.; Fait, A. Network analysis: Tackling complex data to study plant metabolism. Trends Biotechnol. 2013, 31, 29–36. [Google Scholar] [CrossRef]
- Jeong, H.; Tombor, B.; Albert, R.; Oltvai, Z.N.; Barabási, A.L. The large-scale organization of metabolic networks. Nature 2000, 407, 651–654. [Google Scholar] [CrossRef] [Green Version]
- Raghavan Unnithan, S.K.; Kannan, B.; Jathavedan, M. Betweenness centrality in some classes of graphs. Int. J. Comb. 2014, 2014, 241723. [Google Scholar] [CrossRef]
- Sanchez, D.H.; Pieckenstain, F.L.; Escaray, F.; Erban, A.; Kraemer, U.; Udvardi, M.K.; Kopka, J. Comparative ionomics and metabolomics in extremophile and glycophytic Lotus species under salt stress challenge the metabolic pre-adaptation hypothesis. Plant Cell Environ. 2011, 34, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Desbrosses, G.G.; Kopka, J.; Udvardi, M.K. Lotus japonicus metabolic profiling. Development of gas chromatography-mass spectrometry resources for the study of plant-microbe interactions. Plant Physiol. 2005, 137, 1302–1318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocha, M.; Licausi, F.; Araújo, W.L.; Nunes-Nesi, A.; Sodek, L.; Fernie, A.R.; van Dongen, J.T. Glycolysis and the tricarboxylic acid cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of Lotus japonicus. Plant Physiol. 2010, 152, 1501–1513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasmussen, S.; Parsons, A.J.; Jones, C.S. Metabolomics of forage plants: A review. Ann. Bot. 2012, 110, 1281–1290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Sharkey, T.D. The importance of maltose in transitory starch breakdown. Plant Cell Environ. 2006, 29, 353–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lunn, J.E.; Delorge, I.; Figueroa, C.M.; Van Dijck, P.; Stitt, M. Trehalose metabolism in plants. Plant J. 2014, 79, 544–567. [Google Scholar] [CrossRef] [PubMed]
- García, N.A.T.; Iribarne, C.; López, M.; Herrera-Cervera, J.A.; Lluch, C. Physiological implications of trehalase from Phaseolus vulgaris root nodules: Partial purification and characterization. Plant Physiol. Biochem. 2005, 43, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Van Laere, A. Trehalose, reserve and/or stress metabolite? FEMS Microbiol. Rev. 1989, 5, 201–209. [Google Scholar] [CrossRef]
- Lillie, S.H.; Pringle, J.R. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: Responses to nutrient limitation. J. Bacteriol. 1980, 143, 1384–1394. [Google Scholar] [CrossRef] [Green Version]
- Gancedo, C.; Flores, C.-L. The importance of a functional trehalose biosynthetic pathway for the life of yeasts and fungi. FEMS Yeast Res. 2004, 4, 351–359. [Google Scholar] [CrossRef] [Green Version]
- Song, X.-S.; Li, H.-P.; Zhang, J.-B.; Song, B.; Huang, T.; Du, X.-M.; Gong, A.-D.; Liu, Y.-K.; Feng, Y.-N.; Agboola, R.S.; et al. Trehalose 6-phosphate phosphatase is required for development, virulence and mycotoxin biosynthesis apart from trehalose biosynthesis in Fusarium graminearum. Fungal Genet. Biol. 2014, 63, 24–41. [Google Scholar] [CrossRef] [PubMed]
- Hanson, J.; Smeekens, S. Sugar perception and signaling—an update. Curr. Opin. Plant Biol. 2009, 12, 562–567. [Google Scholar] [CrossRef] [PubMed]
- Gazzarrini, S.; McCourt, P. Genetic interactions between ABA, ethylene and sugar signaling pathways. Curr. Opin. Plant Biol. 2001, 4, 387–391. [Google Scholar] [CrossRef]
- Ruan, Y.-L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef]
- Li, L.; Sheen, J. Dynamic and diverse sugar signaling. Curr. Opin. Plant Biol. 2016, 33, 116–125. [Google Scholar] [CrossRef] [Green Version]
- Karthikeyan, A.S.; Varadarajan, D.K.; Jain, A.; Held, M.A.; Carpita, N.C.; Raghothama, K.G. Phosphate starvation responses are mediated by sugar signaling in Arabidopsis. Planta 2007, 225, 907–918. [Google Scholar] [CrossRef]
- Le Roux, M.R.; Khan, S.; Valentine, A.J. Nitrogen and carbon costs of soybean and lupin root systems during phosphate starvation. Symbiosis 2009, 48, 102–109. [Google Scholar] [CrossRef]
- Valentine, A.J.; Kleinert, A.; Benedito, V.A. Adaptive strategies for nitrogen metabolism in phosphate deficient legume nodules. Plant Sci. 2017, 256, 46–52. [Google Scholar] [CrossRef]
- Almeida, J.F.; Hartwig, U.A.; Frehner, M.; Nösberger, J.; Lüscher, A. Evidence that P deficiency induces N feedback regulation of symbiotic N2 fixation in white clover (Trifolium repens L.). J. Exp. Bot. 2000, 51, 1289–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabe, E.; Lovatt, C.J. Increased arginine biosynthesis during phosphorus deficiency: A response to the increased ammonia content of leaves. Plant Physiol. 1986, 81, 774–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Ghazi, Y.; Muller, B.; Pinloche, S.; Tranbarger, T.J.; Nacry, P.; Rossignol, M.; Tardieu, F.; Doumas, P. Temporal responses of Arabidopsis root architecture to phosphate starvation: Evidence for the involvement of auxin signalling. Plant Cell Environ. 2003, 26, 1053–1066. [Google Scholar] [CrossRef] [Green Version]
- Nacry, P.; Canivenc, G.v.; Muller, B.; Azmi, A.; Van Onckelen, H.; Rossignol, M.; Doumas, P. A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol. 2005, 138, 2061–2074. [Google Scholar] [CrossRef] [Green Version]
- Miura, K.; Lee, J.; Gong, Q.; Ma, S.; Jin, J.B.; Yoo, C.Y.; Miura, T.; Sato, A.; Bohnert, H.J.; Hasegawa, P.M. SIZ1 regulation of phosphate starvation-induced root architecture remodeling involves the control of auxin accumulation. Plant Physiol. 2010, 155, 1000–1012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Williams, C.C.; Last, R.L. Induction of Arabidopsis tryptophan pathway enzymes and camalexin by amino acid starvation, oxidative stress, and an abiotic elicitor. Plant Cell 1998, 10, 359–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espasandin, F.D.; Maiale, S.J.; Calzadilla, P.; Ruiz, O.A.; Sansberro, P.A. Transcriptional regulation of 9-cis-epoxycarotenoid dioxygenase (NCED) gene by putrescine accumulation positively modulates ABA synthesis and drought tolerance in Lotus tenuis plants. Plant Physiol. Biochem. 2014, 76, 29–35. [Google Scholar] [CrossRef]
- Calzadilla, P.I.; Gazquez, A.; Maiale, S.J.; Rodriguez, A.A.; Ruiz, O.A.; Menedez, A.B. Polyamines as indicators and modulators of the abiotic stress in plants. In Plant Adaptation to Environmental Change: Significance of Amino Acids and Their Derivatives; Anjum, N.A., Gill, S.S., Gill, R., Eds.; CABI: Wallingford, UK, 2014; pp. 109–128. [Google Scholar]
- Sannazzaro, A.I.; Echeverría, M.; Albertó, E.O.; Ruiz, O.A.; Menéndez, A.B. Modulation of polyamine balance in Lotus glaber by salinity and arbuscular mycorrhiza. Plant Physiol. Biochem. 2007, 45, 39–46. [Google Scholar] [CrossRef]
- Echeverria, M.; Sannazzaro, A.I.; Ruiz, O.A.; Menéndez, A.B. Modulatory effects of Mesorhizobium tianshanense and Glomus intraradices on plant proline and polyamine levels during early plant response of Lotus tenuis to salinity. Plant Soil 2013, 364, 69–79. [Google Scholar] [CrossRef]
- Hernández, G.; Ramírez, M.; Valdés-López, O.; Tesfaye, M.; Graham, M.A.; Czechowski, T.; Schlereth, A.; Wandrey, M.; Erban, A.; Cheung, F.; et al. Phosphorus stress in common bean: Root transcript and metabolic responses. Plant Physiol. 2007, 144, 752–767. [Google Scholar] [CrossRef] [Green Version]
- Hoffland, E.; Van Den Boogaard, R.; Nelemans, J.; Findenegg, G. Biosynthesis and root exudation of citric and malic acids in phosphate-starved rape plants. New Phytol. 1992, 122, 675–680. [Google Scholar] [CrossRef]
- Zhang, F.S.; Ma, J.; Cao, Y.P. Phosphorus deficiency enhances root exudation of low-molecular weight organic acids and utilization of sparingly soluble inorganic phosphates by radish (Raghanus satiuvs L.) and rape (Brassica napus L.) plants. Plant Soil 1997, 196, 261–264. [Google Scholar] [CrossRef]
- Panigrahy, M.; Rao, D.N.; Sarla, N. Molecular mechanisms in response to phosphate starvation in rice. Biotechnol. Adv. 2009, 27, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Stevens, G.G.; Pérez-Fernández, M.A.; Morcillo, R.J.L.; Kleinert, A.; Hills, P.; Brand, D.J.; Steenkamp, E.T.; Valentine, A.J. Roots and nodules response differently to P starvation in the mediterranean-type legume Virgilia divaricata. Front. Plant Sci. 2019, 10, 73. [Google Scholar] [CrossRef] [PubMed]
- Ohwaki, Y.; Hirata, H. Differences in carboxylic acid exudation among p-starved leguminous crops in relation to carboxylic acid contents in plant tissues and phospholipid level in roots. Soil Sci. Plant Nutr. 1992, 38, 235–243. [Google Scholar] [CrossRef] [Green Version]
- Weisskopf, L.; Le Bayon, R.-C.; Kohler, F.; Page, V.; Jossi, M.; Gobat, J.-M.; Martinoia, E.; Aragno, M. Spatio-temporal dynamics of bacterial communities associated with two plant species differing in organic acid secretion: A one-year microcosm study on lupin and wheat. Soil Biol. Biochem. 2008, 40, 1772–1780. [Google Scholar] [CrossRef] [Green Version]
- Touhami, D.; McDowell, R.W.; Condron, L.M. Role of organic anions and phosphatase enzymes in phosphorus acquisition in the rhizospheres of legumes and grasses grown in a low phosphorus pasture soil. Plants 2020, 9, 1185. [Google Scholar] [CrossRef]
- Bolan, N.S.; Naidu, R.; Mahimairaja, S.; Baskaran, S. Influence of low-molecular-weight organic acids on the solubilization of phosphates. Biol. Fertil. Soils 1994, 18, 311–319. [Google Scholar] [CrossRef]
- Randall, P.J.; Hayes, J.E.; Hocking, P.J.; Richardson, A.E. Root exudates in phosphorus acquisition by plants. In Plant Nutrient Acquisition; Ae, N., Arihara, J., Okada, K., Srinivasan, A., Eds.; Springer: Tokyo, Japan, 2001; pp. 71–100. [Google Scholar] [CrossRef]
- Chia, D.W.; Yoder, T.J.; Reiter, W.-D.; Gibson, S.I. Fumaric acid: An overlooked form of fixed carbon in Arabidopsis and other plant species. Planta 2000, 211, 743–751. [Google Scholar] [CrossRef] [Green Version]
- John, D.W.; Dianne, B.J.; Wei-Wen, G.; Pharr, D.M.; Marilyn, E. Sugar alcohols, salt stress, and fungal resistance: Polyols—multifunctional plant protection? J. Am. Soc. Hortic. Sci. 2002, 127, 467–473. [Google Scholar] [CrossRef] [Green Version]
- Dumschott, K.; Richter, A.; Loescher, W.; Merchant, A. Post photosynthetic carbon partitioning to sugar alcohols and consequences for plant growth. Phytochemistry 2017, 144, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Jennings, D.H. Polyol metabolism in fungi. In Advances in Microbial Physiology; Rose, A.H., Tempest, D.W., Eds.; Academic Press: Cambridge, MA, USA, 1985; Volume 25, pp. 149–193. [Google Scholar] [CrossRef]
- Demain, A.L. Regulation of secondary metabolism in fungi. Pure Appl. Chem. 1986, 58, 219–226. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nieva, A.S.; Romero, F.M.; Erban, A.; Carrasco, P.; Ruiz, O.A.; Kopka, J. Metabolic Profiling and Metabolite Correlation Network Analysis Reveal That Fusarium solani Induces Differential Metabolic Responses in Lotus japonicus and Lotus tenuis against Severe Phosphate Starvation. J. Fungi 2021, 7, 765. https://doi.org/10.3390/jof7090765
Nieva AS, Romero FM, Erban A, Carrasco P, Ruiz OA, Kopka J. Metabolic Profiling and Metabolite Correlation Network Analysis Reveal That Fusarium solani Induces Differential Metabolic Responses in Lotus japonicus and Lotus tenuis against Severe Phosphate Starvation. Journal of Fungi. 2021; 7(9):765. https://doi.org/10.3390/jof7090765
Chicago/Turabian StyleNieva, Amira Susana, Fernando Matías Romero, Alexander Erban, Pedro Carrasco, Oscar Adolfo Ruiz, and Joachim Kopka. 2021. "Metabolic Profiling and Metabolite Correlation Network Analysis Reveal That Fusarium solani Induces Differential Metabolic Responses in Lotus japonicus and Lotus tenuis against Severe Phosphate Starvation" Journal of Fungi 7, no. 9: 765. https://doi.org/10.3390/jof7090765
APA StyleNieva, A. S., Romero, F. M., Erban, A., Carrasco, P., Ruiz, O. A., & Kopka, J. (2021). Metabolic Profiling and Metabolite Correlation Network Analysis Reveal That Fusarium solani Induces Differential Metabolic Responses in Lotus japonicus and Lotus tenuis against Severe Phosphate Starvation. Journal of Fungi, 7(9), 765. https://doi.org/10.3390/jof7090765





