Diversity of Colletotrichum Species Associated with Olive Anthracnose Worldwide
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Fungal Isolates
2.2. Phenotypic Characterization
2.2.1. Colonies and Conidial Morphology
2.2.2. Benomyl-Sensitive Assay
2.2.3. Casein-Hydrolysis Assay
2.3. Molecular Characterization
2.3.1. DNA Extraction, PCR, Sequencing, and Nucleotide Alignment
2.3.2. Phylogenetic Analyses and Species Delimitation
2.4. Pathogenicity Test
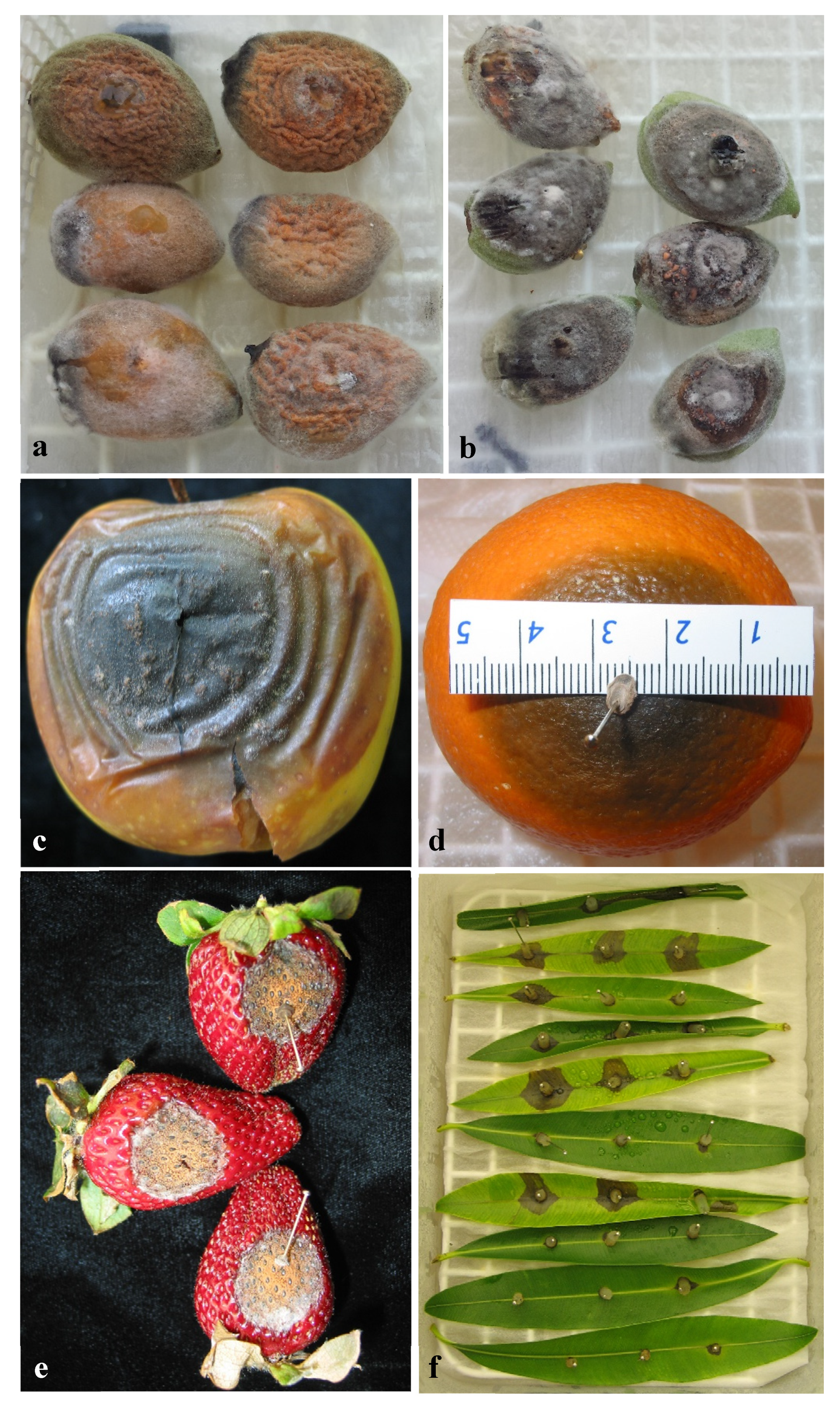
2.4.1. Pathogenicity on Olive Fruit
2.4.2. Pathogenicity on Other Hosts
2.4.3. Disease Assessment and Data Analysis
3. Results
3.1. Collection of Fungal Isolates
3.2. Phenotypic Characterization
3.2.1. Colonies and Conidial Morphology
3.2.2. Benomyl Sensitive Assay
3.2.3. Hydrolysis-Casein Assay
3.3. Molecular Characterization. Phylogenetic Analyses
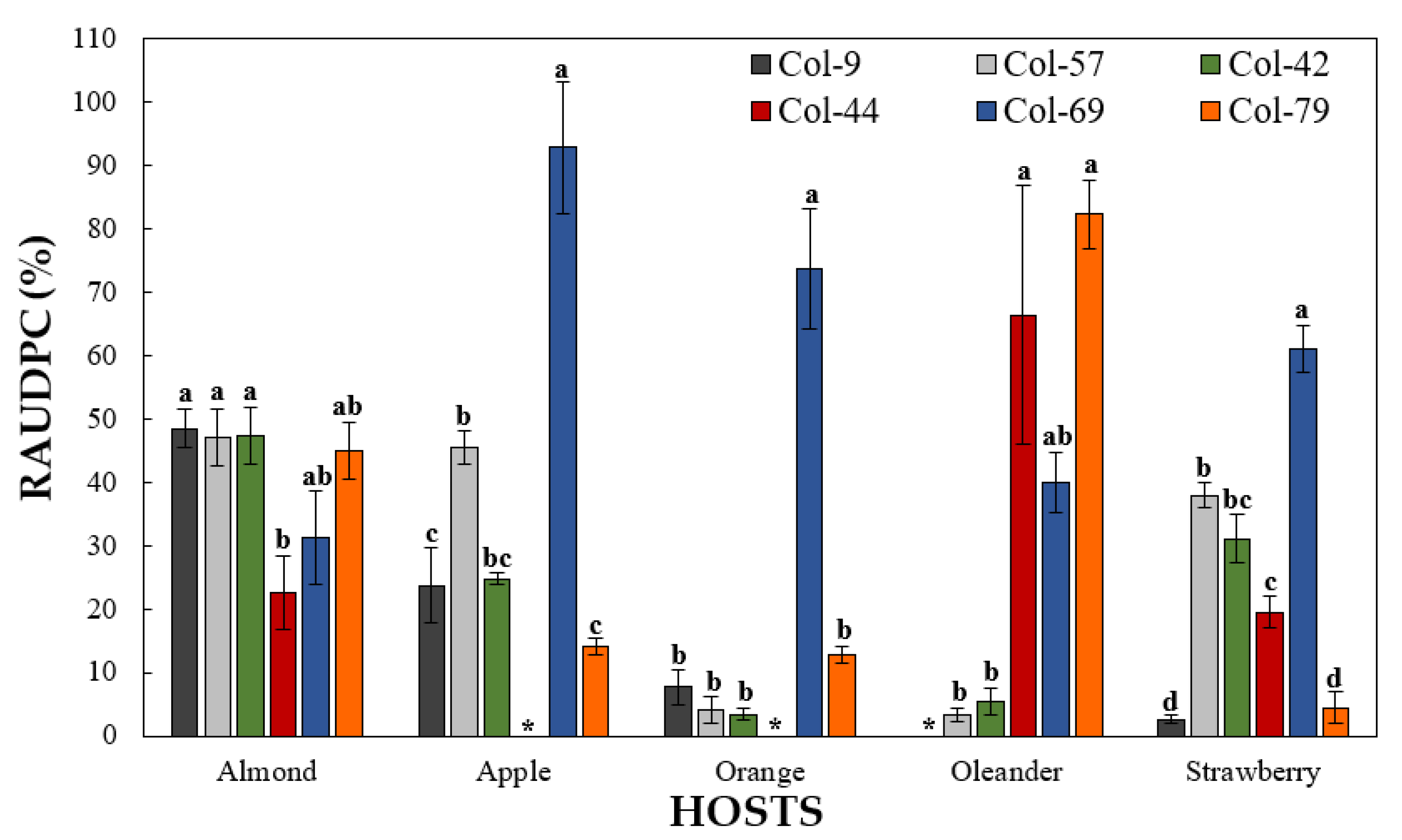
3.4. Pathogenicity Tests
3.4.1. Pathogenicity to Olive Fruit
3.4.2. Pathogenicity on Other Hosts
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agricultural Organisation of the United Nations (FAOSTAT) Website. 2019. Available online: http://www.fao.org/faostat/es/#data/QCL/visualize (accessed on 11 August 2021).
- Lucena, B.; Manrique, T.; Méndez, M.A. La olivicultura en el mundo y en España. In El Cultivo Del Olivo, 7th ed.; Barranco, D., Fernández-Escobar, R., Rallo, L., Eds.; Mundi Prensa: Madrid, Spain, 2017; pp. 3–33. [Google Scholar]
- Moral, J.; Morgan, D.; Trapero, A.; Michailides, T.J. Ecology and epidemiology of diseases of nut crops and olives caused by Botryosphaeriaceae fungi in California and Spain. Plant Dis. 2019, 103, 1809–1827. [Google Scholar] [CrossRef] [Green Version]
- Talhinhas, P.; Sreenivasaprasad, S.; Neves-Martins, J.; Oliveira, H. Molecular and phenotypic analyses reveal association of diverse Colletotrichum acutatum groups and a low level of C. gloeosporioides with olive anthracnose. Appl. Environ. Microbiol. 2005, 71, 2987–2998. [Google Scholar] [CrossRef] [Green Version]
- Talhinhas, P.; Loureiro, A.; Oliveria, H. Olive anthracnose: A yield- and oil quality-degrading disease caused by several species of Colletotrichum that differ in virulence, host preference and geographical distribution. Mol. Plant Pathol. 2018, 19, 1797–1807. [Google Scholar] [CrossRef] [Green Version]
- Cacciola, S.O.; Faedda, R.; Sinatra, F.; Agosteo, G.E.; Schena, L.; Frisullo, S.; Magnano di San Lio, G. Olive anthracnose. J. Plant Pathol. 2012, 94, 29–44. [Google Scholar]
- Moral, J.; Xaviér, C.; Roca, L.F.; Romero, J.; Moreda, W.; Trapero, A. La Antracnosis del olivo y su efecto en la calidad del aceite. Grasas Aceites 2014, 65, e028. [Google Scholar] [CrossRef]
- Moral, J.; Oliveira, R.; Trapero, A. Elucidation of disease cycle of olive anthracnose caused by Colletotrichum acutatum. Phytopathology 2009, 99, 548–556. [Google Scholar] [CrossRef] [Green Version]
- Moral, J.; Bouhmidi, K.; Trapero, A. Influence of fruit maturity, cultivar susceptibility, and inoculation method on infection of olive fruit by Colletotrichum acutatum. Plant Dis. 2008, 92, 1421–1426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moral, J.; Jurado-Bello, J.; Sánchez, M.I.; De Oliveira, R.; Trapero, A. Effect of temperature, wetness duration, and planting density on olive anthracnose caused by Colletotrichum spp. Phytopathology 2012, 102, 974–981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ballio, A.; Bottalico, A.; Bounocore, V.; Carilli, A.; Di Vittorio, V.; Graniti, A. Production and isolation of aspergillomarasmin B (lycomarasmic acid) from cultures of Colletotrichum gloeosporioides (Gloeosporium olivarum). Phytopathol. Mediterr. 1969, 8, 187–196. [Google Scholar]
- Sergeeva, V. The role of epidemiology data in developing integrated management of anthracnose in olives—A review. Acta Hortic. 2014, 1057, 163–168. [Google Scholar] [CrossRef]
- Moreira, V.; Mondino, P.; Alaniz, S. Olive anthracnose caused by Colletotrichum in Uruguay: Field symptoms, species diversity and flowers and fruits pathogenicity. Eur. J. Plant Pathol. 2021, 160, 663–681. [Google Scholar] [CrossRef]
- Moral, J.; Trapero, A. Mummified fruit as a source of inoculum and disease dynamics of olive anthracnose caused by Colletotrichum spp. Phytopathology 2012, 102, 982–989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Almeida, M.J.V. La gaffa des olives en Portugal. Bull. Soc. Mycol. Fr. 1899, 15, 90–94. [Google Scholar]
- Von Arx, J.A. A revision of the fungi classified as Gloeosporium. Bibl. Mycol. 1970, 24, 1–203. [Google Scholar]
- O’Connell, R.J.; Thon, M.R.; Hacquard, S.; Amyotte, S.G.; Kleemann, J.; Torres, M.F.; Altmüller, J. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat. Genet. 2012, 44, 1060. [Google Scholar] [CrossRef] [PubMed]
- Mycobank. 2021. Available online: https://www.mycobank.org/ (accessed on 25 July 2021).
- Bhunjun, C.S.; Phukhamsakda, C.; Jayawardena, R.S.; Jeewon, R.; Promputtha, I.; Hyde, K.D. Investigating species boundaries in Colletotrichum. Fungal Divers. 2021, 107, 107–127. [Google Scholar] [CrossRef]
- Cristóbal-Martínez, A.L.; de Jesús Yáñez-Morales, M.; Solano-Vidal, R.; Segura-León, O.; Hernández-Anguiano, A.M. Diversity of Colletotrichum species in coffee (Coffea arabica) plantations in Mexico. Eur. J. Plant Pathol. 2017, 147, 605–614. [Google Scholar] [CrossRef]
- Sharma, G.; Maymon, M.; Freeman, S. Epidemiology, pathology and identification of Colletotrichum including a novel species associated with avocado (Persea americana) anthracnose in Israel. Sci. Rep. 2017, 7, 15839. [Google Scholar] [CrossRef]
- De Silva, D.D.; Groenewald, J.Z.; Crous, P.W.; Ades, P.K.; Nasruddin, A.; Mongkolporn, O.; Taylor, P.W. Identification, prevalence and pathogenicity of Colletotrichum species causing anthracnose of Capsicum annuum in Asia. IMA Fungus 2019, 10, 8. [Google Scholar] [CrossRef]
- Schena, L.; Mosca, S.; Cacciola, S.O.; Faedda, R.; Sanzani, S.M.; Agosteo, G.E.; Sergeeva, V.; Magnano di San Lio, G. Species of the Colletotrichum gloeosporioides and C. boninense complexes associated with olive anthracnose. Plant Pathol. 2014, 63, 437–446. [Google Scholar] [CrossRef]
- Schena, L.; Abdelfattah, A.; Mosca, S.; Nicosia, M.G.L.D.; Agosteo, G.E.; Cacciola, S.O. Quantitative detection of Colletotrichum godetiae and C. acutatum sensu stricto in the phyllosphere and carposphere of olive during four phenological phases. Eur. J. Plant Pathol. 2017, 149, 337–347. [Google Scholar] [CrossRef]
- Materatski, P.; Varanda, C.; Carvalho, T.; Dias, A.B.; Campos, M.D.; Rei, F.; Félix, M.D.R. Diversity of Colletotrichum species associated with olive anthracnose and new perspectives on controlling the disease in Portugal. Agronomy 2018, 8, 301. [Google Scholar] [CrossRef] [Green Version]
- Faedda, R.; Agosteo, G.E.; Schena, L.; Mosca, S.; Frisullo, S.; Magnano di San Lio, G.; Cacciola, S.O. Colletotrichum clavatum sp. nov. identified as the causal agent of olive anthracnose in Italy. Phytopathol. Mediterr. 2011, 50, 283–302. [Google Scholar]
- Damm, U.; Cannon, P.; Woudenberg, J.; Crous, P.W. The Colletotrichum acutatum species complex. Stud. Mycol. 2012, 73, 37–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosca, S.; Li Destri Nicosia, M.G.; Cacciola, S.O.; Schena, L. Molecular analysis of Colletotrichum species in the carposphere and phyllosphere of olive. PLoS ONE 2014, 9, e114031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelfattah, A.; Nicosia, M.G.L.D.; Cacciola, S.O.; Droby, S.; Schena, L. Metabarcoding analysis of fungal diversity in the phyllosphere and carposphere of olive (Olea europaea). PLoS ONE 2015, 10, e0131069. [Google Scholar] [CrossRef] [Green Version]
- Sergeeva, V.; Schena, L.; Mosca, S.; Mammella, M.A.; Faedda, R.; Cacciola, S.O. Colletotrichum acutatum as causal agent of olive anthracnose in Australia. Petria 2010, 20, 251–252. [Google Scholar]
- Chattaoui, M.; Raya, M.C.; Bouri, M.; Moral, J.; Pérez-Rodríguez, M.; Trapero, A.; Msallem, M.; Rhouma, A. Characterization of a Colletotrichum population causing anthracnose disease on Olive in northern Tunisia. J. Appl. Microbiol. 2016, 120, 1368–1381. [Google Scholar] [CrossRef] [Green Version]
- Iliadi, M.K.; Tjamos, E.C.; Antoniou, P.P.; Tsitsigiannis, D.I. First report of Colletotrichum acutatum causing anthracnose on olives in Greece. Plant Dis. 2018, 102, 820. [Google Scholar] [CrossRef]
- Martín, M.P.; García-Figueres, F. Colletotrichum acutatum and C. gloeosporioides cause anthracnose on olives. Eur. J. Plant Pathol. 1999, 105, 733–741. [Google Scholar] [CrossRef]
- Talhinhas, P.; Neves-Martins, J.; Oliveira, H.; Sreenivasaprasad, S. The distinctive population structure of Colletotrichum species associated with olive anthracnose in the Algarve region of Portugal reflects a host-pathogen diversity hot spot. FEMS Microbiol. Lett. 2009, 296, 31–38. [Google Scholar] [CrossRef]
- Talhinhas, P.; Gonçalves, E.; Sreenivasaprasad, S.; Oliveira, H. Virulence diversity of anthracnose pathogens (Colletotrichum acutatum and C. gloeosporioides species complexes) on eight olive cultivars commonly grown in Portugal. Eur. J. Plant Pathol. 2015, 142, 73–83. [Google Scholar] [CrossRef]
- López-Moral, A.; Raya-Ortega, M.C.; Agustí-Brisach, C.; Roca, L.F.; Lovera, M.; Luque, F.; Arquero, O.; Trapero, A. Morphological, pathogenic and molecular characterization of Colletotrichum acutatum isolates causing almond anthracnose in Spain. Plant Dis. 2017, 101, 2034–2045. [Google Scholar] [CrossRef] [Green Version]
- López-Moral, A.; Agustí-Brisach, C.; Lovera, M.; Arquero, O.; Trapero, A. Almond anthracnose: Current knowledge and future perspectives. Plants 2020, 9, 945. [Google Scholar] [CrossRef]
- Lopes da Silva, L.; Alvarado Moreno, H.L.; Nunes Correia, H.L.; Ferreira Santana, M.; Vieira de Queiroz, M. Colletotrichum: Species complexes, lifestyle, and peculiarities of some sources of genetic variability. Appl. Microbiol. Biotechnol. 2020, 104, 1891–1904. [Google Scholar] [CrossRef]
- Talhinhas, P.; Mota-Capitão, C.; Martins, S.; Ramos, A.P.; Neves-Martins, J.; Guerra-Guimarães, L.; Várzea, V.; Silva, M.C.; Sreenivasaprasad, S.; Oliveira, H. Epidemiology, histopathology, and aetiology of olive anthracnose caused by Colletotrichum acutatum and C. gloeosporioides in Portugal. Plant Pathol. 2011, 60, 483–495. [Google Scholar] [CrossRef] [Green Version]
- Dhingra, O.D.; Sinclair, J.B. Basic Plant Pathology Methods, 2nd ed.; CRC Press: Boca Raton, FL, USA, 1995. [Google Scholar]
- Barnett, H.L.; Hunter, B.B. Illustrated genera of imperfect fungi., 4th ed.; APS Press: St. Paul, MN, USA, 1998. [Google Scholar]
- Rayner, R.W. A mycological colour chart. Mycologia 1972, 64, 230–233. [Google Scholar]
- Peres, N.A.; Souza, N.L.; Peever, T.L.; Timmer, L.W. Benomyl sensitivity of isolates of Colletotrichum acutatum and C. gloeosporioides from citrus. Plant Dis. 2004, 88, 125–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steel, R.G.D.; Torrie, J.H. Bioestadística, 2nd ed.; McGraw-Hill: Bogotá, Colombia, 1985. [Google Scholar]
- Anonimous. Analytical Software, Statistix10; User’s Manual; Anonimous: Tallahassee, FL, USA, 2013. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Snisky, J.J., White, T.J., Eds.; Academic: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Glass, N.L.; Donaldson, G. Development of primer sets designed for use with PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microb. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [Green Version]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Crous, P.W.; Groenewald, J.Z.; Risede, J.M.; Hywel-Jones, N.L. Calonectria species and their Cylindrocladium anamorphs: Species with sphaeropedunculate vesicles. Stud. Mycol. 2004, 50, 415–430. [Google Scholar]
- Guerber, J.C.; Liu, B.; Correll, J.C.; Johnston, P.R. Characterization of diversity in Colletotrichum acutatum sensu lato by sequence analysis of two gene introns, mtDNA and intron RFLPs, and mating compatibility. Mycologia 2003, 95, 872–895. [Google Scholar] [CrossRef]
- Silva, D.N.; Talhinhas, P.; Várzea, V.; Cai, L.; Paulo, O.S.; Batista, D. Application of the Apn2/MAT locus to improve the systematics of the Colletotrichum gloeosporioides complex: An example from coffee (Coffea spp.) hosts. Mycologia 2012, 104, 396–409. [Google Scholar] [CrossRef] [Green Version]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Weir, B.; Johnston, P.; Damm, U. The Colletotrichum gloeosporioides species complex. Stud. Mycol. 2012, 73, 115–180. [Google Scholar] [CrossRef] [Green Version]
- Jayawardena, R.S.; Hyde, K.D.; Damm, U.; Cai, L.; Liu, M.; Li, X.H.; Zhang, W.; Zhao, W.S.; Yan, J.Y. Notes on currently accepted species of Colletotrichum. Mycosphere 2016, 7, 1192–1260. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moral, J.; Xaviér, C.J.; Viruega, J.R.; Roca, L.F.; Caballero, J.; Trapero, A. Variability in susceptibility to anthracnose in the World Collection of Olive Cultivars of Cordoba (Spain). Front. Plant Sci. 2017, 8, 1892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freeman, S.; Katan, T.; Shabi, E. Characterization of Colletotrichum species responsible for anthracnose diseases of various fruits. Plant Dis. 1998, 82, 596–605. [Google Scholar] [CrossRef] [Green Version]
- Baroncelli, R.; Talhinhas, P.; Pensec, F.; Sukno, S.A.; Le Floch, G.; Thon, M.R. The Colletotrichum acutatum species complex as a model system to study evolution and host specialization in plant pathogens. Front. Microbiol. 2017, 8, 02001. [Google Scholar] [CrossRef] [Green Version]
- Dowling, M.; Peres, N.; Villani, S.; Schnabel, G. Managing Colletotrichum on fruit crops: A “complex” challenge. Plant Dis. 2020, 104, 2301–2316. [Google Scholar] [CrossRef]
- Sutton, B.C. The Coelomycetes. In Fungi Imperfecti with Pycnidia, Acervuli and Stromata; Commonwealth Mycological Institute: Kew, UK, 1980. [Google Scholar]
- Freeman, S.; Minz, D.; Jurkevitch, E.; Maymon, M.; Shabi, E. Molecular analyses of Colletotrichum species from almond and other fruits. Phytopathology 2000, 90, 608–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutton, B.C. The genus Glomerella and its anamorph Colletotrichum. In Colletotrichum: Biology, Pathology and Control; Bailey, J.A., Jeger, M.J., Eds.; CAB International: Wallingford, UK, 1992; pp. 1–27. [Google Scholar]
- Simmonds, J.H. A study of the species of Colletotrichum causing ripe fruit rots in Queensland. Qld. J. Agric. Sci. 1965, 22, 361–378. [Google Scholar]
- Hyde, K.D.; Cai, L.; McKenzie, E.H.C.; Yang, Y.L.; Zhang, J.Z.; Prihastuti, H. Colletotrichum: A catalogue of confusion. Fungal Divers. 2009, 39, 1–17. [Google Scholar]
- Cannon, P.F.; Damm, U.; Johnston, P.R.; Weir, B.S. Colletotrichum–current status and future directions. Stud. Mycol. 2012, 73, 181–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernstein, B.; Zehr, E.I.; Deam, R.A.; Shabi, E. Characteristics of Colletotrichum from peach, apple, pecan, and other hosts. Plant Dis. 1995, 79, 478–482. [Google Scholar] [CrossRef]
- Brown, G.E.; Sreenivaprasad, S.; Timmer, L.W. Molecular characterization of slow-growing orange and key lime anthracnose strains of Colletotrichum from citrus as C. acutatum. Phytopathology 1996, 86, 523–527. [Google Scholar] [CrossRef]
- Martín, M.P.; García-Figueres, F.; Trapero, A. Indicadores específicos para detectar las especies de Colletotrichum causantes de la antracnosis de los olivos. Bol. San. Veg. Plagas 2002, 28, 43–50. [Google Scholar]
- Sharma, G.; Pinnaka, A.K.; Shenoy, B.D. Resolving the Colletotrichum siamense species complex using ApMat marker. Fungal Divers. 2015, 71, 247–264. [Google Scholar] [CrossRef]
- Liu, F.; Weir, B.S.; Damm, U.; Crous, P.W.; Wang, Y.; Liu, B.; Wang, M.; Zhang, M.; Cai, L. Unravelling Colletotrichum species associated with Camellia: Employing ApMat and GS loci to resolve species in the C. gloeosporioides complex. Persoonia 2015, 35, 63–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Silva, D.D.; Ades, P.K.; Crous, P.W.; Taylor, P.W.J. Colletotrichum species associated with chili anthracnose in Australia. Plant Pathol. 2017, 66, 254–267. [Google Scholar] [CrossRef]
- Cai, L.; Hyde, K.D.; Taylor, P.W.J.; Weir, B.S.; Waller, J.; Abang, M.M.; Zhang, J.Z.; Yang, Y.L.; Phoulivong, S.; Liu, Z.Y.; et al. A polyphasic approach for studying Colletotrichum. Fungal Divers. 2009, 39, 183–204. [Google Scholar]
- Moral, J.; Agustí-Brisach, C.; Agalliu, G.; de Oliveira, R.; Pérez-Rodríguez, M.; Roca, L.F.; Romero, J.; Trapero, A. Preliminary selection and evaluation of fungicides and natural compounds to control olive anthracnose caused by Colletotrichum species. Crop Prot. 2018, 114, 167–176. [Google Scholar] [CrossRef]
- Shivas, R.G.; Tan, Y.P.; Edwards, J.; Dinh, Q.; Maxwell, A.; Andjic, V.; Liberato, J.R.; Anderson, C.; Beasley, D.R.; Bransgrove, K.; et al. Colletotrichum species in Australia. Australas. Plant Pathol. 2016, 45, 447–464. [Google Scholar] [CrossRef]
- Lichtemberg, P.S.F.; Moral, J.; Morgan, D.P.; Felts, D.G.; Sanders, R.D.; Michailides, T.J. First report of anthracnose caused by Colletotrichum fioriniae and C. karstii in California pistachio orchards. Plant Dis. 2017, 101, 320. [Google Scholar] [CrossRef]
- Cabral, A.; Azinheira, H.G.; Talhinhas, P.; Batista, D.; Ramos, A.P.; Silva, M.C.; Oliveira, H.; Várzea, V. Pathological, morphological, cytogenomic, biochemical and molecular data support the distinction between Colletotrichum cigarro comb. et stat. nov. and C. kahawae. Plants 2020, 9, 502. [Google Scholar] [CrossRef] [Green Version]
- Msairi, S.; Chliyeh, M.; Touhami, A.O.; El Alaoui, A.; Selmaoui, K.; Benkirane, R.; Filali-Maltouf, A.; El Modafar, C.; Douira, A. First report on Colletotrichum lupini causing anthracnose disease in olive fruits in Morocco. Plant Cell Biotechnol. Mol. Biol. 2020, 21, 1–11. [Google Scholar]
- Diao, Y.Z.; Zhang, C.; Liu, F.; Wang, W.Z.; Liu, L.; Cai, L.; Liu, X.L. Colletotrichum species causing anthracnose disease of chili in China. Persoonia 2017, 38, 20–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassan, O.; Lee, Y.S.; Chang, T. Colletotrichum species associated with Japanese plum (Prunus salicina) anthracnose in South Korea. Sci. Rep. 2019, 9, 12089. [Google Scholar] [CrossRef] [PubMed] [Green Version]

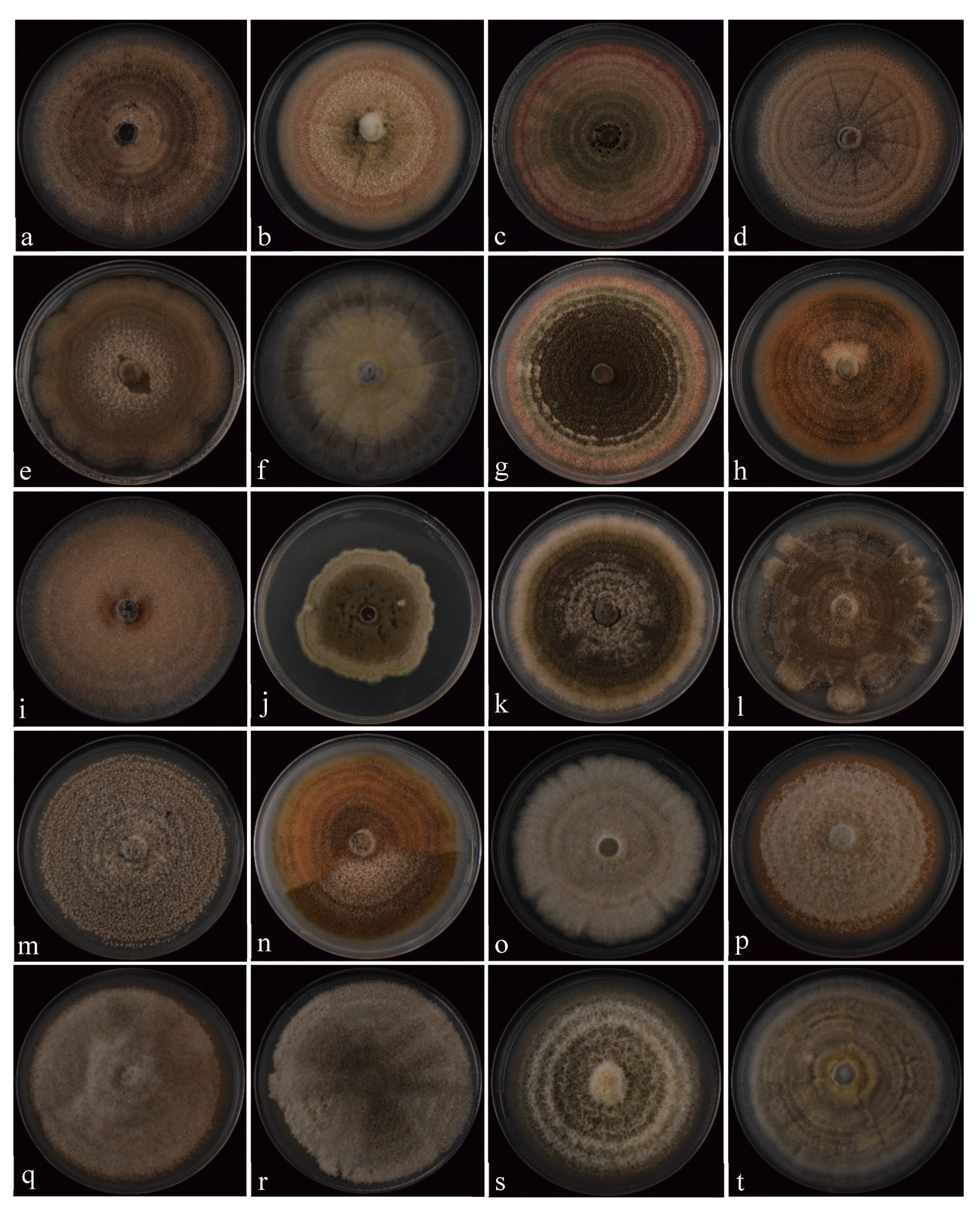
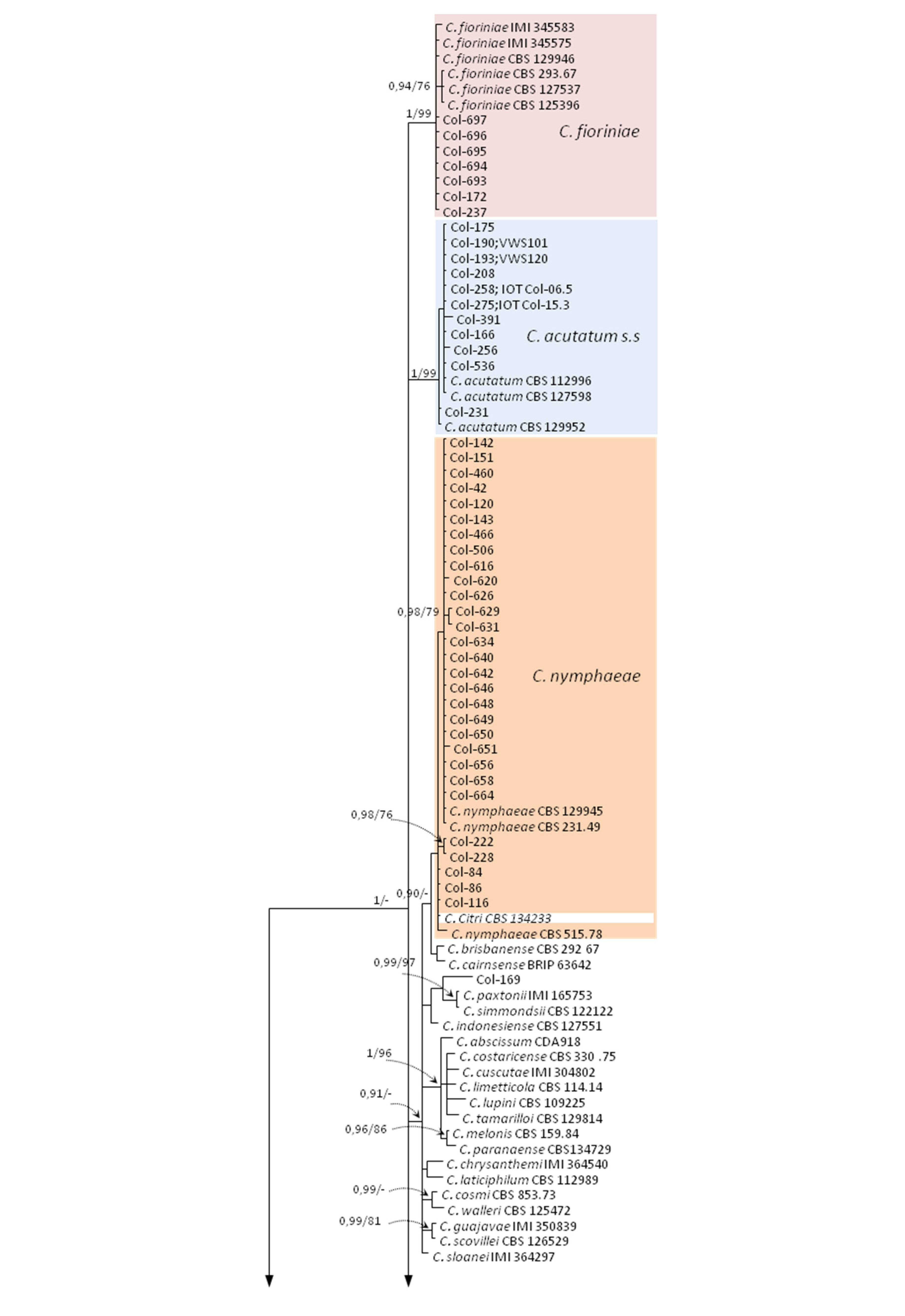
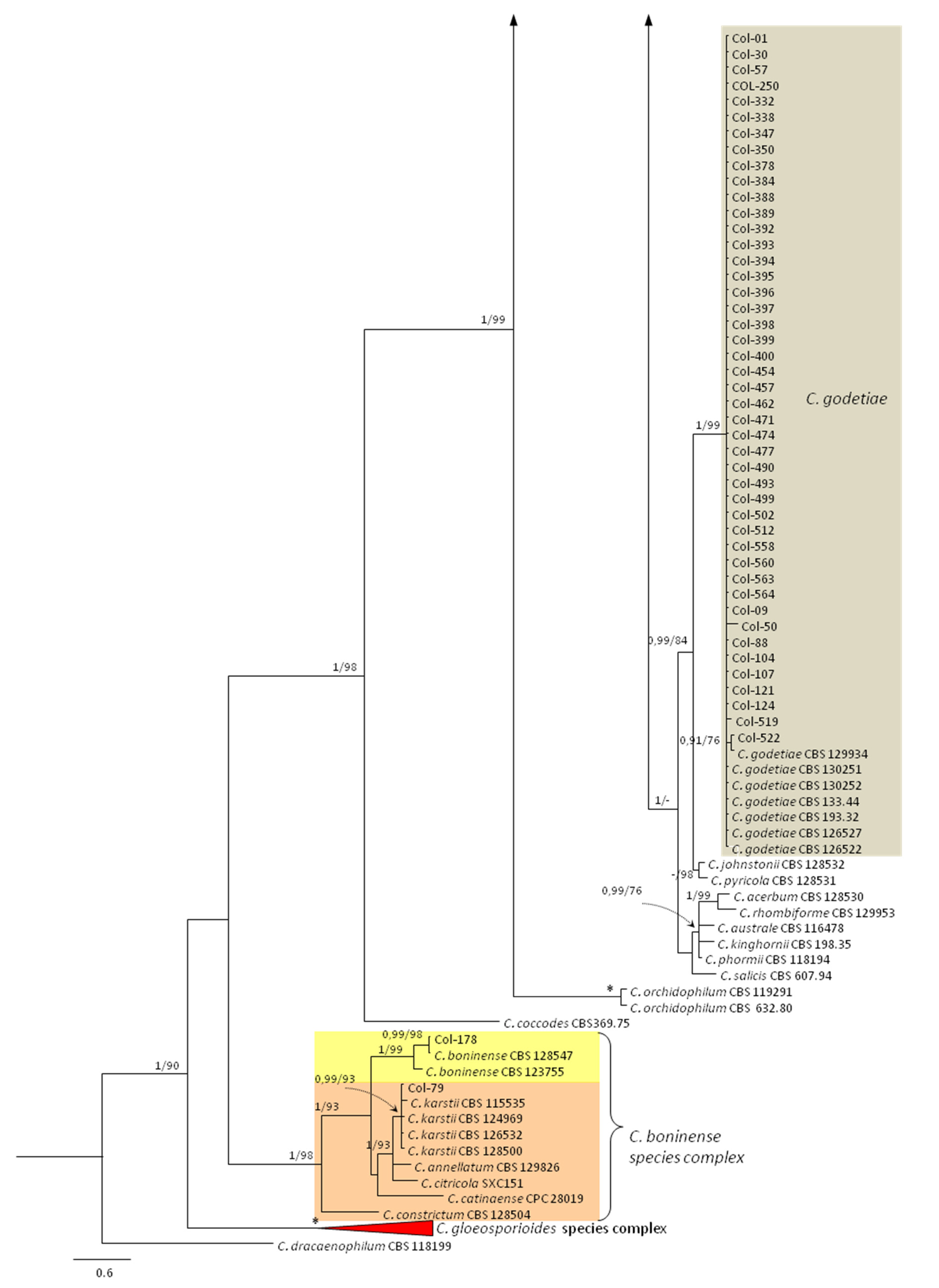
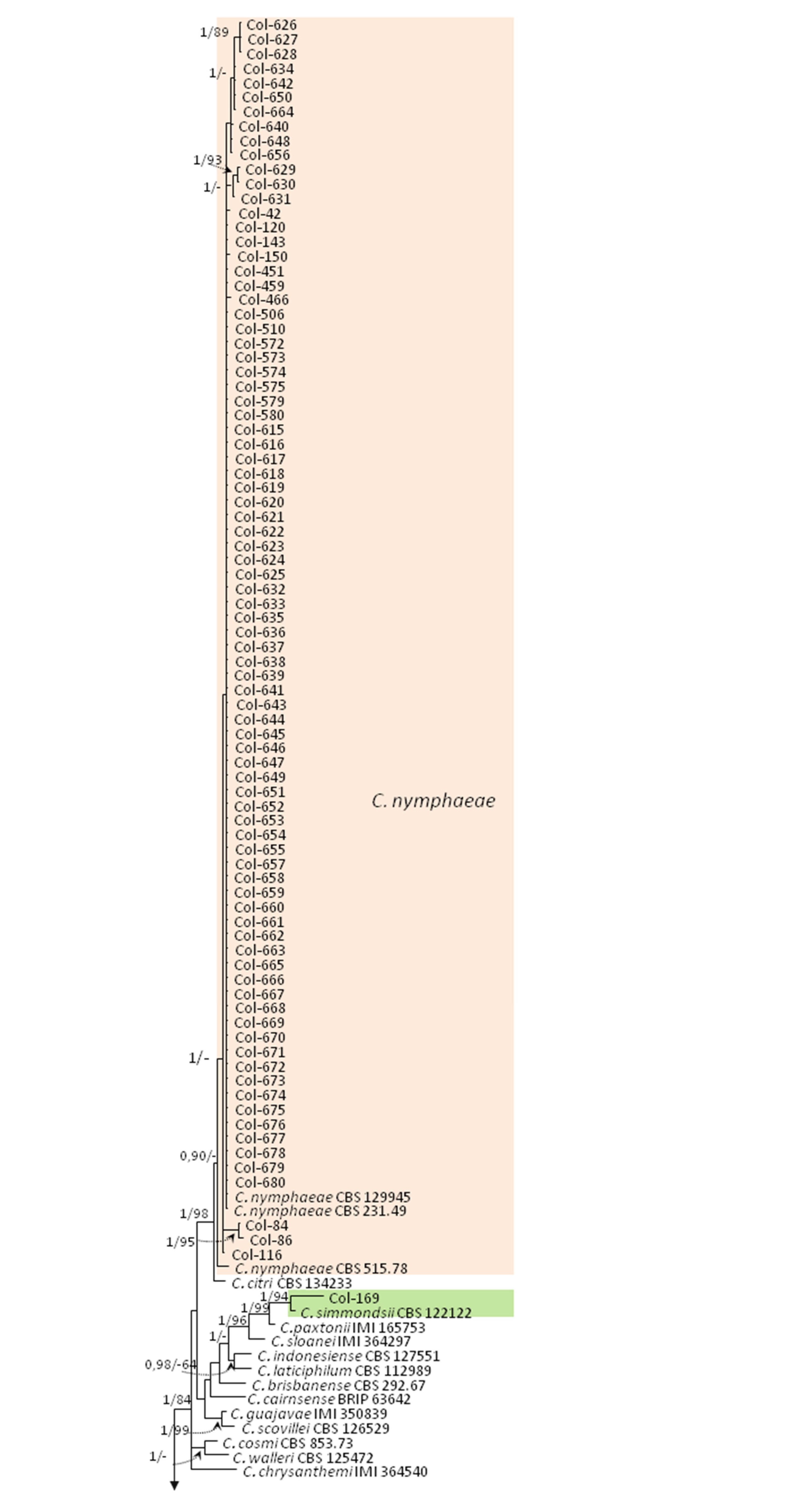
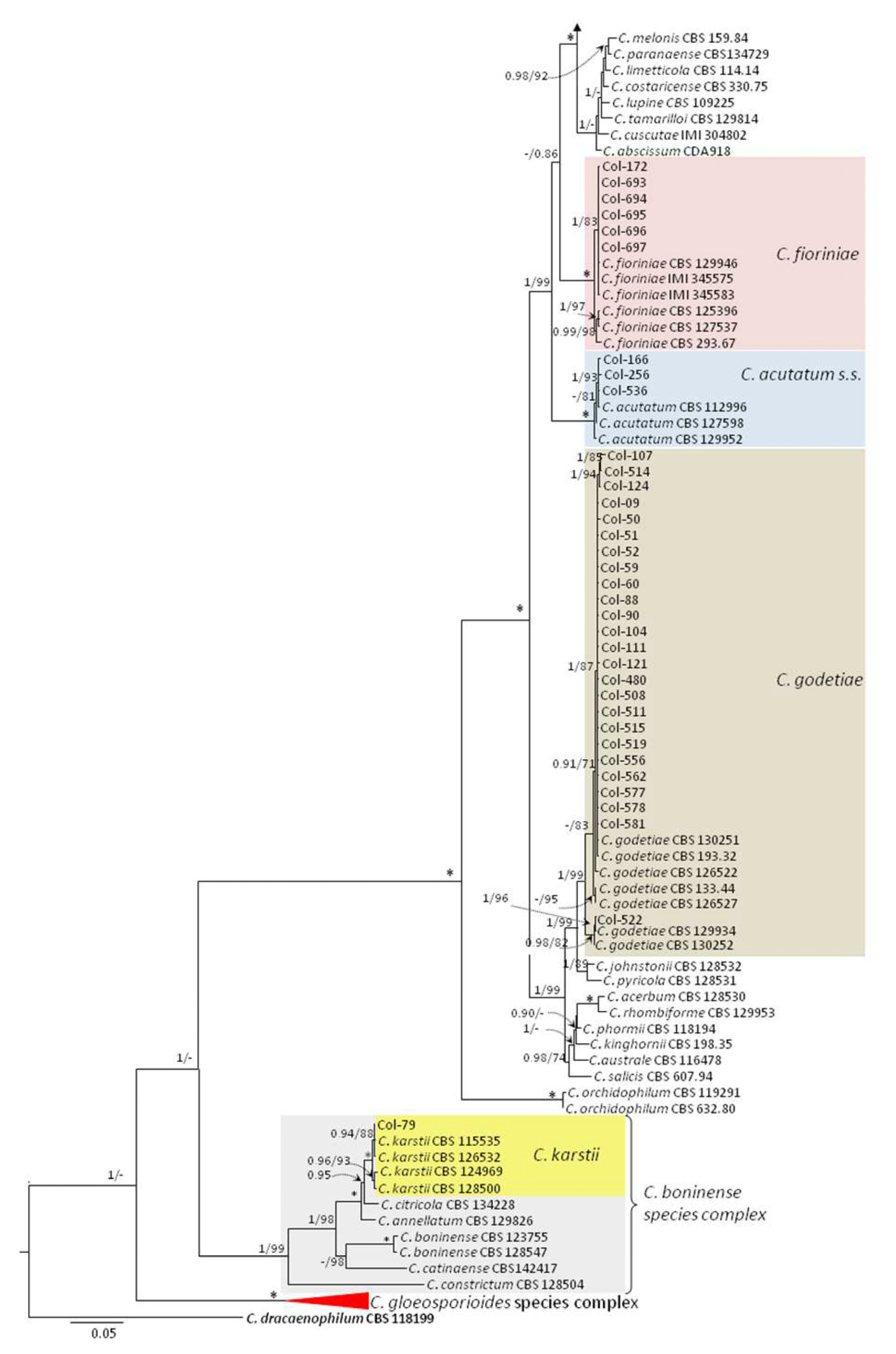
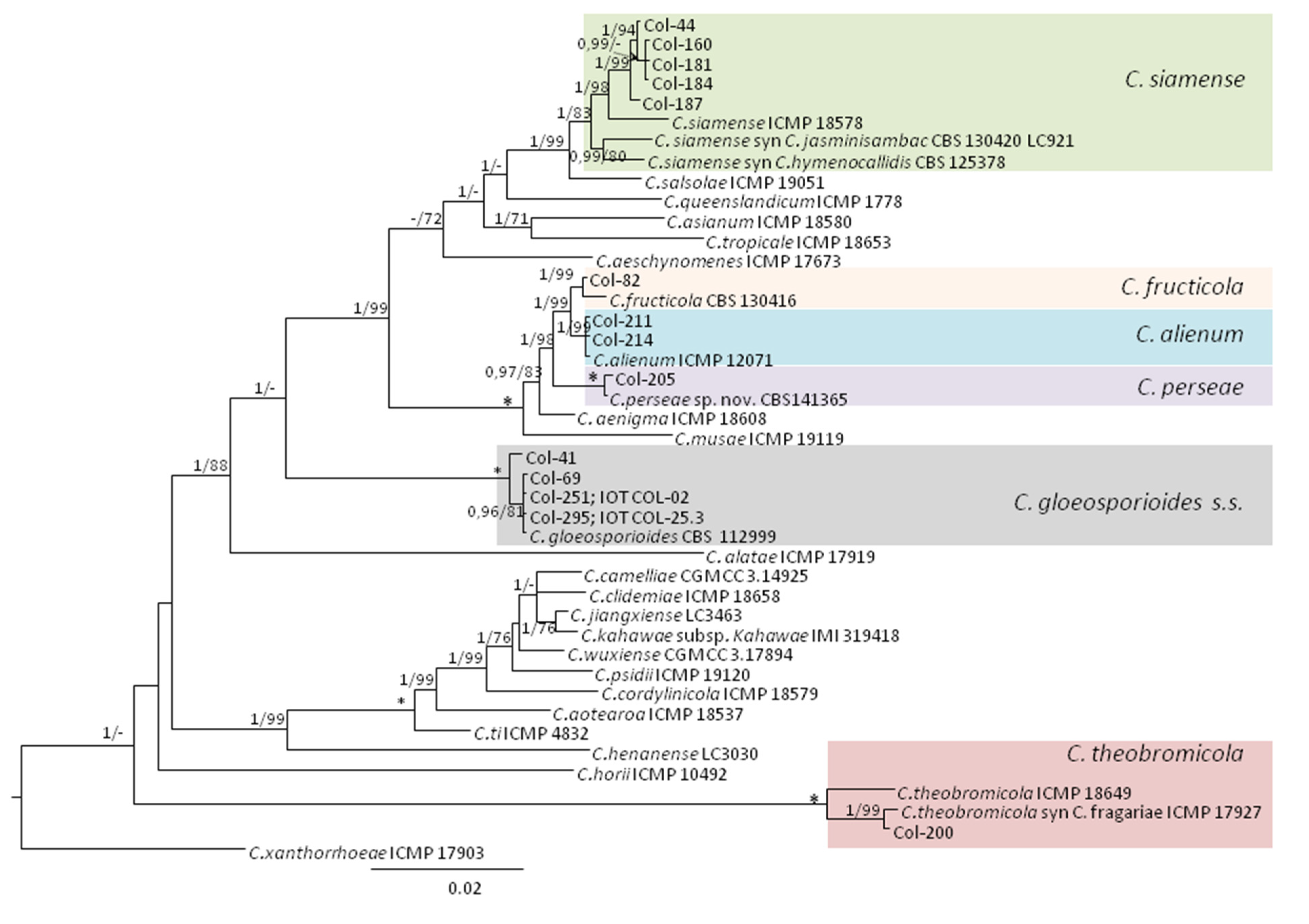
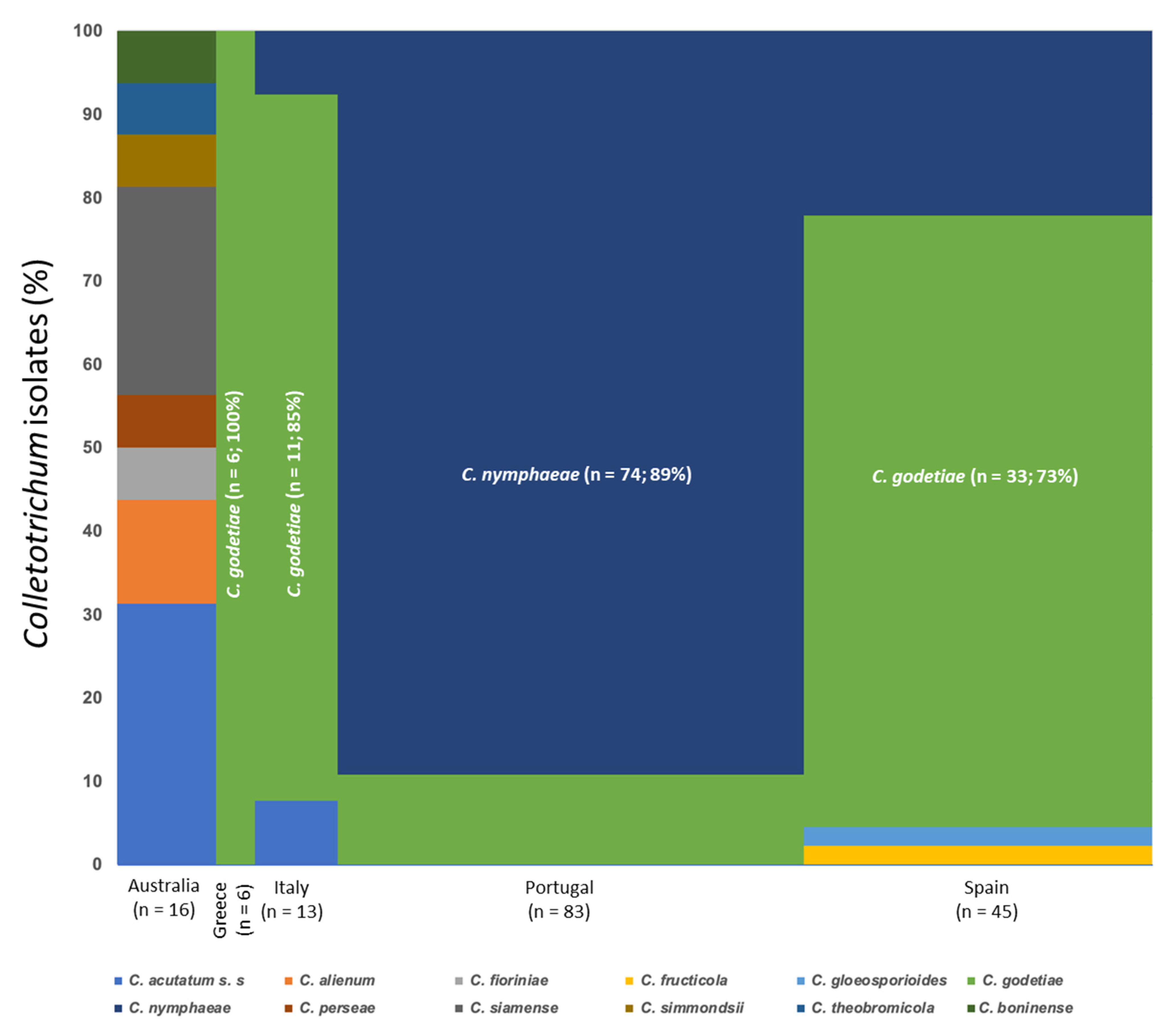
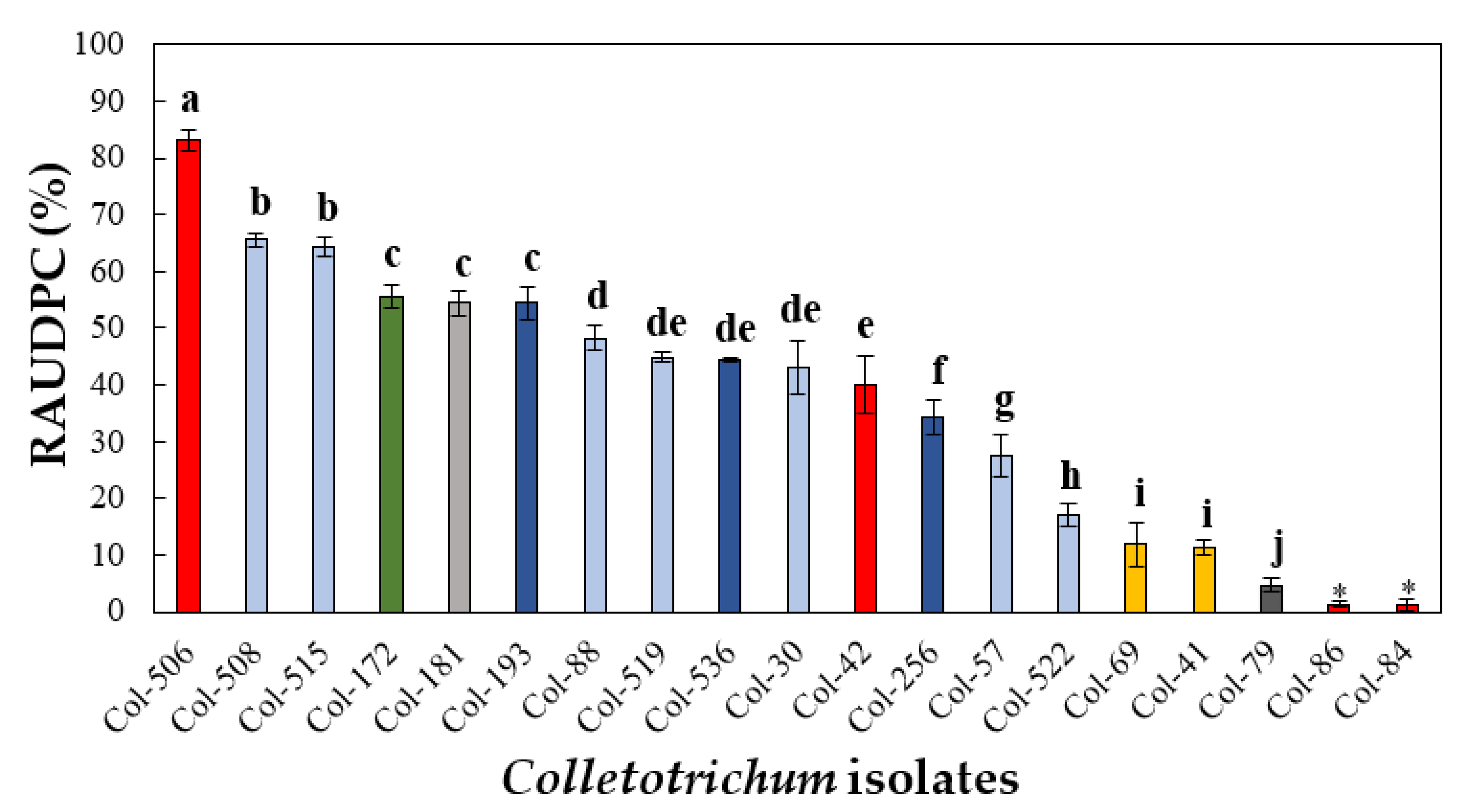

| Species | Isolate b,c | Origin, Year | Substrate, Host | GenBank Accession No. a | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ITS | TUB2 | ACT | CHS-1 | HIS3 | GAPDH | ApMat | ||||
| C. abscissum | COAD 1877T | Brazil, Cafelandia | Psidium guajava | KP843126 | KP843135 | KP843141 | KP843132 | KP843138 | KP843129 | - |
| C. acerbum | CBS 128530T, ICMP 12921, PRJ 1199.3 | New Zealand | Malus domestica | JQ948459 | JQ950110 | JQ949780 | JQ949120 | JQ949450 | JQ948790 | - |
| C. acutatum | Col-166; UWS-65 d,e | Australia; 2009 | Fruit, Olea europaea | MH685231 | MH713165 | MH717594 | MH801883 | MH713299 | MH717458 | - |
| Col-175; UWS-79 d,e | Australia; 2009 | Fruit, Olea europaea | MH685234 | MH713166 | - | - | - | - | - | |
| Col-190; UWS-101 | Australia; 2009 | Fruit, Olea europaea | MH685239 | MH713167 | - | - | - | - | - | |
| Col-193; UWS-120 f | Australia; 2009 | Fruit, Olea europaea | MH685240 | MH713168 | - | - | - | - | - | |
| Col-208;UWS-149 d,e | Australia; 2009 | Fruit, Olea europaea | MH685243 | MH713169 | - | - | - | - | - | |
| Col-231 | Uruguay; 2010 | Fruit, Olea europaea cv. Hojiblanca | MH685249 | MH713170 | - | - | - | - | - | |
| Col-256; IOT COL-04.1 f | Nabeul, Tunisia; 2010 | Fruit, Olea europea cv. Meski | KM594095 | KP197006 | MH717595 | MH801884 | MH713300 | MH717459 | - | |
| Col-258; IOT COL-06.5 | Takelsa, Tunisia; 2010 | Fruit, Olea europea cv. Arbequina | KM594093 | KP185116 | - | - | - | - | - | |
| Col-275; IOT COL-15.3 | Nabeul, Tunisia; 2010 | Fruit, Olea europea cv. Queslati | KM594101 | KP197011 | - | - | - | - | - | |
| Col-391 | Bari, Italy; 2012 | Fruit, Olea europea cv. Arbequina | MH685260 | MH713171 | - | - | - | - | - | |
| Col-536 d,f | Lebrija, Sevilla, Spain; 2014 | Fruit, Prunus dulcis | KY171894 | KY171902 | KY171910 | KY171918 | KY171926 | KY171934 | - | |
| CBS 112996, ATCC 56816, STE-U 5292T | Australia | Carica papaya | JQ005776 | JQ005860 | JQ005839 | JQ005797 | JQ005818 | JQ948677 | - | |
| CBS 129952, PT227, RB015 | Portugal | Olea europaea | JQ948364 | JQ950015 | JQ949685 | JQ949025 | JQ949355 | JQ948695 | - | |
| CBS 127598, 223/09 | South Africa | Olea europaea | JQ948363 | JQ950014 | JQ949684 | JQ949024 | JQ949354 | JQ948694 | - | |
| C. aenigma | ICMP 18608T | Israel | Persea americana | JX010244 | JX010389 | - | - | - | - | KM360143 |
| C. aeschynomenes | ICMP 17673T | USA | Aeschynomene virginica | JX010176 | JX010392 | - | - | - | - | KM360145 |
| C. alatae | ICMP 17919 | India | Dioscorea alata | JX010190 | JX010383 | - | - | - | - | KC888932 |
| C. alienum | Col-211;UWS-152 d,e | Australia; 2009 | Fruit, Olea europaea | MH685244 | MH713162 | - | - | - | - | MH717580 |
| Col-214;UWS-156 d,e | Australia; 2009 | Fruit, Olea europaea | MH685245 | MH713163 | - | - | - | - | MH717581 | |
| ICMP 12071T | New Zealand | Malus domestica | JX010251 | JX010411 | - | - | - | - | KM360144 | |
| C. annellatum | CBS 129826T | Colombia | Leaf, Hevea brasiliensis | JQ0055222 | JQ005656 | JQ005570 | JQ005396 | JQ005483 | JQ005309 | - |
| C. aotearoa | ICMP 18537T | New Zealand | Coprosma sp. | JX010205 | JX010420 | - | - | - | - | KC888930 |
| C. asianum | ICMP 18580T;CBS 130418 | Thailand | Coffea arabica | FJ972612 | JX010406 | - | - | - | - | FR718814 |
| C. australe | CBS 116478T | South Africa | Trachycarpus fortunei | JQ948455 | JQ950106 | JQ949776 | JQ949116 | JQ949446 | JQ948786 | |
| C. boninense | Col-178; UWS-82 d,e | Australia; 2009 | Fruit, Olea europaea | MH685235 | MH713152 | - | - | - | - | - |
| CBS 123755T,MAFF 305972 | Japan | Crinum asiaticum cv. sinicum | JQ005153 | JQ005588 | JQ005501 | JQ005327 | JQ005414 | JQ005240 | - | |
| CBS 128547, ICMP 10338 | New Zealand | Camellia sp. | JQ005159 | JQ005593 | JQ005507 | JQ005333 | JQ005420 | JQ005246 | - | |
| C. brisbanense | CBS 292.67T | Australia | Capsicum annuum | JQ948291 | JQ949942 | JQ949612 | JQ948952 | JQ949282 | JQ948621 | |
| C. cairnsense | BRIP 63642T, CBS 140847 | Australia | Capsicum annuum | KU923672 | KU923688 | KU923716 | KU923710 | KU923722 | KU923704 | - |
| C. catinaense | CBS 142417T; CPC 27978 | Italy, Catania | Citrus reticulata | KY856400 | KY856482 | KY855971 | KY856136 | KY856307 | KY856224 | - |
| C. chrysanthemi | IMI 364540, CPC 18930 | China | Chrysanthemun coronarium | JQ948273 | JQ949924 | JQ949594 | JQ948934 | JQ949264 | JQ948603 | - |
| C. citri | CBS 134233 | China | Citrus aurantiifolia | KC293581 | KC293661 | KY855973 | KY856138 | KY856309 | KC293741 | - |
| C. citricola | CBS 134228 | China | Citrus unchiu | KC293576 | KC293656 | KC293616 | KY856140 | KY856311 | KC293736 | - |
| C. clidemiae | ICMP 18658T | Hawaii, USA | Clidemia hirta | JX010265 | JX010438 | - | - | - | - | KC888929 |
| C. coccodes | CBS 369.75T | The Netherlands | Solanum tuberosum | HM171679 | JX546873 | - | - | - | - | - |
| C. constrictum | CBS 128504 | New Zealand | Citrus limon | JQ005238 | JQ005672 | JQ005586 | JQ005412 | KY856313 | JQ005325 | - |
| C. cordylinicola | ICMP 18579T | Thailand | Cordyline fruticosa | JX010226 | JX010440 | - | - | - | - | JQ899274 |
| C. cosmi | CBS 853.73, PD 73/856T | The Netherlands | Cosmos sp. | JQ948274 | JQ949925 | JQ949595 | JQ948935 | JQ949265 | JQ948604 | - |
| C. costaricense | CBS 330.75T | Costa Rica | Coffea arabica | JQ948180 | JQ949831 | JQ949501 | JQ949120 | JQ949450 | JQ948790 | - |
| C. cuscutae | IMI 304802, CPC 18873T | Dominica | Cuscuta sp. | JQ948195 | JQ949846 | JQ949516 | JQ949025 | JQ949355 | JQ948695 | - |
| C. dracaenophilum | CBS 118199 | China | Dracaena | JX519222 | JX519247 | JX519238 | JX519230 | JX546756 | JX546707 | - |
| C. fioriniae | Col-172;UWS-70 d,e,f | Australia; 2009 | Fruit, Olea europaea | MH685233 | MH713172 | MH717596 | MH801885 | MH713301 | MH717460 | - |
| Col-237 | Uruguay; 2010 | Fruit, Olea europaea cv. Arbequina | MH685250 | MH801882 | - | - | - | - | - | |
| Col-693 d | California, USA; 2017 | Fruit, Olea europaea | MH685372 | MH713173 | MH717597 | MH801886 | MH713302 | MH717461 | - | |
| Col-694 d | California, USA; 2017 | Fruit, Olea europaea | MH685373 | MH713174 | MH717598 | MH801887 | MH713303 | MH717462 | - | |
| Col-695 d | California, USA; 2017 | Fruit, Olea europaea | MH685374 | MH713175 | MH717599 | MH801888 | MH713304 | MH717463 | - | |
| Col-696 d | California, USA; 2017 | Fruit, Olea europaea | MH685375 | MH713176 | MH717600 | MH801889 | MH713305 | MH717464 | - | |
| Col-697 d | California, USA; 2017 | Fruit, Olea europaea | MH685376 | MH713177 | MH717601 | MH801890 | MH713306 | MH717465 | - | |
| IMI 345583, CPC 18889 | USA | Fragaria × ananassa | JQ948333 | JQ949984 | JQ949654 | JQ005797 | JQ005818 | JQ948677 | - | |
| IMI 345575, CPC 18888 | USA | Fragaria × ananassa | JQ948332 | JQ949983 | JQ949653 | JQ949116 | JQ949446 | JQ948786 | - | |
| CBS 125396; GJS 08-140A | USA | Malus domestica | JQ948299 | JQ949950 | JQ949620 | JQ948952 | JQ949282 | JQ948621 | - | |
| CBS 129946, PT170, RB021 | Portugal | Olea europaea | JQ948342 | JQ949993 | JQ949663 | JQ949024 | JQ949354 | JQ948694 | - | |
| CBS 293.67, DPI 13120 | Australia | Persea americana | JQ948310 | JQ949961 | JQ949631 | JQ948934 | JQ949264 | JQ948603 | - | |
| CBS 127537, STE-U 5289 | USA | Vaccinium sp. | JQ948318 | JQ949969 | JQ949639 | JQ948935 | JQ949265 | JQ948604 | - | |
| C. fructicola | Col-82 | Valencia, Spain; 2003 | Leaf, Olea europaea | MH685214 | MH713153 | MH713292 | MH713285 | MH713414 | MH717489 | MH717582 |
| CBS 130416, ICMP 18581 | Thailand | Coffea arabica | JX010165 | JX010405 | - | - | - | - | JQ807838 | |
| C. gloeosporioides | Col-41 d,e,f | Montsia, Tarragona, Spain; 1999 | Fruit, Olea europaea | MH685203 | MH713154 | MH713293 | MH713286 | MH713415 | MH717490 | MH717583 |
| Col-69 d,e,f | Fuente la Palomera, Córdoba, Spain; 2001 | Citrus sinensis | MH685212 | MH713155 | MH713294 | MH713287 | MH713416 | MH717491 | MH717584 | |
| Col-251; IOT COL-02 | Nabeul, Tunisia; 2010 | Fruit, Olea europaea | KM594085 | KP176441 | - | - | - | - | MH717585 | |
| Col-295; IOT COL-25.3 | Nabeul, Tunisia; 2010 | Fruit, Olea europea cv. Meski | KM594112 | KP197021 | - | - | - | - | MH717586 | |
| CBS 112999 | Italy | Citrus sinensis | JQ005152 | JQ005587 | JQ005500 | JQ005326 | JQ005413 | JQ005239 | JQ807843 | |
| C. godetiae | Col-1 d | Almodóvar, Córdoba, Spain; 1998 | Fruit, Olea europaea cv. Hojiblanca | MH685200 | MH713003 | - | - | - | - | - |
| Col-9 d,e,f | Antequera, Málaga, Spain: 1998 | Fruit, Olea europea cv. Hojiblanca | MH685201 | MH713004 | MH717602 | MH713178 | MH713307 | MH717466 | - | |
| Col-30 d,e,f | Llanos D. Juan, Córdoba, Spain; 1998 | Fruit, Olea europaea cv. Hojiblanca | MH685202 | MH713005 | - | - | - | - | - | |
| Col-50 d,e | Lucena, Córdoba, Spain, 1999 | Fruit, Olea europea | MH685206 | MH713006 | MH717603 | MH713179 | MH713308 | MH717467 | - | |
| Col-51 d,e | Lucena, Córdoba, Spain, 1999 | Fruit, Olea europea | MH685207 | MH713007 | MH717604 | MH713180 | MH713309 | MH717468 | - | |
| Col-52 d | Antequera, Málaga, Spain; 1999 | Fruit, Olea europea | MH685208 | MH713008 | MH717605 | MH713181 | MH713310 | MH717469 | - | |
| Col-57 d,e,f | Archidona, Málaga, Spain; 2002 | Fruit, Olea europaea | MH685209 | MH713009 | - | - | - | - | - | |
| Col-59 d,e | Archidona, Málaga, Spain; 2001 | Fruit, Olea europaea cv. Hojiblanca | MH685210 | MH713010 | MH717606 | MH713182 | MH713311 | MH717470 | - | |
| Col-60 d,e | Archidona, Málaga, Spain; 2001 | Fruit, Olea europaea cv. Hojiblanca | MH685211 | MH713011 | MH717607 | MH713183 | MH713312 | MH717471 | - | |
| Col-88 d,e | Montilla, Córdoba Spain; 2004 | Fruit, Olea europaea cv. Picudo | MH685217 | MH713012 | MH717608 | MH713184 | MH713313 | MH717472 | - | |
| Col-90 | CIFA Cabra, Córdoba Spain; 2004 | Fruit, Olea europaea cv. Picudo | MH685218 | MH713013 | MH717609 | MH713185 | MH713314 | MH717473 | - | |
| Col-104 | Cabra, Córdoba, Spain; 2014 | Fruit, Olea europaea cv. Picudo | MH685219 | MH713014 | MH717610 | MH713186 | MH713315 | MH717474 | - | |
| Col-107 | La Rambla, Córdoba, Spain; | Fruit, Olea europaea cv. Hojiblanca | MH685220 | MH713015 | MH717611 | MH713187 | MH713316 | MH717475 | - | |
| Col-111 | Mengíbar, Jaén, Spain; 2014 | Fruit, Olea europaea cv. Ocal | MH685221 | MH713016 | MH717612 | MH713188 | MH713317 | MH717476 | - | |
| Col-121 | Montilla, Córdoba, Spain; 2014 | Fruit, Olea europaea cv. Hojiblanca | MH685224 | MH713017 | MH717613 | MH713189 | MH713318 | MH717477 | - | |
| Col-124 | Puente Genil, Córdoba, Spain; 2014 | Fruit, Olea europaea | MH685225 | MH713018 | MH717614 | MH713190 | MH713319 | MH717478 | - | |
| Col-250 | El Pedroso, Sevilla, Spain; 2011 | Fruit, Pistacia terebinthus | MH685251 | MH713019 | - | - | - | - | - | |
| Col-332 | Parga, Greece; 2012 | Fruit, Olea europaea | MH685252 | MH713020 | - | - | - | - | - | |
| Col-338 | Parga, Greece; 2012 | Fruit, Olea europaea | MH685253 | MH713021 | - | - | - | - | - | |
| Col-347 | Parga, Greece; 2012 | Fruit, Olea europaea | MH685254 | MH713022 | - | - | - | - | - | |
| Col-350 | Parga, Greece; 2012 | Fruit, Olea europaea | MH685255 | MH713023 | - | - | - | - | - | |
| Col-378 | Parga, Greece; 2012 | Fruit, Olea europaea | MH685256 | MH713024 | - | - | - | - | - | |
| Col-384 | Parga, Greece; 2012 | Fruit, Olea europaea | MH685257 | MH713025 | - | - | - | - | - | |
| Col-388 | Bari, Italy; 2012 | Fruit, Olea europea cv. Arbosana | MH685258 | MH713026 | - | - | - | - | - | |
| Col-389 | Bari, Italy; 2012 | Fruit, Olea europea cv. Arbequina | MH685259 | MH713027 | - | - | - | - | - | |
| Col-392 | Bari, Italy; 2012 | Fruit, Olea europea cv. Cellina di Nardò | MH685261 | MH713028 | - | - | - | - | - | |
| Col-393 | Bari, Italy; 2012 | Fruit, Olea europea cv. Cellina di Nardò | MH685262 | MH713029 | - | - | - | - | ||
| Col-394 | Bari, Italy; 2012 | Fruit, Olea europea cv. Cellina di Nardò | MH685263 | MH713030 | - | - | - | - | - | |
| Col-395 | Bari, Italy; 2012 | Fruit, Olea europea cv. Ogliarola Salentina | MH685264 | MH713031 | - | - | - | - | - | |
| Col-396 | Bari, Italy; 2012 | Fruit, Olea europea cv. Ogliarola Salentina | MH685265 | MH713032 | - | - | - | - | - | |
| Col-397 | Bari, Italy; 2012 | Fruit, Olea europea cv. Cellina di Nardò | MH685266 | MH713033 | - | - | - | - | - | |
| Col-398 | Bari, Italy; 2012 | Fruit, Olea europea cv. Ogliarola Salentina | MH685267 | MH713034 | - | - | - | - | - | |
| Col-399 | Bari, Italy; 2012 | Fruit, Olea europea cv. Ogliarola Salentina | MH685268 | MH713035 | - | - | - | - | - | |
| Col-400 | Bari, Italy; 2012 | Fruit, Olea europea cv. Cellina di Nardò | MH685269 | MH713036 | - | - | - | - | - | |
| Col-454 | Jerez, Cádiz, Spain; 2013 | Fruit, Olea europaea cv. Arbequina | MH685271 | MH713037 | - | - | - | - | - | |
| Col-457 | Jerez, Cádiz, Spain; 2013 | Fruit, Olea europaea cv. Hojiblanca | MH685272 | MH713038 | - | - | - | - | -- | |
| Col-462 | Jerez, Cádiz, Spain; 2013 | Fruit, Olea europaea cv. Arbequina | MH685275 | MH713039 | - | - | - | - | - | |
| Col-471 | Montilla, Córdoba, Spain; 2013 | Fruit, Olea europaea cv. Picudo | MH685277 | MH713040 | - | - | - | - | - | |
| Col-474 | Montilla, Córdoba, Spain; 2013 | Fruit, Olea europaea cv. Hojiblanca | MH685278 | MH713041 | - | - | - | - | - | |
| Col-477 | Castro del Río, Córdoba, Spain; 2013 | Fruit, Olea europaea cv. Picudo | MH685279 | MH713042 | - | - | - | - | - | |
| Col-480 | Castro del Río, Córdoba, Spain; 2013 | Fruit, Olea europaea cv. Picudo | MH685280 | MH713043 | MH717615 | MH713191 | MH713320 | MH717479 | - | |
| Col-490 | Jerez, Cádiz, Spain; 2013 | Fruit, Olea europaea cv. Hojiblanca | MH685281 | MH713044 | - | - | - | - | - | |
| Col-493 | Jerez, Cádiz, Spain; 2013 | Fruit, Olea europaea cv. Hojiblanca | MH685282 | MH713045 | - | - | - | - | - | |
| Col-499 | Montilla, Córdoba, Spain; 2013 | Fruit, Olea europaea cv. Hojiblanca | MH685283 | MH713046 | - | - | - | - | - | |
| Col-502 | Fuente la Palomera, Córdoba, Spain; 2013 | Fruit, Olea europaea | MH685284 | MH713047 | - | - | - | - | - | |
| Col-508d,f | Hornachuelos, Córdoba, Spain; 2014 | Fruit, Olea europaea cv. Arbequina | KY171892 | KY171900 | KY171908 | KY171916 | KY171924 | KY171932 | - | |
| Col-511 | Hornachuelos, Córdoba, Spain; 2014 | Fruit, Olea europaea cv. Picual | MH685286 | MH713048 | MH717616 | MH713192 | MH713321 | MH717480 | - | |
| Col-512 | Hornachuelos, Córdoba, Spain; 2014 | Fruit, Olea europaea cv. Picual | MH685287 | MH713049 | - | - | - | - | - | |
| Col-514 | Córdoba, Spain; 2014 | Fruit, Olea europaea cv. Picual | MH685288 | MH713050 | MH717617 | MH713193 | MH713322 | MH717481 | - | |
| Col-515 f | Córdoba, Spain; 2014 | Fruit, Olea europaea cv. Picual | MH685289 | MH713051 | MH717618 | MH713194 | MH713323 | MH717482 | - | |
| Col-519 f | Córdoba, Spain; 2014 | Fruit, Olea europaea cv. Hojiblanca | MH685290 | MH713052 | MH717619 | MH713195 | MH713324 | MH717483 | - | |
| Col-522 d,f | Lebrija, Sevilla, Spain; 2014 | Fruit, Prunus dulcis | KY171893 | KY171901 | KY171909 | KY171917 | KY171925 | KY171933 | - | |
| Col-556 | Beja, Portugal; 2014 | Fruit, Olea europaea | MH685291 | MH713053 | MH717620 | MH713196 | MH713325 | MH717484 | - | |
| Col-558 | Beja, Portugal; 2014 | Fruit, Olea europaea | MH685292 | MH713054 | - | - | - | - | - | |
| Col-560 | Beja, Portugal; 2014 | Fruit, Olea europaea | MH685293 | MH713055 | - | - | - | - | - | |
| Col-562 | Beja, Portugal; 2014 | Fruit, Olea europaea | MH685294 | MH713056 | MH717621 | MH713197 | MH713326 | MH717485 | - | |
| Col-563 | Beja, Portugal; 2014 | Fruit, Olea europaea | MH685295 | MH713057 | - | - | - | - | - | |
| Col-564 | Beja, Portugal; 2014 | Fruit, Olea europaea | MH685296 | MH713058 | - | - | - | - | - | |
| Col-577 | Montesandinha, Portugal; 2014 | Fruit, Olea europaea cv. Arbequina | MH685301 | MH713059 | MH717622 | MH713198 | MH713327 | MH717486 | - | |
| Col-578 | Capela, Portugal; 2014 | Fruit, Olea europaea cv. Arbequina | MH685302 | MH713060 | MH717623 | MH713199 | MH713328 | MH717487 | - | |
| Col-581 | Montesandinha, Portugal; 2014 | Fruit, Olea europaea cv. Arbequina | MH685305 | MH713061 | MH717624 | MH713200 | MH713329 | MH717488 | - | |
| CBS 133.44T | Denmark | Clarkia hybrida | JQ948402 | JQ950053 | JQ949723 | JQ949063 | JQ949393 | JQ948733 | - | |
| CBS 130251, OL 10, IMI 398854 | Italy | Olea europaea | JQ948413 | JQ950064 | JQ949734 | JQ949074 | JQ949404 | JQ948744 | - | |
| CBS 193.32 | Greece | Olea euroapea | JQ948415 | JQ950066 | JQ949736 | JQ949076 | JQ949406 | JQ948746 | - | |
| CBS 130252, IMI 398855, OL 20 | Italy | Olea europaea | JQ948414 | JQ950065 | JQ949735 | JQ949075 | JQ949405 | JQ948745 | - | |
| CBS 126527, PD 93/1748 | United Kingdom | Prunus avium | JQ948408 | JQ950059 | JQ949729 | JQ949069 | JQ949399 | JQ948739 | - | |
| CBS 126522, PD 88/472, BBA 70345 | The Netherlands | Prunus cerasus | JQ948411 | JQ950062 | JQ949732 | JQ949072 | JQ949402 | JQ948742 | - | |
| CBS 129934, ALM-IKS-7Q | Israel | Prunus dulcis | JQ948431 | JQ950082 | JQ949752 | JQ949092.1 | JQ949422 | JQ948762 | - | |
| C. guajavae | IMI 350839, CPC 18893T | India | Psidium guajava | JQ948270 | JQ949921 | JQ949591 | JQ948931 | JQ949261 | JQ948600 | - |
| C. henanense | LC3030, CGMCC 3.17354, LF238T | China | Camellia sinensis | KJ955109 | KJ955257 | - | - | - | - | KJ954524 |
| C. horii | ICMP 10492T | Japan | Diospyros kaki | GQ329690 | JX010450 | - | - | - | - | JQ807840 |
| C. indonesiense | CBS 127551, CPC 14986T | Indonesia | Eucalyptus sp. | JQ948288 | JQ949939 | JQ949609 | JQ948949 | JQ949279 | JQ948618 | - |
| C. jiangxiense | LC3463, CGMCC 3.17363, LF687T | China | Camellia. sinensis | KJ955201 | KJ955348 | - | - | - | - | KJ954607 |
| C. johnstonii | CBS 128532, ICMP 12926, PRJ 1139.3T | New Zealand | Solanum lycopersicum | JQ948444 | JQ950095 | JQ949435 | JQ949105 | JQ949105 | JQ948775 | - |
| C. kahawae subsp. kahawae | IMI 319418, ICMP 17816 | Kenya | Coffea arabica | JX010231 | JX010444 | - | - | - | - | JQ894579 |
| C. karstii | Col-79 d,e | Huelva, Spain | Citrus sp. | MH685213 | MH713151 | MH713295 | MH713288 | MH713417 | MH717492 | - |
| CBS 126532 | South Africa | Citrus sp. | JQ005209 | JQ005643 | JQ005557 | JQ005383 | JQ005470 | JQ005296 | - | |
| CBS 128500, ICMP 18585 | New Zealand | Fruit, Annona cherimola | JQ005202 | JQ005636 | JQ005550 | JQ005376 | JQ005463 | JQ005289 | - | |
| CBS 124969, LCM 232 | Panama | Leaf, Quercus salicifolia | JQ005179 | JQ005613 | JQ005527 | JQ005353 | JQ005440 | JQ005266 | - | |
| CBS 115535, STE-U 5210 | Portugal, Madeira | Protea obtusifolia | JQ005214 | JQ005648 | JQ005562 | JQ005388 | JQ005475 | JQ005301 | - | |
| C. kinghornii | CBS 198.35T | United Kingdom | Phormium sp. | JQ948454 | JQ950105 | JQ949775 | JQ949115 | JQ949445 | JQ948785 | - |
| C. laticiphilum | CBS 112989, IMI 383015, STE-U 5303T | India | Hevea basiliensis | JQ948289 | JQ949940 | JQ949610 | JQ948950 | JQ949280 | JQ948619 | - |
| C. limetticola | CBS 114.14T | Florida, USA | Citrus aurantifolia | JQ948193 | JQ949844 | JQ949514 | JQ948854 | JQ949184 | JQ948523 | - |
| C. lupini | CBS 109225; BBA 70884T | Ukraine | Lupinus albus | JQ948155 | JQ949806 | JQ949476 | JQ948816 | JQ949146 | JQ948485 | - |
| C. melonis | CBS 159.84T | Brazil | Cucumis melo | JQ948194 | JQ949845 | JQ949515 | JQ948855 | JQ949185 | JQ948524 | - |
| C. musae | ICMP 19119, CBS 116870 | USA | Musa sp. | JX010146 | HQ596280 | - | - | - | - | KC888926 |
| C. nymphaeae | Col-42 d,e | Tarragona, Spain, 1999 | Fruit, Olea europaea | MH685204 | MH713062 | MH717625 | MH713201 | MH713330 | MH717496 | - |
| Col-84 d,e,f | Sevilla, Spain; 2004 | Fruit, Fragaria × ananassa | MH685215 | MH713063 | MH717626 | MH713202 | MH713331 | MH717497 | - | |
| Col-86 d,e,f | Sevilla, Spain; 2004 | Fruit, Fragaria × ananassa | MH685216 | MH713064 | MH717627 | MH713203 | MH713332 | MH717498 | - | |
| Col-116 | Montefalco, Perugia, Italy; 2014 | Fruit, Olea europaea cv. Moraiolo | MH685222 | MH713065 | MH717628 | MH713204 | MH713333 | MH717499 | - | |
| Col-120 | Navalvillar de Pela, Badajoz, Spain; 2014 | Fruit, Olea europaea cv. Verdial de Badajoz | MH685223 | MH713066 | MH717629 | MH713205 | MH713334 | MH717500 | - | |
| Col-142 | Elvas, Portugal; 2008 | Fruit, Olea euroapea | MH685226 | MH713067 | - | - | - | - | - | |
| Col-143 | Elvas, Portugal; 2008 | Fruit, Olea europaea | MH685227 | MH713068 | MH717630 | MH713206 | MH713335 | MH717501 | - | |
| Col-150 | Puebla de Guzman, Huelva, Spain; 2008 | Fruit, Olea europaea | MH685228 | MH713069 | MH717631 | MH713207 | MH713336 | MH717502 | - | |
| Col-151 | Puebla de Guzman, Huelva, Spain; 2008 | Fruit, Olea europaea | MH685229 | MH713070 | - | - | - | - | - | |
| Col-222 | Caçapava, Brasil; 2010 | Fruit, Olea europaea cv. Arbequina | MH685246 | MH713071 | - | - | - | - | - | |
| Col-228 | Uruguay; 2010 | Fruit, Olea europaea | MH685248 | MH713072 | - | - | - | - | - | |
| Col-451 | Jerez, Cádiz, Spain; 2013 | Fruit, Olea europaea cv. Arbequina | MH685270 | MH713073 | MH717632 | MH713208 | MH713337 | MH717503 | - | |
| Col-459 | Jerez, Cádiz, Spain; 2013 | Fruit, Olea europaea cv. Hojiblanca | MH685273 | MH713074 | MH717633 | MH713209 | MH713338 | MH717504 | - | |
| Col-460 | Jerez, Cádiz, Spain; 2013 | Fruit, Olea europaea cv. Hojiblanca | MH685274 | MH713075 | - | - | - | - | - | |
| Col-466 | Jerez, Cádiz, Spain; 2013 | Fruit, Olea europaea cv. Arbequina | MH685276 | MH713076 | MH717634 | MH713210 | MH713339 | MH717505 | - | |
| Col-506 d,f | Hornachuelos, Córdoba, Spain; 2014 | Fruit, Olea europaea cv. Arbequina | KY171891 | KY171899 | KY171907 | KY171915 | KY171923 | KY171931 | - | |
| Col-510 | Hornachuelos, Córdoba, Spain; 2014 | Fruit, Olea europaea cv. Picual | MH685285 | MH713077 | MH717635 | MH713211 | MH713340 | MH717506 | - | |
| Col-572 | Montesardinha,Portugal; 2014 | Fruit, Olea europaea cv. Picual | MH685297 | MH713078 | MH717636 | MH713212 | MH713341 | MH717507 | - | |
| Col-573 | Capela, Portugal; 2014 | Fruit, Olea europaea cv. Picual | MH685298 | MH713079 | MH717637 | MH713213 | MH713342 | MH717508 | - | |
| Col-574 | Montesardinha,Portugal; 2014 | Fruit, Olea europaea cv. Arbequina | MH685299 | MH713080 | MH717638 | MH713214 | MH713343 | MH717509 | - | |
| Col-575 | Montesardinha,Portugal; 2014 | Fruit, Olea europaea cv. Picual | MH685300 | MH713081 | MH717639 | MH713215 | MH713344 | MH717510 | - | |
| Col-579 | Capela, Portugal; 2014 | Fruit, Olea europaea cv. Picual | MH685303 | MH713082 | MH717640 | MH713216 | MH713345 | MH717511 | - | |
| Col-580 | Montesardinha,Portugal; 2014 | Fruit, Olea europaea cv. Picual | MH685304 | MH713083 | MH717641 | MH713217 | MH713346 | MH717512 | - | |
| Col-615 | Portugal; 2016 | Fruit, Olea europaea | MH685306 | MH713084 | MH717642 | MH713218 | MH713347 | MH717513 | - | |
| Col-616 | Portugal; 2016 | Fruit, Olea europaea | MH685307 | MH713085 | MH717643 | MH713219 | MH713348 | MH717514 | - | |
| Col-617 | Portugal; 2016 | Fruit, Olea europaea | MH685308 | MH713086 | MH717644 | MH713220 | MH713349 | MH717515 | - | |
| Col-618 | Portugal; 2016 | Fruit, Olea europaea | MH685309 | MH713087 | MH717645 | MH713221 | MH713350 | MH717516 | - | |
| Col-619 | Portugal; 2016 | Fruit, Olea europaea | MH685310 | MH713088 | MH717646 | MH713222 | MH713351 | MH717517 | -- | |
| Col-620 | Portugal; 2016 | Fruit, Olea europaea | MH685311 | MH713089 | MH717647 | MH713223 | MH713352 | MH717518 | - | |
| Col-621 | Portugal; 2016 | Fruit, Olea europaea | MH685312 | MH713090 | MH717648 | MH713224 | MH713353 | MH717519 | - | |
| Col-622 | Portugal; 2016 | Fruit, Olea europaea | MH685313 | MH713091 | MH717649 | MH713225 | MH713354 | MH717520 | - | |
| Col-623 | Portugal; 2016 | Fruit, Olea europaea | MH685314 | MH713092 | MH717650 | MH713226 | MH713355 | MH717521 | - | |
| Col-624 | Portugal; 2016 | Fruit, Olea europaea | MH685315 | MH713093 | MH717651 | MH713227 | MH713356 | MH717522 | - | |
| Col-625 | Portugal; 2016 | Fruit, Olea europaea | MH685316 | MH713094 | MH717652 | MH713228 | MH713357 | MH717523 | - | |
| Col-626 | Portugal; 2016 | Fruit, Olea europaea | MH685317 | MH713095 | MH717653 | MH713229 | MH713358 | MH717524 | - | |
| Col-627 | Portugal; 2016 | Fruit, Olea europaea | MH685318 | MH713096 | MH717654 | MH713230 | MH713359 | MH717525 | - | |
| Col-628 | Portugal; 2016 | Fruit, Olea europaea | MH685319 | MH713097 | MH717655 | MH713231 | MH713360 | MH717526 | - | |
| Col-629 | Portugal; 2016 | Fruit, Olea europaea | MH685320 | MH713098 | MH717656 | MH713232 | MH713361 | MH717527 | - | |
| Col-630 | Portugal; 2016 | Fruit, Olea europaea | MH685321 | MH713099 | MH717657 | MH713233 | MH713362 | MH717528 | - | |
| Col-631 | Portugal; 2016 | Fruit, Olea europaea | MH685322 | MH713100 | MH717658 | MH713234 | MH713363 | MH717529 | - | |
| Col-632 | Portugal; 2016 | Fruit, Olea europaea | MH685323 | MH713101 | MH717659 | MH713235 | MH713364 | MH717530 | - | |
| Col-633 | Portugal; 2016 | Fruit, Olea europaea | MH685324 | MH713102 | MH717660 | MH713236 | MH713365 | MH717531 | - | |
| Col-634 | Portugal; 2016 | Fruit, Olea europaea | MH685325 | MH713103 | MH717661 | MH713237 | MH713366 | MH717532 | - | |
| Col-635 | Portugal; 2016 | Fruit, Olea europaea | MH685326 | MH713104 | MH717662 | MH713238 | MH713367 | MH717533 | - | |
| Col-636 | Portugal; 2016 | Fruit, Olea europaea | MH685327 | MH713105 | MH717663 | MH713239 | MH713368 | MH717534 | -- | |
| Col-637 | Portugal; 2016 | Fruit, Olea europaea | MH685328 | MH713106 | MH717664 | MH713240 | MH713369 | MH717535 | - | |
| Col-638 | Portugal; 2016 | Fruit, Olea europaea | MH685329 | MH713107 | MH717665 | MH713241 | MH713370 | MH717536 | - | |
| Col-639 | Portugal; 2016 | Fruit, Olea europaea | MH685330 | MH713108 | MH717666 | MH713242 | MH713371 | MH717537 | - | |
| Col-640 | Portugal; 2016 | Fruit, Olea europaea | MH685331 | MH713109 | MH717667 | MH713243 | MH713372 | MH717538 | - | |
| Col-641 | Portugal; 2016 | Fruit, Olea europaea | MH685332 | MH713110 | MH717668 | MH713244 | MH713373 | MH717539 | - | |
| Col-642 | Portugal; 2016 | Fruit, Olea europaea | MH685333 | MH713111 | MH717669 | MH713245 | MH713374 | MH717540 | - | |
| Col-643 | Portugal; 2016 | Fruit, Olea europaea | MH685334 | MH713112 | MH717670 | MH713246 | MH713375 | MH717541 | - | |
| Col-644 | Portugal; 2016 | Fruit, Olea europaea | MH685335 | MH713113 | MH717671 | MH713247 | MH713376 | MH717542 | - | |
| Col-645 | Portugal; 2016 | Fruit, Olea europaea | MH685336 | MH713114 | MH717672 | MH713248 | MH713377 | MH717543 | - | |
| Col-646 | Portugal; 2016 | Fruit, Olea europaea | MH685337 | MH713115 | MH717673 | MH713249 | MH713378 | MH717544 | - | |
| Col-647 | Portugal; 2016 | Fruit, Olea europaea | MH685338 | MH713116 | MH717674 | MH713250 | MH713379 | MH717545 | - | |
| Col-648 | Portugal; 2016 | Fruit, Olea europaea | MH685339 | MH713117 | MH717675 | MH713251 | MH713380 | MH717546 | - | |
| Col-649 | Portugal; 2016 | Fruit, Olea europaea | MH685340 | MH713118 | MH717676 | MH713252 | MH713381 | MH717547 | - | |
| Col-650 | Portugal; 2016 | Fruit, Olea europaea | MH685341 | MH713119 | MH717677 | MH713253 | MH713382 | MH717548 | - | |
| Col-651 | Portugal; 2016 | Fruit, Olea europaea | MH685342 | MH713120 | MH717678 | MH713254 | MH713383 | MH717549 | - | |
| Col-652 | Portugal; 2016 | Fruit, Olea europaea | MH685343 | MH713121 | MH717679 | MH713255 | MH713384 | MH717550 | - | |
| Col-653 | Portugal; 2016 | Fruit, Olea europaea | MH685344 | MH713122 | MH717680 | MH713256 | MH713385 | MH717551 | - | |
| Col-654 | Portugal; 2016 | Fruit, Olea europaea | MH685345 | MH713123 | MH717681 | MH713257 | MH713386 | MH717552 | - | |
| Col-655 | Portugal; 2016 | Fruit, Olea europaea | MH685346 | MH713124 | MH717682 | MH713258 | MH713387 | MH717553 | - | |
| Col-656 | Portugal; 2016 | Fruit, Olea europaea | MH685347 | MH713125 | MH717683 | MH713259 | MH713388 | MH717554 | - | |
| Col-657 | Portugal; 2016 | Fruit, Olea europaea | MH685348 | MH713126 | MH717684 | MH713260 | MH713389 | MH717555 | - | |
| Col-658 | Portugal; 2016 | Fruit, Olea europaea | MH685349 | MH713127 | MH717685 | MH713261 | MH713390 | MH717556 | - | |
| Col-659 | Portugal; 2016 | Fruit, Olea europaea | MH685350 | MH713128 | MH717686 | MH713262 | MH713391 | MH717557 | - | |
| Col-660 | Portugal; 2016 | Fruit, Olea europaea | MH685351 | MH713129 | MH717687 | MH713263 | MH713392 | MH717558 | - | |
| Col-661 | Portugal; 2016 | Fruit, Olea europaea | MH685352 | MH713130 | MH717688 | MH713264 | MH713393 | MH717559 | - | |
| Col-662 | Portugal; 2016 | Fruit, Olea europaea | MH685353 | MH713131 | MH717689 | MH713265 | MH713394 | MH717560 | - | |
| Col-663 | Portugal; 2016 | Fruit, Olea europaea | MH685354 | MH713132 | MH717690 | MH713266 | MH713395 | MH717561 | - | |
| Col-664 | Portugal; 2016 | Fruit, Olea europaea | MH685355 | MH713133 | MH717691 | MH713267 | MH713396 | MH717562 | - | |
| Col-665 | Portugal; 2016 | Fruit, Olea europaea | MH685356 | MH713134 | MH717692 | MH713268 | MH713397 | MH717563 | - | |
| Col-666 | Portugal; 2016 | Fruit, Olea europaea | MH685357 | MH713135 | MH717693 | MH713269 | MH713398 | MH717564 | - | |
| Col-667 | Portugal; 2016 | Fruit, Olea europaea | MH685358 | MH713136 | MH717694 | MH713270 | MH713399 | MH717565 | - | |
| Col-668 | Portugal; 2016 | Fruit, Olea europaea | MH685359 | MH713137 | MH717695 | MH713271 | MH713400 | MH717566 | - | |
| Col-669 | Portugal; 2016 | Fruit, Olea europaea | MH685360 | MH713138 | MH717696 | MH713272 | MH713401 | MH717567 | - | |
| Col-670 | Portugal; 2016 | Fruit, Olea europaea | MH685361 | MH713139 | MH717697 | MH713273 | MH713402 | MH717568 | - | |
| Col-671 | Portugal; 2016 | Fruit, Olea europaea | MH685362 | MH713140 | MH717698 | MH713274 | MH713403 | MH717569 | - | |
| Col-672 | Portugal; 2016 | Fruit, Olea europaea | MH685363 | MH713141 | MH717699 | MH713275 | MH713404 | MH717570 | - | |
| Col-673 | Portugal; 2016 | Fruit, Olea europaea | MH685364 | MH713142 | MH717700 | MH713276 | MH713405 | MH717571 | - | |
| Col-674 | Portugal; 2016 | Fruit, Olea europaea | MH685365 | MH713143 | MH717701 | MH713277 | MH713406 | MH717572 | - | |
| Col-675 | Portugal; 2016 | Fruit, Olea europaea | MH685366 | MH713144 | MH717702 | MH713278 | MH713407 | MH717573 | - | |
| Col-676 | Portugal; 2016 | Fruit, Olea europaea | MH685367 | MH713145 | MH717703 | MH713279 | MH713408 | MH717574 | - | |
| Col-677 | Portugal; 2016 | Fruit, Olea europaea | MH685368 | MH713146 | MH717704 | MH713280 | MH713409 | MH717575 | - | |
| Col-678 | Portugal; 2016 | Fruit, Olea europaea | MH685369 | MH713147 | MH717705 | MH713281 | MH713410 | MH717576 | - | |
| Col-679 | Portugal; 2016 | Fruit, Olea europaea | MH685370 | MH713148 | MH717706 | MH713282 | MH713411 | MH717577 | - | |
| Col-680 | Portugal; 2016 | Fruit, Olea europaea | MH685371 | MH713149 | MH717707 | MH713283 | MH713412 | MH717578 | - | |
| CBS 515.78T | The Netherlands | Nymphaea alba | JQ948197 | JQ949848 | JQ949518 | JQ948858 | JQ949188 | JQ948527 | - | |
| CBS 231.49 | Portugal | Olea europaea | JQ948202 | JQ949853 | JQ949523 | JQ948863 | JQ949193 | JQ948532 | - | |
| CBS 129945, PT135, RB012 | Portugal | Olea europaea | JQ948201 | JQ949852 | JQ949522 | JQ948862 | JQ949192 | JQ948531 | - | |
| C. orchidophilum | CBS 119291, MEP 1545 | Panama | Cycnoches aureum | JQ948154 | JQ949805 | JQ949475 | JQ948815 | JQ949145 | JQ948484 | - |
| CBS 632.80T | USA | Dendrobium sp. | JQ948151 | JQ949802 | JQ949472 | JQ948812 | JQ949142 | JQ948481 | - | |
| C. paranaense | CBS 134729T | Brazil, Parana | Malus domestica | KC204992 | KC205060 | KC205077 | KC205043 | KC205004 | KC205026 | - |
| C. paxtonii | IMI 165753, CPC 18868T | Saint Lucia | Musa sp. | JQ948285 | JQ949936 | JQ949606 | JQ948946 | JQ949276 | JQ948615 | - |
| C. perseae | Col-205; UWS-139 | Australia; 2009 | Fruit, Olea europaea | MH685242 | MH713156 | - | - | - | - | MH717588 |
| CBS141365 | Israel | Persea americana | KX620308 | KX620341 | - | - | - | - | KX620177 | |
| C. phormii | CBS 118194, AR 3546T | Germany | Phormium sp. | JQ948446 | JQ950097 | JQ949767 | JQ949107 | JQ949437 | JQ948777 | - |
| C. psidii | ICMP 19120T | Italy | Psidium sp. | JX010219 | JX010443 | - | - | - | - | KC888931 |
| C. pyricola | CBS 128531, ICMP 12924, PRJ 977.1T | New Zealand | Pyrus communis | JQ948445 | JQ950096 | JQ949766 | JQ949106 | JQ949436 | JQ948776 | - |
| C. queenslandicum | ICMP 1778T | Australia | Carica papaya | JX010276 | JX010414 | - | - | - | - | KC888928 |
| C. rhombiforme | CBS 129953, PT250, RB011T | Portugal | Olea europaea | JQ948457 | JQ950108 | JQ949778 | JQ949115 | JQ949448 | JQ948788 | - |
| C. salicis | CBS 607.94T | The Netherlands | Salix sp. | JQ948460 | JQ950111 | JQ949781 | JQ949121 | JQ949451 | JQ948791 | - |
| C. salsolae | ICMP 19051T | Hungary | Salsola tragus | JX010242 | JX010403 | - | - | - | - | KC888925 |
| C. scovillei | CBS 126529, PD 94/921-3, BBA 70349T | Indonesia | Capsicum sp. | JQ978267 | JQ949918 | JQ949588 | JQ948928 | JQ948928 | JQ948597 | - |
| C. siamense | Col-44; IMI-345047 d,e,f | Spain 1999 | Fragaria vesca | MH685205 | MH713157 | MH713296 | MH713289 | MH713418 | MH717493 | MH717589 |
| Col-160-UWS-13 d,e | Australia 2009 | Fruit, Olea europaea | MH685230 | MH713158 | MH713297 | MH713290 | MH713419 | MH717494 | MH717590 | |
| Col-181; UWS-90 f | Australia; 2009 | Fruit, Olea europaea | MH685236 | MH713159 | - | - | - | - | MH717591 | |
| Col-184; UWS-92 d,e | Australia; 2009 | Fruit, Olea europaea | MH685237 | MH713160 | - | - | - | - | MH717592 | |
| Col-187; UWS-94 d,e | Australia; 2009 | Fruit, Olea europaea | MH685238 | MH713161 | - | - | - | - | MH717587 | |
| ICMP 18578, CBS-130417 | Thailand | Coffea arabica | JX010171 | JX010404 | - | - | - | - | JQ899289 | |
| C. siamense (syn. C. jasminisambac) | CBS 130420, ICMP 19118 | Vietnam | Jasminum sambac | HM131511 | JX010415 | - | - | - | - | JQ807841 |
| C. siamense (syn. C. hymenocallidis) | CBS 125378, ICMP 18642, LC0043 | China | Hymenocallis americana | JX010278 | JX010410 | - | - | - | - | JQ899283 |
| C. simmondsii | Col-169-UWS-68 d,e | Australia; 2009 | Fruit, Olea europaea | MH685232 | MH713150 | MH717708 | MH713284 | MH713413 | MH717579 | - |
| CBS 122122, BRIP 28519T | Australia | Carica papaya | JQ948276 | JQ949927 | JQ949597 | JQ948937 | JQ949267 | JQ948606 | - | |
| C. sloanei | IMI 364297, CPC 18929T | Malaysia | Theobroma cacao | JQ948287 | JQ949938 | JQ949608 | JQ948948 | JQ949278 | JQ948617 | - |
| C. tamarilloi | CBS 129814, T.A.6T | Colombia | Solanum betaceum | JQ948184 | JQ949835 | JQ949505 | JQ948845 | JQ949175 | JQ948514 | - |
| C. theobromicola | Col-200;UWS-131 d,e | Australia; 2009 | Fruit, Olea europaea | MH685241 | MH713164 | MH713298 | MH713291 | MH713420 | MH717495 | MH717593 |
| CBS 124945T, ICMP 18649 | Panama | Theobroma cacao | JX010294 | JX010447 | - | - | - | - | KC790726 | |
| C. theobromicola (syn. C. fragariae) | CBS 142.31T, ICMP 17927 | USA | Fragaria ananassa | JX010286 | JX010373 | - | - | - | - | JQ807844 |
| C. ti | ICMP 4832 | New Zealand | Cordyline sp. | JX010269 | JX010442 | - | - | - | - | KM360146 |
| C. tropicale | CBS 124949, ICMP 18653 | Panama | Theobroma cacao | JX010264 | JX010407 | - | - | - | - | KC790728 |
| C. walleri | CBS 125472, BMT(HL)19T | Vietnam | Coffea sp. | JQ948275 | JQ949926 | JQ949596 | JQ948936 | JQ949266 | JQ948605 | - |
| C. wuxiense | CGMCC 3.17894T | China | Camellia sinensis | KU251591 | KU252200 | - | - | - | - | KU251722 |
| C. xanthorrhoeae | ICMP 17903T | Australia | Xanthorrhoea preissii | JX010261 | JX010448 | - | - | - | - | KC790689 |
| Species Complex/Fungal Species | Isolate | Mycelium | Conidia d | |||||
|---|---|---|---|---|---|---|---|---|
| Color a | Benomyl Inhibition (%) b | Casein Hydrolysis c | Length (µm) | Width (µm) | Length/Width | Type | ||
| Colletotrichum acutatum complex | ||||||||
| Colletotrichum acutatum | Col-166/UWS-65 | Pink white | 33.5 | + | 11.1 ± 2.18 | 3.2 ± 0.80 | 3.6 ± 0.86 | Ellipsoid |
| Col-175/UWS-79 | Pink-orange | 57.4 | + | 8.3 ± 1.51 | 2.7 ± 0.58 | 3.1 ± 0.53 | Ellipsoid | |
| Col-208/UWS-149 | Pink gray | 67.4 | + | 13.2 ± 2.28 | 4.7 ± 1.45 | 3.0 ± 0.71 | Clavate | |
| Col-536 | Pink-orange | N/D | N/D | 10.4 ± 1.19 | 3.2 ± 0.62 | 3.3 ± 0.53 | Fusiform | |
| Colletotrichum fioriniae | Col-172/UWS-70 | Light gray | 71.1 | - | 13.4 ± 1.16 | 4.4 ± 0.49 | 3.1 ± 0.43 | Fusiform |
| Col-693 | White | N/D | N/D | 10.4 ± 0.87 | 3.3 ± 0.23 | 3.3 ± 0.32 | Fusiform | |
| Col-694 | White | N/D | N/D | 10.4 ± 0.62 | 3.4 ± 0.23 | 3.2 ± 0.19 | Fusiform | |
| Col-695 | Orange gray | N/D | N/D | 9.3 ± 0.55 | 3.2 ± 0.15 | 3.0 ± 0.28 | Fusiform | |
| Col-696 | Pink-orange | N/D | N/D | 10.3 ± 0.45 | 3.8 ± 0.31 | 2.8 ± 0.13 | Fusiform | |
| Col-697 | White | N/D | N/D | 9.9 ± 0.66 | 3.4 ± 0.23 | 3.0 ± 0.15 | Fusiform | |
| Colletotrichum godetiae | Col-1 | Dark gray | N/D | N/D | 14.8 ± 0.85 | 5.0 ± 0.00 | 3.0 ± 0.17 | Clavate |
| Col-9 | Dark gray | 55.7 | ++ | 14.8 ± 1.45 | 4.9 ± 0.18 | 3.0 ± 0.33 | Clavate | |
| Col-30 | Dark gray | 63.7 | ++ | 12.5 ± 1.75 | 5.1 ± 0.38 | 2.4 ± 0.38 | Clavate | |
| Col-50 | Dark gray | 59.8 | ++ | 13.9 ± 1.23 | 5.0 ± 0.12 | 2.8 ± 0.25 | Clavate | |
| Col-51 | Dark gray | 67.6 | ++ | 13.5 ± 1.40 | 5.0 ± 0.18 | 2.7 ± 0.30 | Clavate | |
| Col-52 | Dark gray | N/D | N/D | 13.1 ± 1.38 | 3.8 ± 0.19 | 3.5 ± 0.35 | Clavate | |
| Col-57 | Dark gray | 57.2 | ++ | 13.5 ± 1.43 | 5.0 ± 0.18 | 2.7 ± 0.31 | Clavate | |
| Col-59 | Dark gray | 69.0 | ++ | 13.8 ± 1.51 | 5.0 ± 0.18 | 2.8 ± 0.37 | Clavate | |
| Col-60 | Dark gray | 67.0 | ++ | 14.0 ± 1.72 | 4.9 ± 0.38 | 2.9 ± 0.42 | Clavate | |
| Col-88 | Dark gray | 64.5 | ++ | 12.9 ± 1.19 | 5.0 ± 0.04 | 2.6 ± 0.28 | Ellipsoid | |
| Col-508 | Dark gray | N/D | N/D | 14.4 ± 1.25 | 3.8 ± 0.41 | 3.8 ± 0.47 | Clavate | |
| Col-522 | Light gray | N/D | N/D | 12.8 ± 1.29 | 3.7 ± 0.33 | 3.5 ± 0.93 | Fusiform | |
| Colletotrichum nymphaeae | Col-42 | Light gray | 41.4 | ++ | 13.9 ± 1.56 | 3.5 ± 0.54 | 4.2 ± 1.06 | Fusiform |
| Col-84 | Light gray | 60.5 | ++ | 13.5 ± 1.40 | 3.6 ± 0.89 | 3.9 ± 0.35 | Clavate | |
| Col-86 | Light gray | 58.7 | + | 14.0 ± 1.22 | 3.6 ± 0.43 | 3.9 ± 0.92 | Clavate | |
| Col-506 | Light gray | N/D | N/D | 12.1 ± 1.44 | 3.4 ± 0.62 | 3.6 ± 0.57 | Clavate | |
| Colletotrichum simmondsii | Col-169/UWS-68 | Whitish gray | 64.6 | + | 12.4 ± 1.12 | 3.9 ± 0.67 | 3.2 ± 0.49 | Fusiform |
| Colletotrichum boninense complex | ||||||||
| Colletotrichum boninense | Col-178/UWS-82 | Whitish gray | 95.0 | - | 13.2 ± 1.02 | 4.8 ± 0.37 | 2.9 ± 0.32 | Clavate |
| Colletotrichum karstii | Col-79 | Pink-orange | 99.4 | - | 12.6 ± 1.31 | 5.0 ± 0.02 | 2.6 ± 0.26 | Ellipsoid |
| Colletotrichum gloeosporioides complex | ||||||||
| Colletotrichum alienum | Col-211/UWS-152 | White | 94.0 | ++ | 14.1 ± 1.22 | 4.6 ± 0.71 | 3.2 ± 0.55 | Ellipsoid |
| Col-214/UWS-156 | Pink White | 95.1 | - | 13.9 ± 1.14 | 4.6 ± 0.43 | 3.1 ± 0.33 | Ellipsoid | |
| Colletotrichum fructicola | Col-82 | Light gray | 95.0 | - | 12.0 ± 1.6 | 3.7 ± 0.63 | 3.3 ± 0.56 | Ellipsoid |
| Colletotrichum gloeosporioides | Col-41 | Whitish gray | 100 | - | 14.8 ± 1.45 | 4.4 ± 0.72 | 3.5 ± 0.80 | Ellipsoid |
| Col-69 | Light gray | 99.7 | - | 13.2 ± 1.05 | 5.1 ± 0.23 | 2.6 ± 0.24 | Ellipsoid | |
| Colletotrichum persease | Col-205/UWS-139 | Light gray | 98.2 | - | 14.8 ± 1.38 | 4.8 ± 0.86 | 3.2 ± 0.55 | Ellipsoid |
| Colletotrichum siamense | Col-44 | Green gray | 96.5 | - | 13.9 ± 1.41 | 4.6 ± 0.70 | 3.1 ± 0.66 | Clavate |
| Col-160/UWS-13 | Whitish gray | 93.8 | - | 12.3 ± 1.01 | 4.5 ± 0.67 | 2.8 ± 0.50 | Ellipsoid | |
| Col-184/UWS-92 | Whitish gray | 93.9 | + | 11.9 ± 1.38 | 3.5 ± 0.54 | 3.4 ± 0.61 | Clavate | |
| Col-187/UWS-94 | Whitish gray | 96.0 | ++ | 13.3 ± 1.03 | 3.8 ± 0.43 | 3.5 ± 0.49 | Ellipsoid | |
| Colletotrichum theobromicola | Col-200/UWS-131 | White | 94.4 | - | 13.3 ± 2.55 | 4.9 ± 0.4 | 2.8 ± 0.48 | Ellipsoid |
| HSD0.05 e | - | - | 5.67 | - | 1.7 | 0.64 | 0.63 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moral, J.; Agustí-Brisach, C.; Raya, M.C.; Jurado-Bello, J.; López-Moral, A.; Roca, L.F.; Chattaoui, M.; Rhouma, A.; Nigro, F.; Sergeeva, V.; et al. Diversity of Colletotrichum Species Associated with Olive Anthracnose Worldwide. J. Fungi 2021, 7, 741. https://doi.org/10.3390/jof7090741
Moral J, Agustí-Brisach C, Raya MC, Jurado-Bello J, López-Moral A, Roca LF, Chattaoui M, Rhouma A, Nigro F, Sergeeva V, et al. Diversity of Colletotrichum Species Associated with Olive Anthracnose Worldwide. Journal of Fungi. 2021; 7(9):741. https://doi.org/10.3390/jof7090741
Chicago/Turabian StyleMoral, Juan, Carlos Agustí-Brisach, Maria Carmen Raya, José Jurado-Bello, Ana López-Moral, Luis F. Roca, Mayssa Chattaoui, Ali Rhouma, Franco Nigro, Vera Sergeeva, and et al. 2021. "Diversity of Colletotrichum Species Associated with Olive Anthracnose Worldwide" Journal of Fungi 7, no. 9: 741. https://doi.org/10.3390/jof7090741
APA StyleMoral, J., Agustí-Brisach, C., Raya, M. C., Jurado-Bello, J., López-Moral, A., Roca, L. F., Chattaoui, M., Rhouma, A., Nigro, F., Sergeeva, V., & Trapero, A. (2021). Diversity of Colletotrichum Species Associated with Olive Anthracnose Worldwide. Journal of Fungi, 7(9), 741. https://doi.org/10.3390/jof7090741









