Identification of Clinical Isolates of Candida albicans with Increased Fitness in Colonization of the Murine Gut
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Genetic Procedures
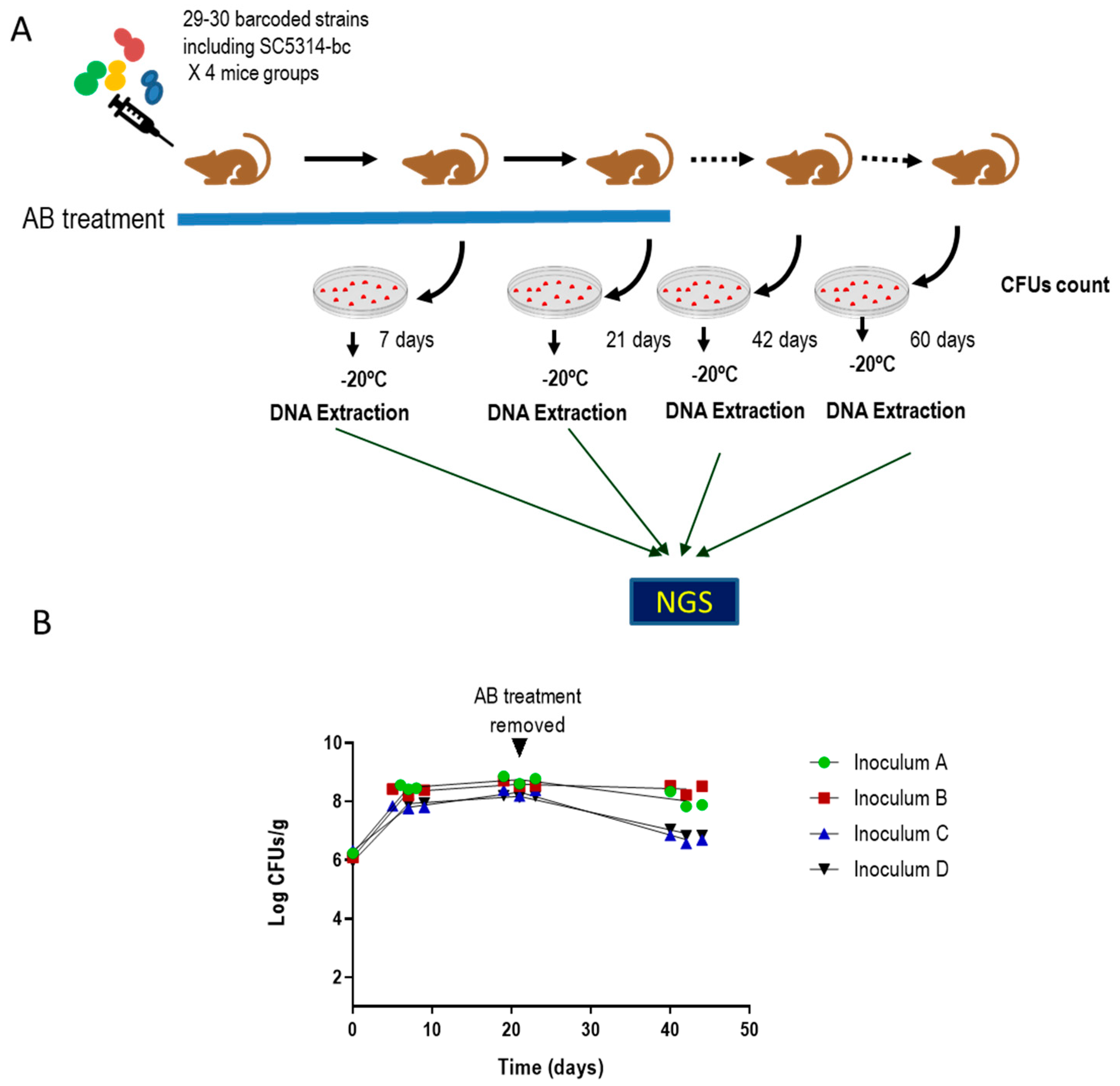
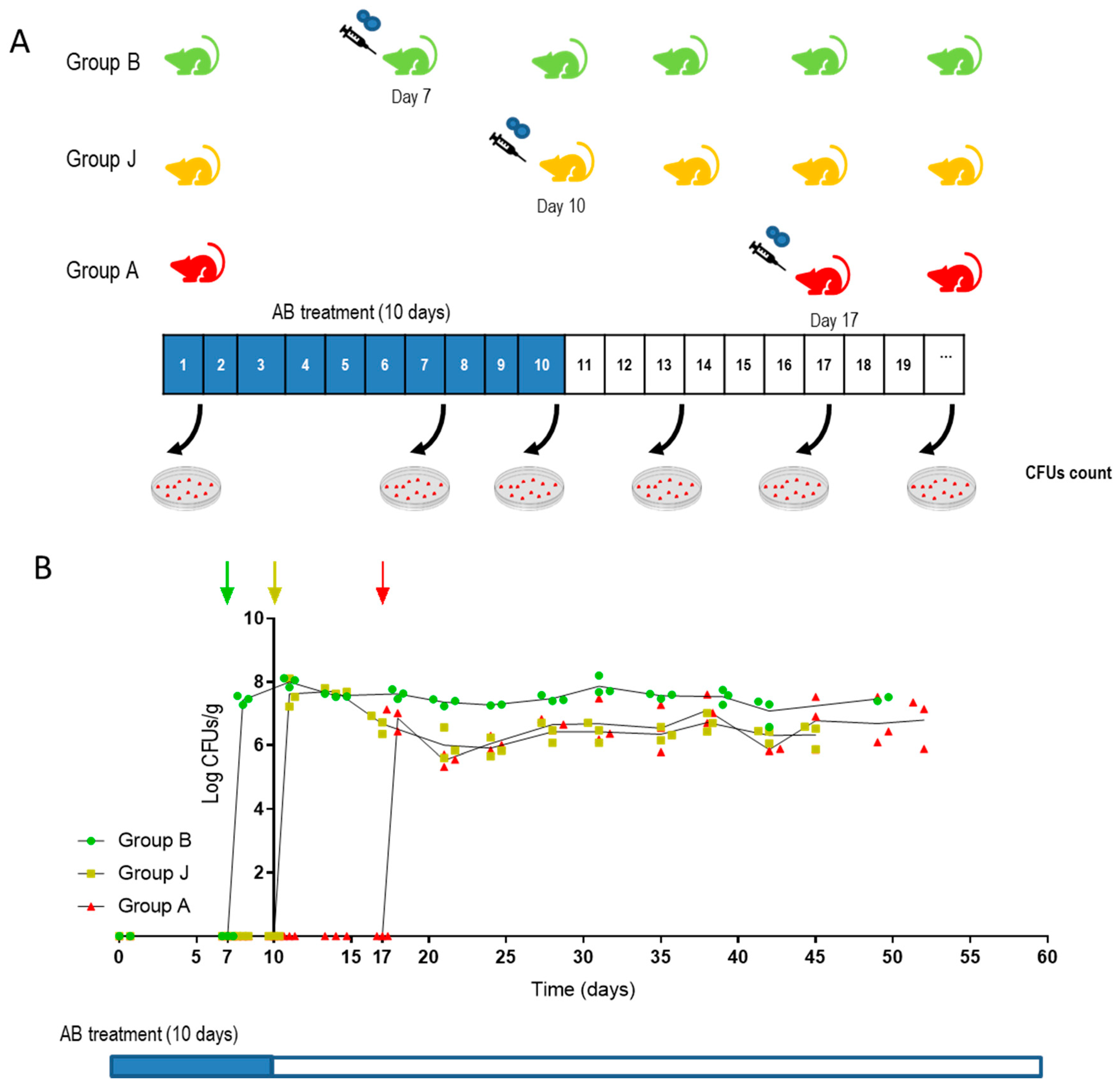
2.3. In Vivo Procedures
2.4. NGS Assay
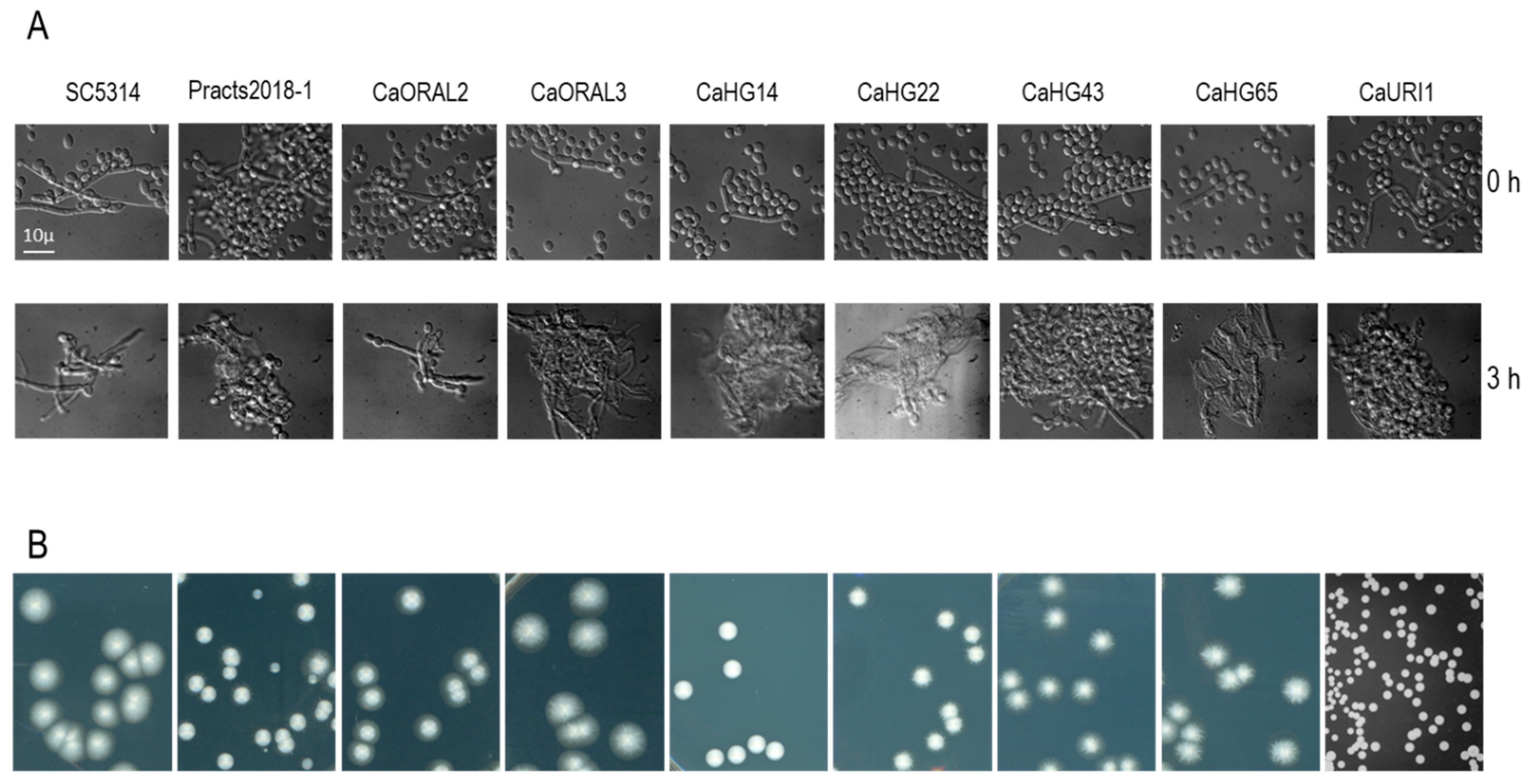
2.5. C. albicans Filamentation Assay
2.6. Supplementary Material and Methods
2.6.1. Susceptibility Assays
2.6.2. Adhesion Assay
2.6.3. Phospholipase Activity
2.7. C. albicans Susceptibility Testing
3. Results
3.1. Identification of Clinical Isolates of Candida albicans with Increased Gut Colonization Load
3.2. Selected C. albicans Strains Displayed Enhanced Fitness Compared to SC5314
3.3. Strains with Increased Fitness Are Able to Form True Hyphae
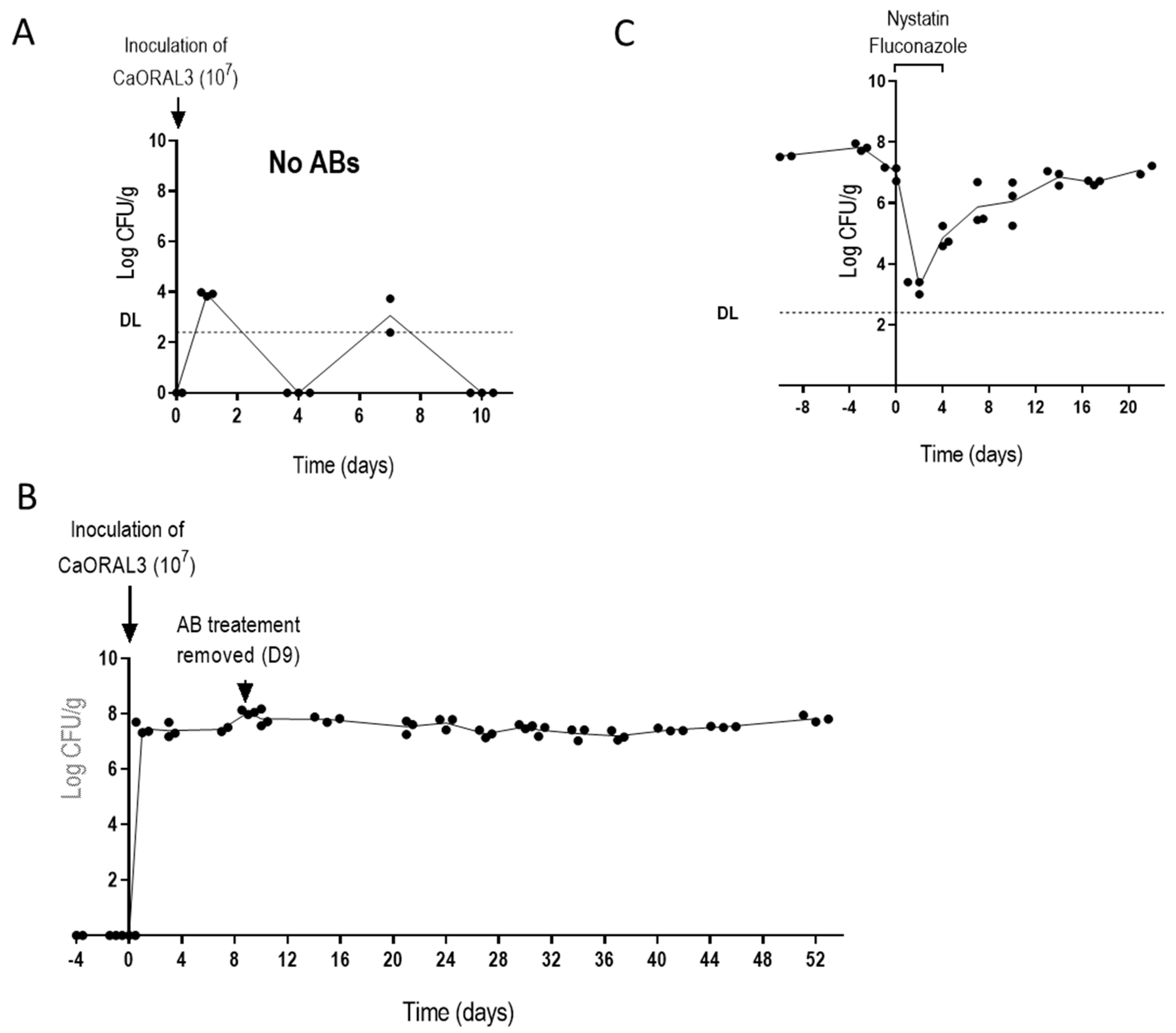
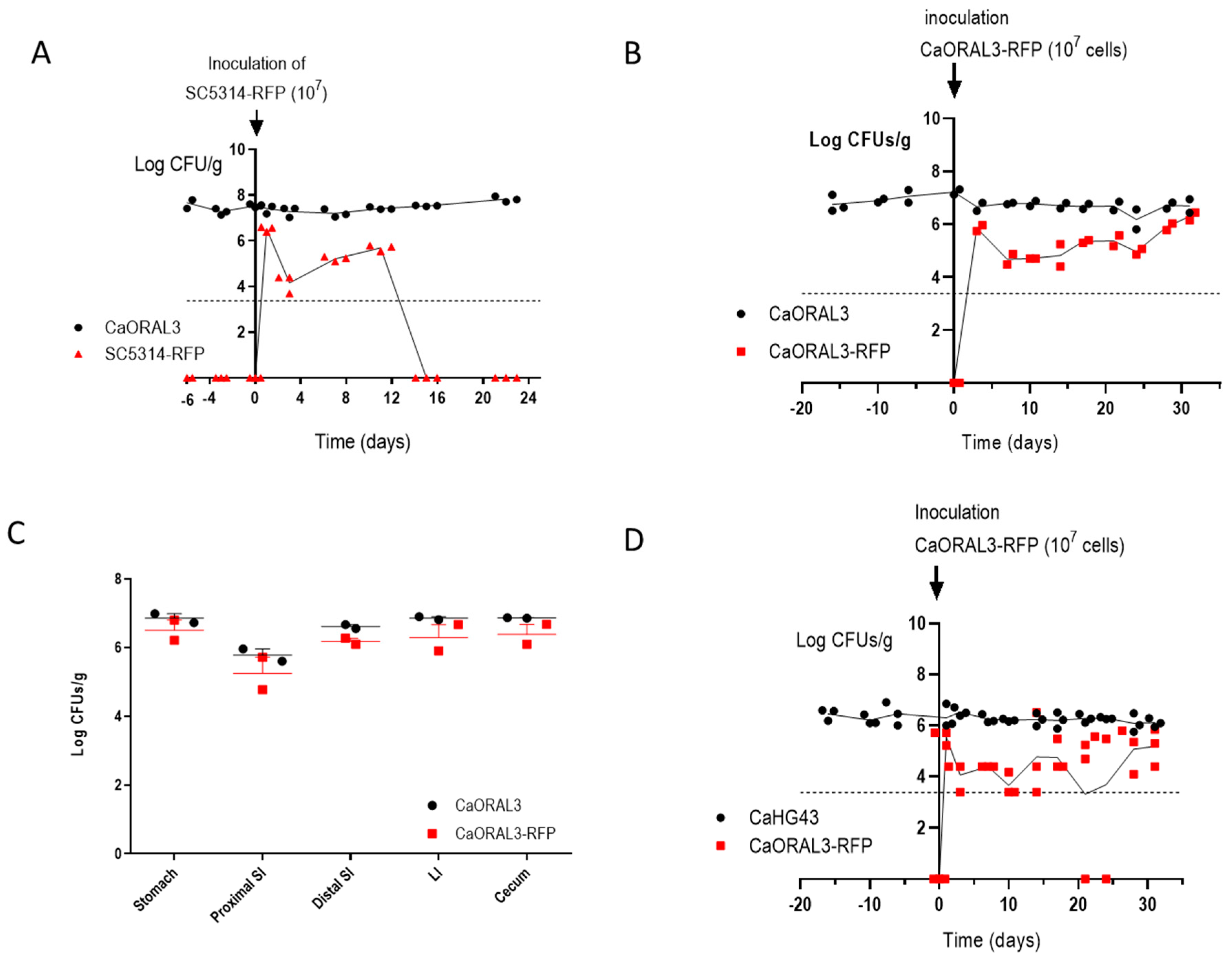
3.4. CaORAL3 Strain Still Requires Antibiotic Treatment to Colonize Murine Gut
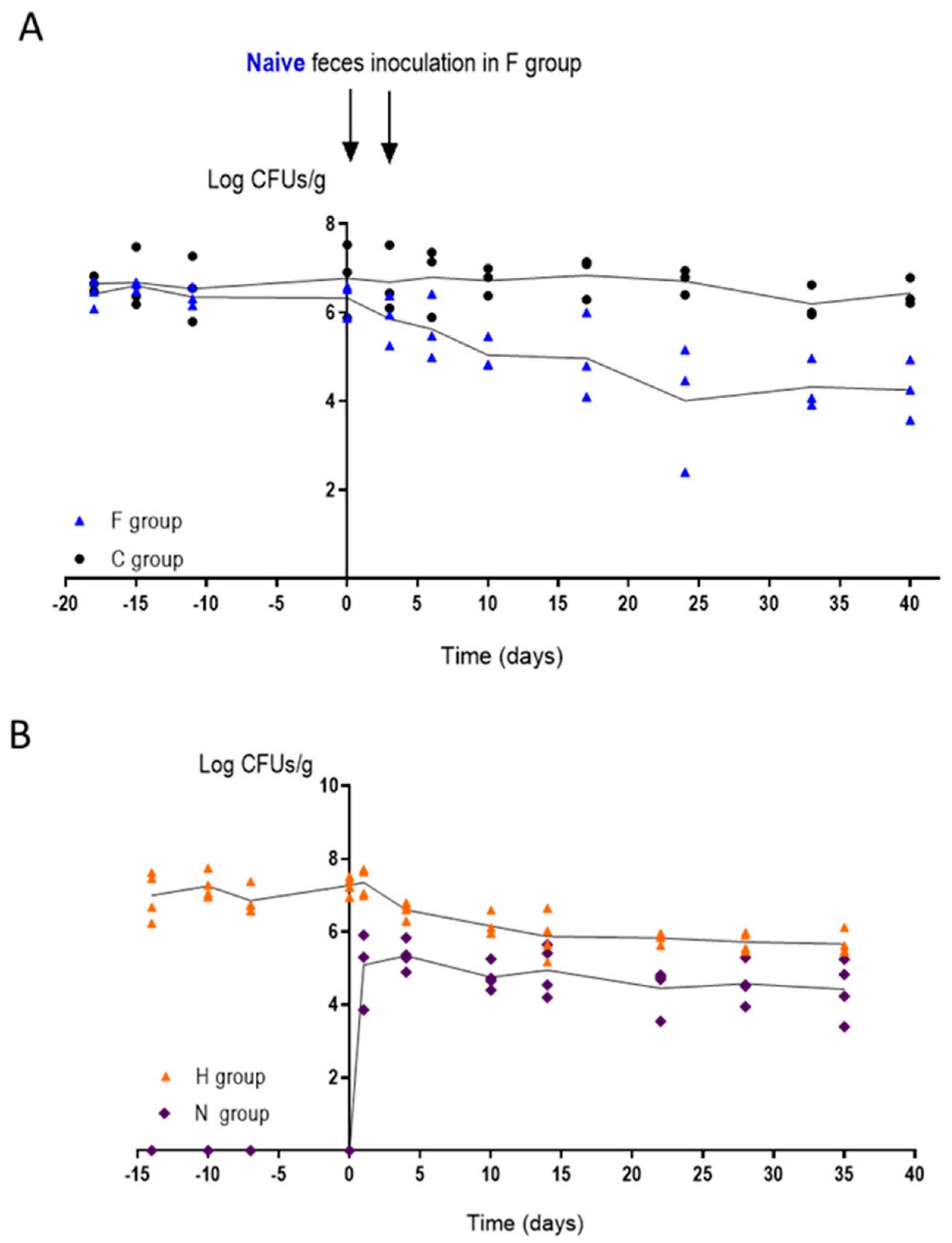
3.5. Changes in the Intestinal Microbiota Allow CaORAL3 Colonization
3.6. CaORAL3 Is Able to Colonize Murine Gut Previously Colonized with C. albicans
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pappas, P.G.; Lionakis, M.S.; Arendrup, M.C.; Ostrosky-Zeichner, L.; Kullberg, B.J. Invasive candidiasis. Nat. Rev. Dis. Primers 2018, 4, 18026. [Google Scholar] [CrossRef]
- Odds, F.C. Candida and Candidosis; Baillière Tindall: London, UK, 1988; Volume 2. [Google Scholar]
- McCarty, T.P.; Pappas, P.G. Invasive Candidiasis. Infect. Dis. Clin. N. Am. 2016, 30, 103–124. [Google Scholar] [CrossRef]
- Vergidis, P.; Clancy, C.J.; Shields, R.K.; Park, S.Y.; Wildfeuer, B.N.; Simmons, R.L.; Nguyen, M.H. Intra-Abdominal Candidiasis: The Importance of Early Source Control and Antifungal Treatment. PLoS ONE 2016, 11, e0153247. [Google Scholar] [CrossRef] [Green Version]
- Russell, C.; Lay, K.M. Natural history of Candida species and yeasts in the oral cavities of infants. Arch. Oral Biol. 1973, 18, 957–962. [Google Scholar] [CrossRef]
- Odds, F.C.; Davidson, A.D.; Jacobsen, M.D.; Tavanti, A.; Whyte, J.A.; Kibbler, C.C.; Ellis, D.H.; Maiden, M.C.; Shaw, D.J.; Gow, N.A. Candida albicans strain maintenance, replacement, and microvariation demonstrated by multilocus sequence typing. J. Clin. Microbiol. 2006, 44, 3647–3658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miranda, L.N.; van der Heijden, I.M.; Costa, S.F.; Sousa, A.P.; Sienra, R.A.; Gobara, S.; Santos, C.R.; Lobo, R.D.; Pessoa, V.P., Jr.; Levin, A.S. Candida colonisation as a source for candidaemia. J. Hosp. Infect. 2009, 72, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Nucci, M.; Anaissie, E. Revisiting the source of candidemia: Skin or gut? Clin. Infect. Dis. 2001, 33, 1959–1967. [Google Scholar] [CrossRef] [PubMed]
- Koh, A.Y. Murine Models of Candida Gastrointestinal Colonization and Dissemination. Eukaryot. Cell 2013, 12, 1416–1422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koh, A.Y.; Kohler, J.R.; Coggshall, K.T.; van Rooijen, N.; Pier, G.B. Mucosal damage and neutropenia are required for Candida albicans dissemination. PLoS Pathog. 2008, 4, e35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohm, L.; Torsin, S.; Tint, S.H.; Eckstein, M.T.; Ludwig, T.; Perez, J.C. The yeast form of the fungus Candida albicans promotes persistence in the gut of gnotobiotic mice. PLoS Pathog. 2017, 13, e1006699. [Google Scholar] [CrossRef] [Green Version]
- Phillips, A.W.; Balish, E. Growth and invasiveness of Candida albicans in the germ-free and conventional mouse after oral challenge. Appl. Microbiol. 1966, 14, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, S.M.; Jechorek, R.P.; Garni, R.M.; Bendel, C.M.; Wells, C.L. Gastrointestinal colonization by Candida albicans mutant strains in antibiotic-treated mice. Clin. Diagn. Lab. Immunol. 2001, 8, 192–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prieto, A.D.; Román, E.; Correia, I.; Pla, J. The HOG pathway is critical for the colonization of the mouse gastrointestinal tract by Candida albicans. PLoS ONE 2014, 9, e87128. [Google Scholar] [CrossRef] [Green Version]
- Perez, J.C.; Kumamoto, C.A.; Johnson, A.D. Candida albicans commensalism and pathogenicity are intertwined traits directed by a tightly knit transcriptional regulatory circuit. PLoS Biol. 2013, 11, e1001510. [Google Scholar] [CrossRef] [Green Version]
- White, S.J.; Rosenbach, A.; Lephart, P.; Nguyen, D.; Benjamin, A.; Tzipori, S.; Whiteway, M.; Mecsas, J.; Kumamoto, C.A. Self-Regulation of Candida albicans Population Size during GI Colonization. PLoS Pathog. 2007, 3, e184. [Google Scholar] [CrossRef] [Green Version]
- Prieto, D.; Correia, I.; Pla, J.; Roman, E. Adaptation of Candida albicans to commensalism in the gut. Future Microbiol. 2016, 11, 567–583. [Google Scholar] [CrossRef]
- Neville, B.A.; d’Enfert, C.; Bougnoux, M.E. Candida albicans commensalism in the gastrointestinal tract. FEMS Yeast Res. 2015, 15. [Google Scholar] [CrossRef] [Green Version]
- Perez, J.C. Candida albicans dwelling in the mammalian gut. Curr. Opin. Microbiol. 2019, 52, 41–46. [Google Scholar] [CrossRef]
- Mamouei, Z.; Zeng, G.; Wang, Y.M.; Wang, Y. Candida albicans possess a highly versatile and dynamic high-affinity iron transport system important for its commensal-pathogenic lifestyle. Mol. Microbiol. 2017, 106, 986–998. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Pande, K.; French, S.D.; Tuch, B.B.; Noble, S.M. An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe 2011, 10, 118–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pande, K.; Chen, C.; Noble, S.M. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat. Genet. 2013, 45, 1088–1091. [Google Scholar] [CrossRef] [Green Version]
- Roman, E.; Huertas, B.; Prieto, D.; Diez-Orejas, R.; Pla, J. TUP1-mediated filamentation in Candida albicans leads to inability to colonize the mouse gut. Future Microbiol. 2018, 13, 857–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierce, J.V.; Kumamoto, C.A. Variation in Candida albicans EFG1 expression enables host-dependent changes in colonizing fungal populations. mBio 2012, 3, e00117-12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tso, G.H.W.; Reales-Calderon, J.A.; Tan, A.S.M.; Sem, X.; Le, G.T.T.; Tan, T.G.; Lai, G.C.; Srinivasan, K.G.; Yurieva, M.; Liao, W.; et al. Experimental evolution of a fungal pathogen into a gut symbiont. Science 2018, 362, 589–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Znaidi, S.; van Wijlick, L.; Hernandez-Cervantes, A.; Sertour, N.; Desseyn, J.L.; Vincent, F.; Atanassova, R.; Gouyer, V.; Munro, C.A.; Bachellier-Bassi, S.; et al. Systematic gene overexpression in Candida albicans identifies a regulator of early adaptation to the mammalian gut. Cell Microbiol. 2018, 20, e12890. [Google Scholar] [CrossRef] [Green Version]
- Prieto, D.; Roman, E.; Alonso-Monge, R.; Pla, J. Overexpression of the Transcriptional Regulator WOR1 Increases Susceptibility to Bile Salts and Adhesion to the Mouse Gut Mucosa in Candida albicans. Front. Cell Infect. Microbiol. 2017, 7, 389. [Google Scholar] [CrossRef]
- Armstrong, A.E.; Rossoff, J.; Hollemon, D.; Hong, D.K.; Muller, W.J.; Chaudhury, S. Cell-free DNA next-generation sequencing successfully detects infectious pathogens in pediatric oncology and hematopoietic stem cell transplant patients at risk for invasive fungal disease. Pediatr. Blood Cancer 2019, 66, e27734. [Google Scholar] [CrossRef] [Green Version]
- Biswas, C.; Chen, S.C.; Halliday, C.; Kennedy, K.; Playford, E.G.; Marriott, D.J.; Slavin, M.A.; Sorrell, T.C.; Sintchenko, V. Identification of genetic markers of resistance to echinocandins, azoles and 5-fluorocytosine in Candida glabrata by next-generation sequencing: A feasibility study. Clin. Microbiol. Infect. 2017, 23, 676.e7–676.e10. [Google Scholar] [CrossRef] [Green Version]
- Gerami-Nejad, M.; Zacchi, L.F.; Matter, K.; McClellan, M.; Berman, J. Shuttle vectors for facile gap repair cloning and integration into a neutral locus in Candida albicans. Microbiology 2013, 159 Pt 3, 565. [Google Scholar] [CrossRef] [Green Version]
- Köhler, G.A.; White, T.C.; Agabian, N. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J. Bacteriol. 1997, 179, 2331–2338. [Google Scholar] [CrossRef] [Green Version]
- Prieto, A.D.; Román, E.; Correia, I.; Pla, J. Role of Candida albicans MAPK Pathways in Colonization of the Mouse Gastrointestinal Tract. In Proceedings of the 12th ASM Conference on Candida and Candidiasis, New Orleans, LA, USA, 26–30 March 2014. [Google Scholar]
- Illumina. 16S Metagenomic Sequencing Library Preparation; Part#15044223 Rev. B; Illumina: San Diego, CA, USA, 2013. [Google Scholar]
- Echevarria, A.; Durante, A.G.; Arechaval, A.; Negroni, R. Comparative study of two culture media for the detection of phospholipase activity of Candida albicans and Cryptococcus neoformans strains. Rev. Iberoam. Micol. 2002, 19, 95–98. [Google Scholar]
- Macfarlane, M.G.; Knight, B.C. The biochemistry of bacterial toxins: The lecithinase activity of Cl. welchii toxins. Biochem. J. 1941, 35, 884–902. [Google Scholar] [CrossRef] [Green Version]
- Ellepola, A.N.; Samaranayake, L.P.; Khan, Z.U. Extracellular phospholipase production of oral Candida albicans isolates from smokers, diabetics, asthmatics, denture wearers and healthy individuals following brief exposure to polyene, echinocandin and azole antimycotics. Braz. J. Microbiol. 2016, 47, 911–916. [Google Scholar] [CrossRef] [Green Version]
- Wayne, P. M27 Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, 4th ed.; Clinical and Laboratory Standards Institute: Wayne, NY, USA, 2017; Available online: https://clsi.org/media/1897/m27ed4_sample.pdf (accessed on 26 July 2021).
- Znaidi, S.; Barker, K.S.; Weber, S.; Alarco, A.M.; Liu, T.T.; Boucher, G.; Rogers, P.D.; Raymond, M. Identification of the Candida albicans Cap1p regulon. Eukaryot. Cell 2009, 8, 806–820. [Google Scholar] [CrossRef] [Green Version]
- Pierce, J.V.; Dignard, D.; Whiteway, M.; Kumamoto, C.A. Normal adaptation of Candida albicans to the murine gastrointestinal tract requires Efg1p-dependent regulation of metabolic and host defense genes. Eukaryot. Cell 2013, 12, 37–49. [Google Scholar] [CrossRef] [Green Version]
- Prieto, D.; Pla, J. Distinct stages during colonization of the mouse gastrointestinal tract by Candida albicans. Front. Microbiol. 2015, 6, 792. [Google Scholar] [CrossRef]
- Matsuo, K.; Haku, A.; Bi, B.; Takahashi, H.; Kamada, N.; Yaguchi, T.; Saijo, S.; Yoneyama, M.; Goto, Y. Fecal microbiota transplantation prevents Candida albicans from colonizing the gastrointestinal tract. Microbiol. Immunol. 2019, 63, 155–163. [Google Scholar] [CrossRef]
- Ribeiro, F.C.; de Barros, P.P.; Rossoni, R.D.; Junqueira, J.C.; Jorge, A.O. Lactobacillus rhamnosus inhibits Candida albicans virulence factors in vitro and modulates immune system in Galleria mellonella. J. Appl. Microbiol. 2017, 122, 201–211. [Google Scholar] [CrossRef]
- Fan, D.; Coughlin, L.A.; Neubauer, M.M.; Kim, J.; Kim, M.S.; Zhan, X.; Simms-Waldrip, T.R.; Xie, Y.; Hooper, L.V.; Koh, A.Y. Activation of HIF-1alpha and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat. Med. 2015, 21, 808–814. [Google Scholar] [CrossRef] [Green Version]

| Strain Name | Strain Background and Genotype | Reference |
|---|---|---|
| SC5314-RFP | SC5314 ADH1/adh1::SAT1-dTOM2 | This work |
| CaORAL3-RFP | CaORAL3 ADH1/adh1::SAT1-dTOM2 | This work |
| Strain | Origin | Sample |
|---|---|---|
| Practs 2018-1 | Commensal | Stool culture |
| CaORAL2 | Hospital | Oral cavity |
| CaORAL3 | Hospital | Oral cavity |
| CaHG14 | Hospital | Bronchoalveolar lavage |
| CaHG22 | Hospital | Pharyngeal exudate |
| CaHG43 | Hospital | Pharyngeal exudate |
| CaHG65 | Hospital | Urine culture |
| CaURI1 | Hospital | Urine culture |
| Strain | Invasiveness in Spider Medium | Adhesion to Polyestyrene | Bile Salts Sensitivity | Menadione | Phospholipase Activity (SEA) |
|---|---|---|---|---|---|
| Pract. 2018-1 | n.s. | n.s. | R | R | n.s. |
| CaORAL2 | n.s. | n.s. | R | n.s. | n.s. |
| CaORAL3 | n.s. | < | R | n.s. | n.s. |
| CaHG14 | Non invasive | n.s. | n.s. | R | n.s. |
| CaHG22 | n.s. | n.s. | S | n.s. | ↓ |
| CaHG43 | n.s. | n.s. | n.s. | n.s. | +++ |
| CaHG65 | n.s. | n.s. | n.s. | n.s. | ↓ |
| CaURI1 | Non invasive | n.s. | n.s. | R | ++ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alonso-Monge, R.; Prieto, D.; Coman, I.; Rochas, S.; Arana, D.M.; Hidalgo-Vico, S.; Román, E.; Pla, J. Identification of Clinical Isolates of Candida albicans with Increased Fitness in Colonization of the Murine Gut. J. Fungi 2021, 7, 695. https://doi.org/10.3390/jof7090695
Alonso-Monge R, Prieto D, Coman I, Rochas S, Arana DM, Hidalgo-Vico S, Román E, Pla J. Identification of Clinical Isolates of Candida albicans with Increased Fitness in Colonization of the Murine Gut. Journal of Fungi. 2021; 7(9):695. https://doi.org/10.3390/jof7090695
Chicago/Turabian StyleAlonso-Monge, Rebeca, Daniel Prieto, Ioana Coman, Sara Rochas, David M. Arana, Susana Hidalgo-Vico, Elvira Román, and Jesús Pla. 2021. "Identification of Clinical Isolates of Candida albicans with Increased Fitness in Colonization of the Murine Gut" Journal of Fungi 7, no. 9: 695. https://doi.org/10.3390/jof7090695
APA StyleAlonso-Monge, R., Prieto, D., Coman, I., Rochas, S., Arana, D. M., Hidalgo-Vico, S., Román, E., & Pla, J. (2021). Identification of Clinical Isolates of Candida albicans with Increased Fitness in Colonization of the Murine Gut. Journal of Fungi, 7(9), 695. https://doi.org/10.3390/jof7090695







