Essential Role of COP9 Signalosome Subunit 5 (Csn5) in Insect Pathogenicity and Asexual Development of Beauveria bassiana
Abstract
1. Introduction
2. Materials and Methods
2.1. Recognition and Bioinformatic Analysis of Fungal Csn5 Orthologues
2.2. Subcellular Localisation of Csn5 in B. bassiana
2.3. Generation of csn5 Mutants
2.4. Western Blot
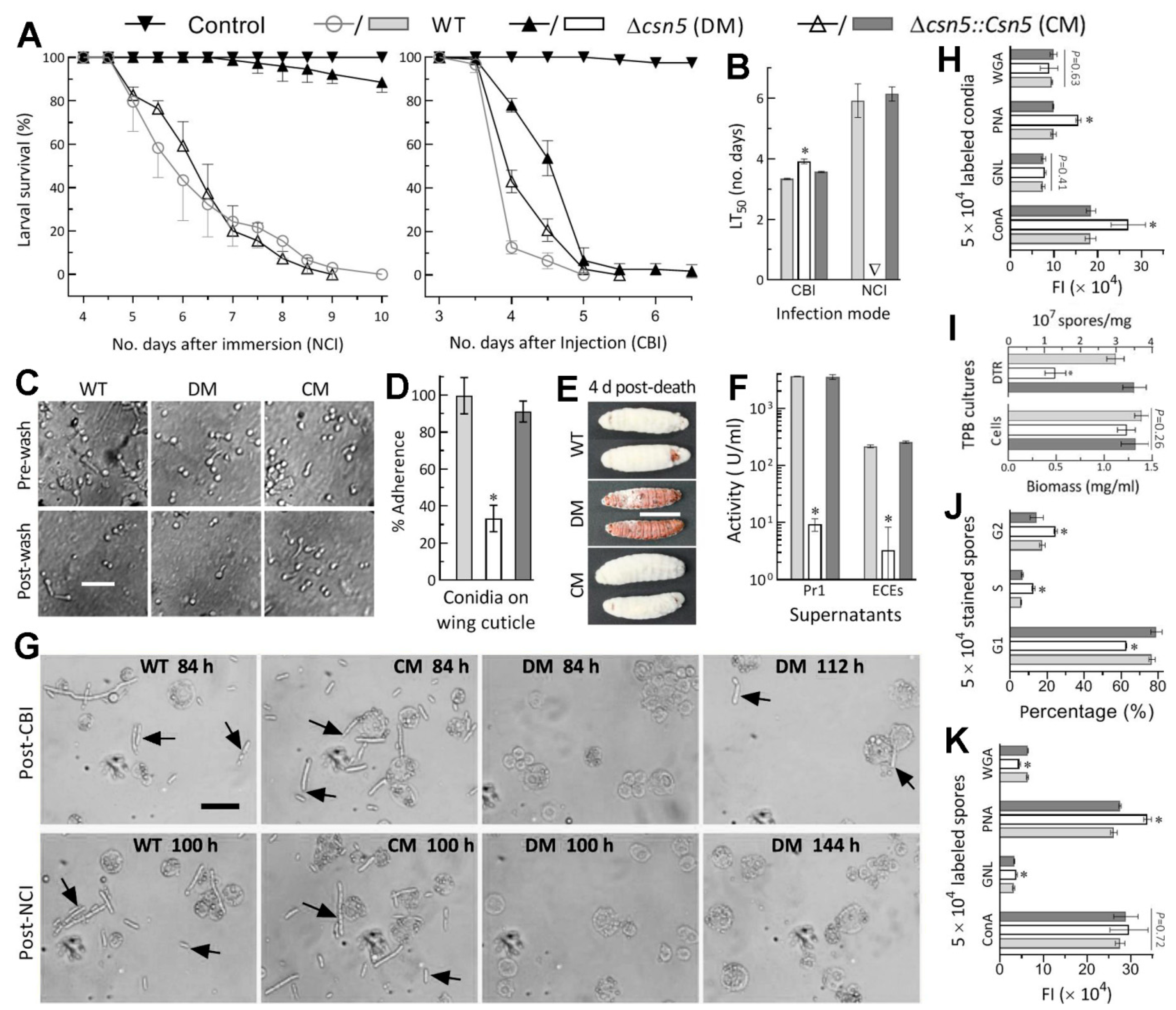
2.5. Bioassays for Fungal Virulence
2.6. Examination of Cellular Events Associated with NCI and Virulence
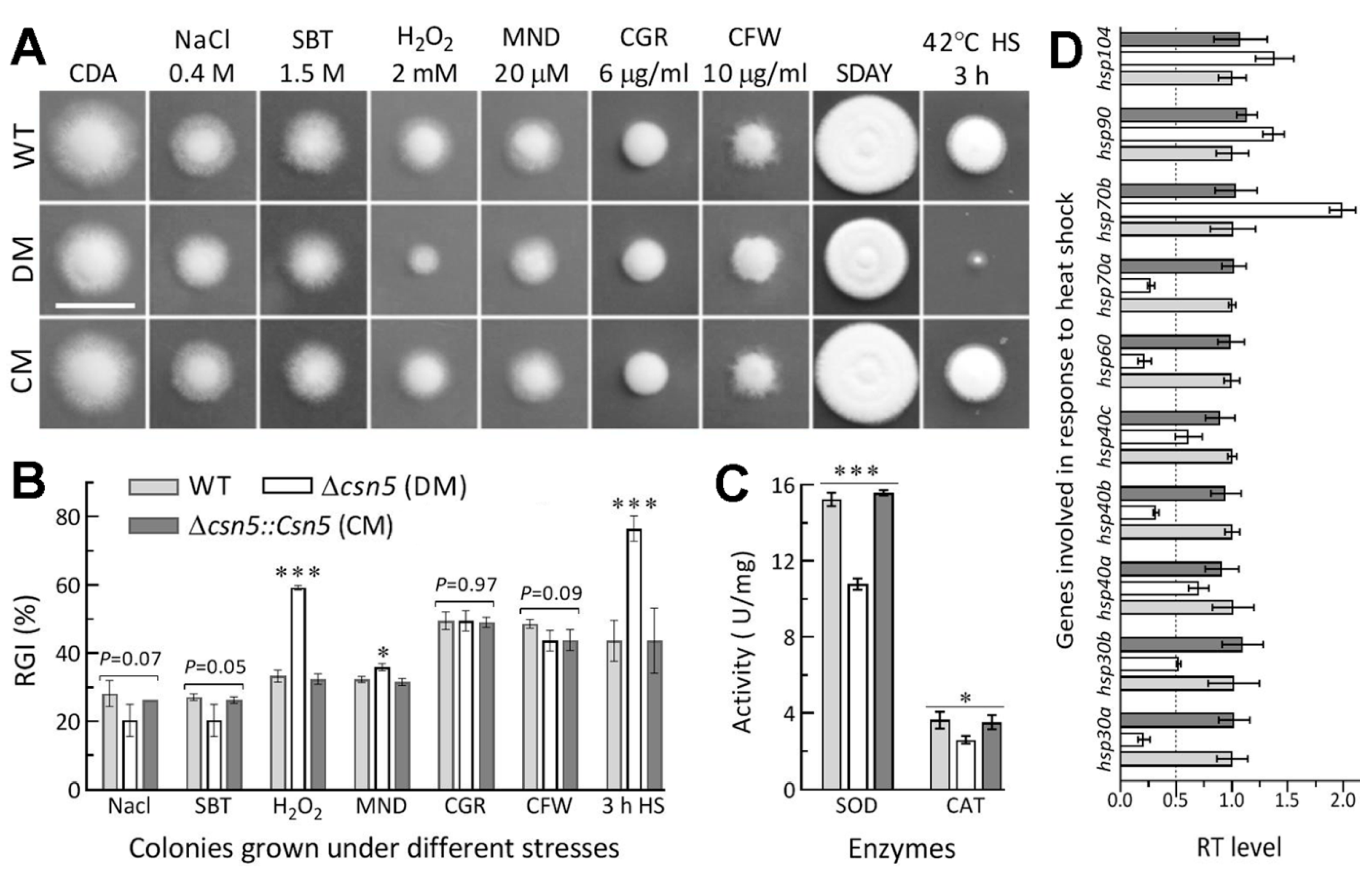
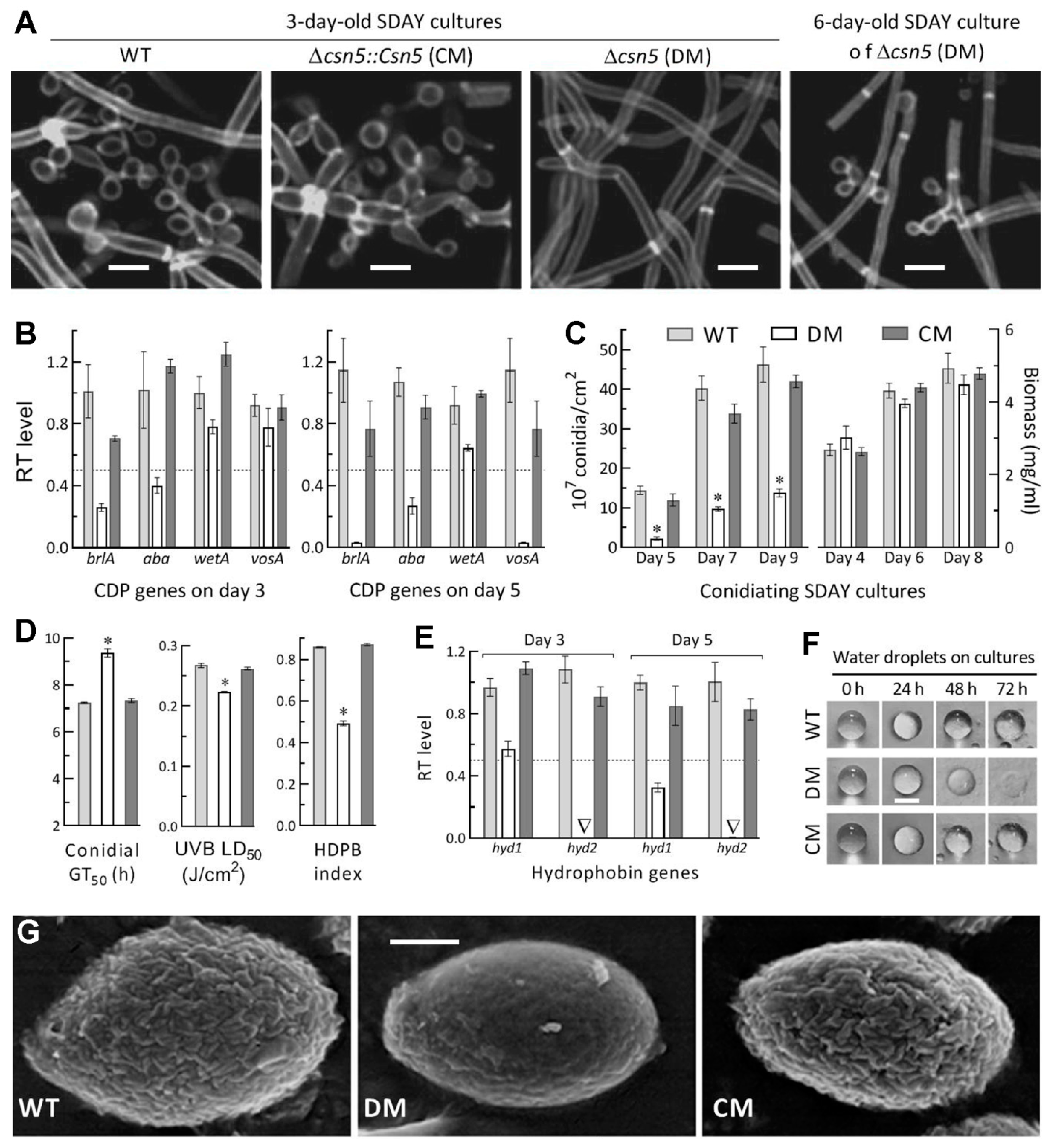
2.7. Assays for Radial Growth, Stress Response, Conidiation Capacity, and Conidial Quality
2.8. Transcription Profiling
3. Results
3.1. Sequence Comparison of Fungal Csn5 Orthologues
3.2. Subcellular Localisation of Csn5 and Its Essentiality for Ubiquitination
3.3. Indispensability of Csn5 for Fungal Insect Pathogenicity
3.4. Roles of Csn5 in Radial Growth and Stress Response
3.5. Essential Roles of Csn5 in Asexual Development and Cell Hydrophobiciy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, C.S.; Wang, S.B. Insect pathogenic fungi: Genomics, molecular interactions, and genetic improvements. Annu. Rev. Entomol. 2017, 62, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.X.; Tong, S.M.; Zeng, D.Y.; Xia, Y.X.; Feng, M.G. Colony heating protects honey bee populations from a risk of contact with wide-spectrum Beauveria bassiana insecticides applied in the field. Pest Manag. Sci. 2020, 76, 2627–2634. [Google Scholar] [CrossRef]
- Zhang, L.B.; Feng, M.G. Antioxidant enzymes and their contributions to biological control potential of fungal insect pathogens. Appl. Microbiol. Biotechnol. 2018, 102, 4995–5004. [Google Scholar] [CrossRef]
- Tong, S.M.; Feng, M.G. Insights into regulatory roles of MAPK-cascaded pathways in multiple stress responses and life cycles of insect and nematode mycopathogens. Appl. Microbiol. Biotechnol. 2019, 103, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.M.; Feng, M.G. Phenotypic and molecular insights into heat tolerance of formulated cells as active ingredients of fungal insecticides. Appl. Microbiol. Biotechnol. 2020, 104, 5711–5724. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.H.; Feng, M.G. Insight into vital role of autophagy in sustaining biological control potential of fungal pathogens against pest insects and nematodes. Virulence 2019, 10, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.Y.; Ren, K.; Tong, S.M.; Ying, S.H.; Feng, M.G. Pleiotropic effects of Ubi4, a polyubiquitin precursor required for ubiquitin accumulation, conidiation and pathogenicity of a fungal insect pathogen. Environ. Microbiol. 2020, 22, 2564–2580. [Google Scholar] [CrossRef]
- Wang, D.Y.; Mou, Y.N.; Du, X.; Guan, Y.; Feng, M.G. Ubr1-mediated ubiquitylation orchestrates developmental pathway, polar growth and virulence-related cellular events in Beauveria bassiana. Appl. Microbiol. Biotechnol. 2021, 105, 2747–2758. [Google Scholar] [CrossRef]
- Jin, J.; Li, X.; Gygi, S.P.; Harper, J.W. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature 2007, 447, 1135–1138. [Google Scholar] [CrossRef]
- Schulman, B.A.; Harper, J.W. Ubiquitin-like protein activation by E1 enzymes: The apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009, 10, 319–331. [Google Scholar] [CrossRef]
- Rape, M. Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 59–70. [Google Scholar] [CrossRef]
- Seeger, M.; Kraft, R.; Ferrell, K.; Bech-Otschir, D.; Dumdey, R.; Schade, R.; Gordon, C.; Naumann, M.; Dubiel, W. A novel protein complex involved in signal transduction possessing similarities to 26S proteasome subunits. FASEB J. 1998, 12, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Groisman, R.; Polanowska, J.; Kuraoka, I.; Sawada, J.; Saijo, M.; Drapkin, R.; Kisselev, A.F.; Tanaka, K.; Nakatani, Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 2003, 113, 357–367. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Deng, X.W. COP9 signalosome revisited: A novel mediator of protein degradation. Trends Cell Biol. 2001, 11, 420–426. [Google Scholar] [CrossRef]
- Cope, G.A.; Deshaies, R.J. COP9 signalosome: A multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 2003, 114, 663–671. [Google Scholar] [CrossRef]
- Chamovitz, D.A. Revisiting the COP9 signalosome as a transcriptional regulator. EMBO Rep. 2009, 10, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Braus, G.H.; Irniger, S.; Bayram, O. Fungal development and the COP9 signalosome. Curr. Opin. Microbiol. 2010, 13, 672–676. [Google Scholar] [CrossRef]
- Nie, X.Y.; Li, B.W.; Wang, S.H. Epigenetic and posttranslational modifications in regulating the biology of Aspergillus species. Adv. Appl. Microbiol. 2018, 105, 191–226. [Google Scholar] [PubMed]
- Wu, G.; Xu, G.; Schulman, B.A.; Jeffrey, P.D.; Harper, J.W.; Pavletich, N.P. Structure of a β-TrCP1-Skp1-β-catenin complex: Destruction motif binding and lysine specificity of the SCFβ-TrCP1 ubiquitin ligase. Mol. Cell 2003, 11, 1445–1456. [Google Scholar] [CrossRef]
- Pintard, L.; Willems, A.; Peter, M. Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 2004, 23, 1681–1687. [Google Scholar] [CrossRef]
- Petroski, M.D.; Deshaies, R.J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005, 6, 9–20. [Google Scholar] [CrossRef]
- Deshaies, R.J.; Emberley, E.D.; Saha, A. Control of cullin-ring ubiquitin ligase activity by nedd8. Subcell. Biochem. 2010, 54, 41–56. [Google Scholar]
- Mundt, K.E.; Porte, J.; Murray, J.M.; Brikos, C.; Christensen, P.U.; Caspari, T.; Hagan, I.M.; Millar, J.B.A.; Simanis, V.; Hofmann, K.; et al. The COP9/signalosome complex is conserved in fission yeast and has a role in S phase. Curr. Biol. 1999, 9, 1427–1430. [Google Scholar] [CrossRef]
- Mundt, K.E.; Liu, C.; Carr, A.M. Deletion mutants in COP9/signalosome subunits in fission yeast Schizosaccharomyces pombe display distinct phenotypes. Mol. Biol. Cell 2002, 13, 493–502. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wee, S.; Hetfeld, B.; Dubiel, W.; Wolf, D.A. Conservation of the COP9/signalosome in budding yeast. BMC Genet. 2002, 3, 15. [Google Scholar] [CrossRef]
- Sinha, A.; Israeli, R.; Cirigliano, A.; Gihaz, S.; Trabelcy, B.; Braus, G.H.; Gerchman, Y.; Fishman, A.; Negri, R.; Rinaldi, T.; et al. The COP9 signalosome mediates the Spt23 regulated fatty acid desaturation and ergosterol biosynthesis. FASEB J. 2020, 34, 4870–4889. [Google Scholar] [CrossRef] [PubMed]
- Sela, N.; Atir-Lande, A.; Kornitzer, D. Neddylation and CAND1 independently stimulate SCF ubiquitin ligase activity in Candida albicans. Eukaryot. Cell 2012, 11, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Cope, G.A.; Suh, G.S.; Aravind, L.; Schwarz, S.E.; Zipursky, S.L.; Koonin, E.V.; Deshaies, R.J. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 2002, 298, 608–611. [Google Scholar] [CrossRef]
- Zhou, Z.P.; Wang, Y.; Cai, G.H.; He, Q. Neurospora COP9 signalosome integrity plays major roles for hyphal growth, conidial development, and circadian function. PLoS Genet. 2012, 8, e1002712. [Google Scholar] [CrossRef]
- Busch, S.; Eckert, S.E.; Krappmann, S.; Braus, G.H. The COP9 signalosome is an essential regulator of development in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 2003, 49, 717–730. [Google Scholar] [CrossRef]
- Busch, S.; Schwier, E.U.; Nahlik, K.; Bayram, O.; Helmstaedt, K.; Draht, O.W.; Krappmann, S.; Valerius, O.; Lipscomb, W.N.; Braus, G.H. An eight-subunit COP9 signalosome with an intact JAMM motif is required for fungal fruit body formation. Proc. Natl. Acad. Sci. USA 2007, 104, 8089–8094. [Google Scholar] [CrossRef]
- Lima, J.F.; Malavazi, I.; Kress, M.R.V.; Savoldi, M.; Goldman, M.H.; Schwier, E.; Braus, G.H.; Goldman, G.H. The csnD/csnE signalosome genes are involved in the Aspergillus nidulans DNA damage response. Genetics 2005, 171, 1003–1015. [Google Scholar] [CrossRef]
- Kress, M.R.V.; Harting, R.; Bayram, O.; Christmann, M.; Irmer, H.; Valerius, O.; Schinke, J.; Goldman, G.H.; Braus, G.H. The COP9 signalosome counteracts the accumulation of cullin SCF ubiquitin E3 RING ligases during fungal development. Mol. Microbiol. 2012, 83, 1162–1177. [Google Scholar] [CrossRef]
- Nahlik, K.; Dumkow, M.; Bayram, O.; Helmstaedt, K.; Busch, S.; Valerius, O.; Gerke, J.; Hoppert, M.; Schwier, E.; Opitz, L.; et al. The COP9 signalosome mediates transcriptional and metabolic response to hormones, oxidative stress protection and cell wall rearrangement during fungal development. Mol. Microbiol. 2010, 78, 964–979. [Google Scholar] [CrossRef]
- Christmann, M.; Schmaler, T.; Gordon, C.; Huang, X.H.; Bayram, O.; Schinke, J.; Stumpf, S.; Dubiel, W.; Braus, G.H. Control of multicellular development by the physically interacting deneddylases DEN1/DenA and COP9 signalosome. PLoS Genet. 2013, 9, e1003275. [Google Scholar] [CrossRef]
- Beckmann, E.A.; Kohler, A.M.; Meister, C.; Christmann, M.; Draht, O.W.; Rakebrandt, N.; Valerius, O.; Braus, G.H. Integration of the catalytic subunit activates deneddylase activity in vivo as final step in fungal COP9 signalosome assembly. Mol. Microbiol. 2015, 97, 110–124. [Google Scholar] [CrossRef] [PubMed]
- Meister, C.; Thieme, K.G.; Thieme, S.; Kohler, A.M.; Schmitt, K.; Valerius, O.; Braus, G.H. COP9 signalosome interaction with UspA/Usp15 deubiquitinase controls VeA-mediated fungal multicellular development. Biomolecules 2019, 9, 238. [Google Scholar] [CrossRef] [PubMed]
- Dubiel, W.; Chaithongyot, S.; Dubiel, D.; Naumann, M. The COP9 signalosome: A multi-DUB complex. Biomolecules 2020, 10, 1082. [Google Scholar] [CrossRef]
- Zheng, Y.J.; Wang, X.N.; Zhang, X.L.; Li, W.; Liu, G.; Wang, S.H.; Yan, X.F.; Zou, H.X.; Yin, W.B. COP9 signalosome subunit PfCsnE regulates secondary metabolism and conidial formation in Pestalotiopsis fici. Sci. China Life Sci. 2017, 60, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.S.; Yang, X.; Ruan, R.X.; Fu, H.L.; Li, H.Y. Csn5 is required for the conidiogenesis and pathogenesis of the Alternaria alternata tangerine pathotype. Front. Microbiol. 2018, 9, 508. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.Y.; Fu, B.; Tong, S.M.; Ying, S.H.; Feng, M.G. Two photolyases repair distinct DNA lesions and reactivate UVB-inactivated conidia of an insect mycopathogen under visible light. Appl. Environ. Microbiol. 2019, 85, e02459-18. [Google Scholar] [CrossRef] [PubMed]
- Mou, Y.N.; Fu, B.; Ren, K.; Ying, S.H.; Feng, M.G. A small cysteine-free protein acts as a novel regulator of fungal insect-patho- genic lifecycle and genomic expression. Msystems 2021, 6, e00098-21. [Google Scholar] [CrossRef]
- Zhou, Q.; Yu, L.; Ying, S.H.; Feng, M.G. Comparative roles of three adhesin genes (adh1–3) in insect-pathogenic lifecycle of Beauveria bassiana. Appl. Microbiol. Biotechnol. 2021, 105, 5491–5502. [Google Scholar] [CrossRef]
- Ortiz-Urquiza, A.; Keyhani, N.O. Action on the surface: Entomopathogenic fungi versus the insect cuticle. Insects 2013, 4, 357–374. [Google Scholar] [CrossRef]
- Gao, B.J.; Mou, Y.N.; Tong, S.M.; Ying, S.H.; Feng, M.G. Subtilisin-like Pr1 proteases marking evolution of pathogenicity in a wide-spectrum insect-pathogenic fungus. Virulence 2020, 11, 365–380. [Google Scholar] [CrossRef]
- Wang, J.; Ying, S.H.; Hu, Y.; Feng, M.G. Mas5, a homologue of bacterial DnaJ, is indispensable for the host infection and environmental adaptation of a filamentous fungal insect pathogen. Environ. Microbiol. 2016, 18, 1037–1047. [Google Scholar] [CrossRef]
- Ren, K.; Mou, Y.N.; Tong, S.M.; Ying, S.H.; Feng, M.G. DIM5/KMT1 controls fungal insect pathogenicity and genome stability by methylation of histone H3K4, H3K9 and H3K36. Virulence 2021, 12, 1306–1322. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.M.; Wang, D.Y.; Cai, Q.; Ying, S.H.; Feng, M.G. Opposite nuclear dynamics of two FRH-dominated frequency proteins orchestrate non-rhythmic conidiation of Beauveria bassiana. Cells 2020, 9, 626. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.M.; Zhang, A.X.; Guo, C.T.; Ying, S.H.; Feng, M.G. Daylight length-dependent translocation of VIVID photoreceptor in cells and its essential role in conidiation and virulence of Beauveria bassiana. Environ. Microbiol. 2018, 20, 169–185. [Google Scholar] [CrossRef]
- Shao, W.; Cai, Q.; Tong, S.M.; Ying, S.H.; Feng, M.G. Nuclear Ssr4 is required for the in vitro and in vivo asexual cycles and global gene activity of Beauveria bassiana. Msystems 2020, 5, e00677-19. [Google Scholar] [CrossRef]
- Wojda, I. Immunity of the greater wax moth Galleria mellonella. Insect Sci. 2017, 24, 342–357. [Google Scholar] [CrossRef]
- Xie, X.Q.; Li, F.; Ying, S.H.; Feng, M.G. Additive contributions of two manganese-cored superoxide dismutases (MnSODs) to anti-oxidation, UV tolerance and virulence of Beauveria bassiana. PLoS ONE 2012, 7, e30298. [Google Scholar]
- Wang, Z.L.; Zhang, L.B.; Ying, S.H.; Feng, M.G. Catalases play differentiated roles in the adaptation of a fungal entomopathogen to environmental stresses. Environ. Microbiol. 2013, 15, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Shi, H.Q.; Ying, S.H.; Feng, M.G. Distinct contributions of one Fe- and two Cu/Zn-cofactored superoxide dismutases to antioxidation, UV tolerance and virulence of Beauveria bassiana. Fungal Genet. Biol. 2015, 81, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ying, S.H.; Hu, Y.; Feng, M.G. Vital role for the J-domain protein Mdj1 in asexual development, multiple stress tolerance and virulence of Beauveria bassiana. Appl. Microbiol. Biotechnol. 2017, 101, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, J.W.; Hu, Y.; Ying, S.H.; Feng, M.G. Roles of six Hsp70 genes in virulence, cell wall integrity, antioxidant activity and multiple stress tolerance of Beauveria bassiana. Fungal Genet. Biol. 2020, 144, 103437. [Google Scholar] [CrossRef]
- Li, F.; Shi, H.Q.; Ying, S.H.; Feng, M.G. WetA and VosA are distinct regulators of conidiation capacity, conidial quality, and biological control potential of a fungal insect pathogen. Appl. Microbiol. Biotechnol. 2015, 99, 10069–10081. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.X.; Mouhoumed, A.Z.; Tong, S.M.; Ying, S.H.; Feng, M.G. BrlA and AbaA govern virulence-required dimorphic switch, conidiation and pathogenicity in a fungal insect pathogen. Msystems 2019, 4, e00140-19. [Google Scholar] [CrossRef]
- Zhang, S.Z.; Xia, Y.X.; Kim, B.; Keyhani, N.O. Two hydrophobins are involved in fungal spore coat rodlet layer assembly and each play distinct roles in surface interactions, development and pathogenesis in the entomopathogenic fungus, Beauveria bassiana. Mol. Microbiol. 2011, 80, 811–826. [Google Scholar] [CrossRef]
- Wösten, H.A.B. Hydrophobins: Multipurpose proteins. Annu. Rev. Microbiol. 2001, 55, 625–646. [Google Scholar] [CrossRef]
- Clancy, L.M.; Jones, R.; Cooper, A.L.; Griffith, G.W.; Santer, R.D. Dose-dependent behavioural fever responses in desert locusts challenged with the entomopathogenic fungus Metarhizium acridum. Sci. Rep. 2018, 8, 14222. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mou, Y.-N.; Ren, K.; Tong, S.-M.; Ying, S.-H.; Feng, M.-G. Essential Role of COP9 Signalosome Subunit 5 (Csn5) in Insect Pathogenicity and Asexual Development of Beauveria bassiana. J. Fungi 2021, 7, 642. https://doi.org/10.3390/jof7080642
Mou Y-N, Ren K, Tong S-M, Ying S-H, Feng M-G. Essential Role of COP9 Signalosome Subunit 5 (Csn5) in Insect Pathogenicity and Asexual Development of Beauveria bassiana. Journal of Fungi. 2021; 7(8):642. https://doi.org/10.3390/jof7080642
Chicago/Turabian StyleMou, Ya-Ni, Kang Ren, Sen-Miao Tong, Sheng-Hua Ying, and Ming-Guang Feng. 2021. "Essential Role of COP9 Signalosome Subunit 5 (Csn5) in Insect Pathogenicity and Asexual Development of Beauveria bassiana" Journal of Fungi 7, no. 8: 642. https://doi.org/10.3390/jof7080642
APA StyleMou, Y.-N., Ren, K., Tong, S.-M., Ying, S.-H., & Feng, M.-G. (2021). Essential Role of COP9 Signalosome Subunit 5 (Csn5) in Insect Pathogenicity and Asexual Development of Beauveria bassiana. Journal of Fungi, 7(8), 642. https://doi.org/10.3390/jof7080642








