Aflatoxin Biosynthesis, Genetic Regulation, Toxicity, and Control Strategies: A Review
Abstract
1. Introduction
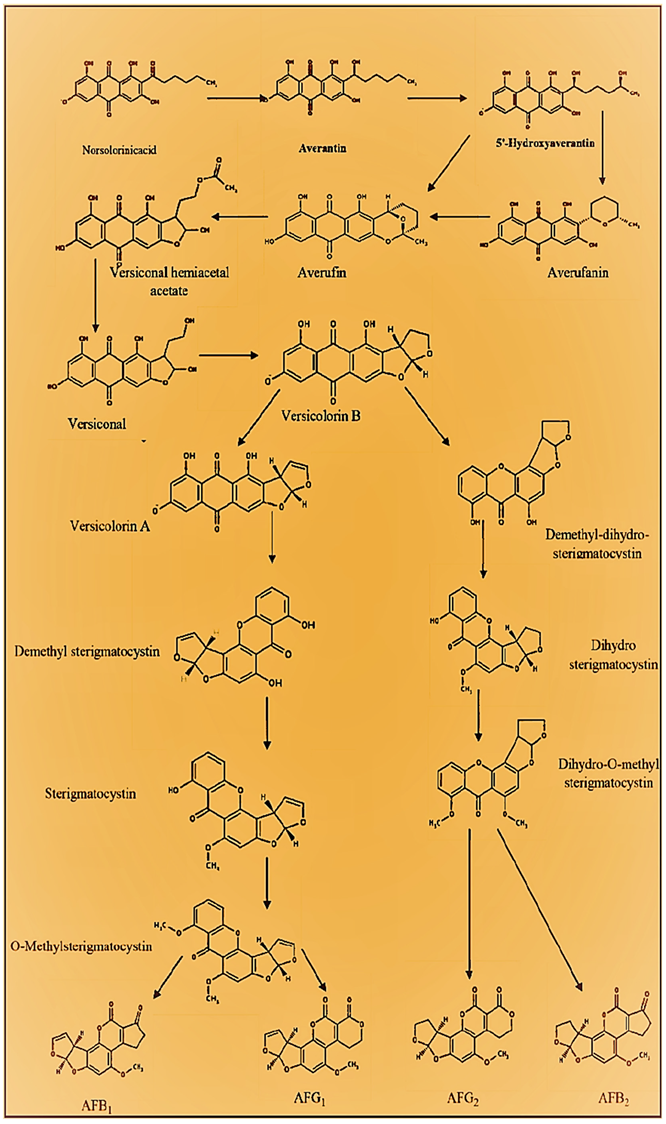
2. Biosynthetic Pathway of Aflatoxins
2.1. Synthesis of Norsolorinic Acid (NOR)
2.2. NOR Conversion to Averantin (AVN)
2.3. AVN Conversion to 5′-Hydroxyaverantin (HAVN)
2.4. HAVN Conversion to Averufin (AVF)
2.5. AVF Conversion to Versiconal Hemiacetal Acetate (VHA)
2.6. VHA Conversion to Versiconal (VAL)
2.7. VAL Conversion to Versicolorin-B (VERB)
2.8. VERB Conversion to Versicolorin A (VERA)-AFB1-AFG1 Pathway
2.9. VERA Conversion to Demethylsterigmatocystin (DMST) and VERB Conversion to Dihydro Demethylsterigmatocystin (DHDMST)
2.10. DMST Conversion to Sterigmatocystin (ST) and DHDMST Conversion to Dihydrosterigmatocystin (DHST)
2.11. ST Conversion to O-Methylsterigmatocystin (OMST) and DHST Conversion to Dihydro-O-Methylsterigmatocystin (DHOMST)
2.12. OMST Conversion to AFB1 and DHOMST Conversion to AFB2
2.13. Bis. OMST Conversion to AFG1 and DHOMST Conversion to AFG2
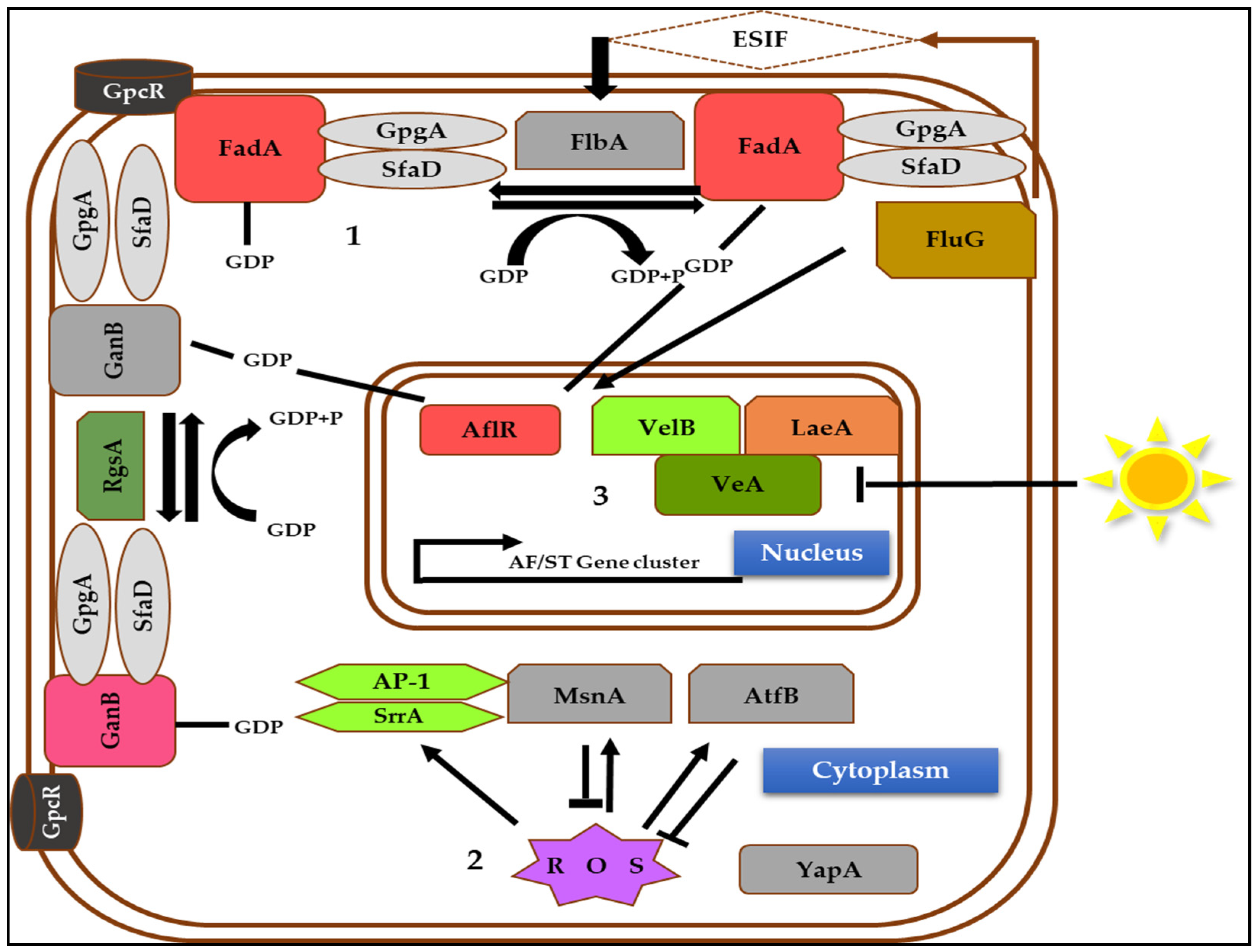
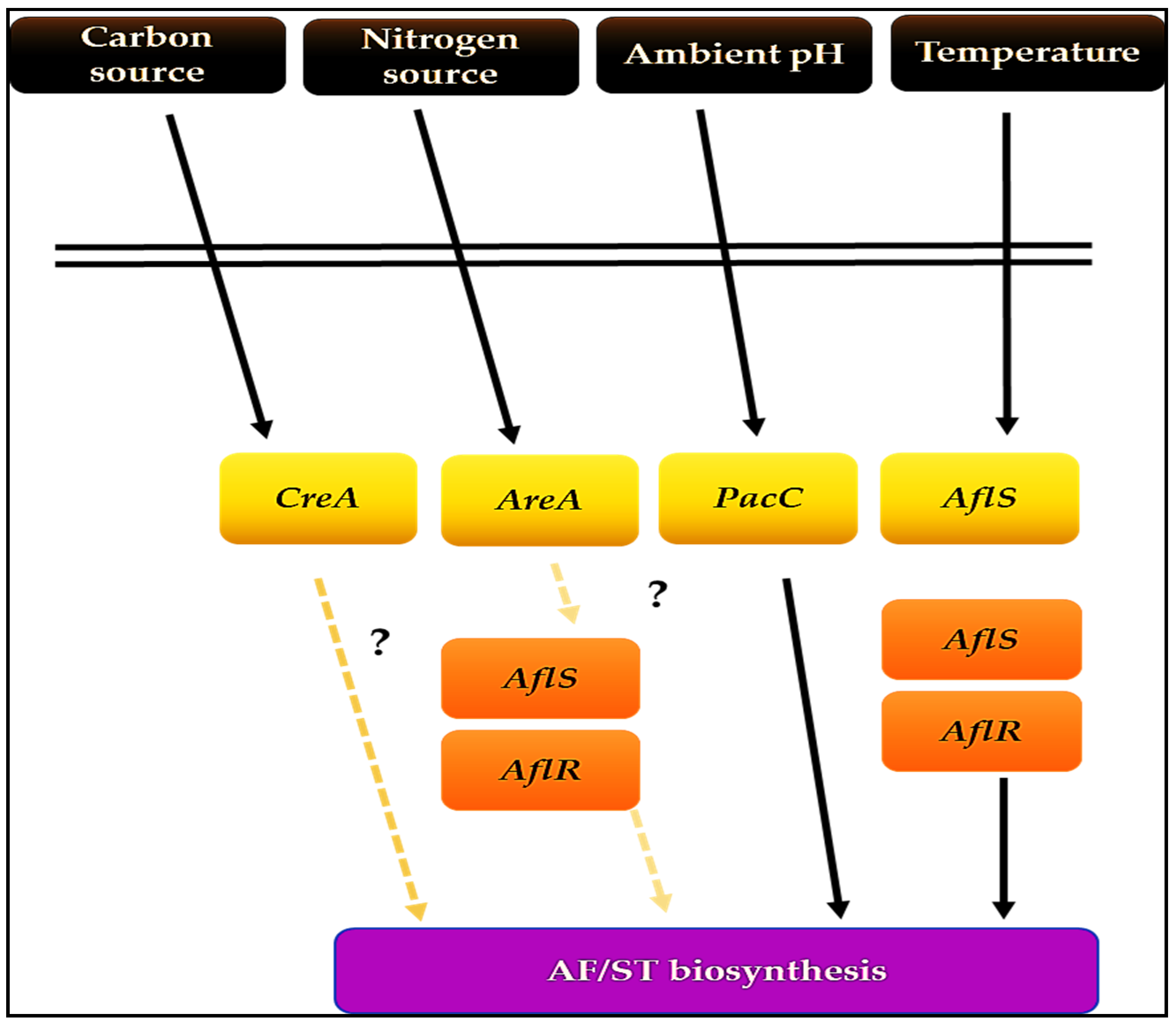
3. Genetic Regulation of Aflatoxin Biosynthesis
3.1. AflR, a Specific Transcription Factor
3.2. AflS, a Putative Transcription Factor
- It may operate as an aflR coactivator [44], although its deletion has little effect on aflR transcript levels.
- It strongly affects the early genes involved in AF production [45].
- AflS mutants inhibit the aflC, aflD, aflM, and aflP’s transcription by up to 20 times yet do not affect the expression of aflR. In contrast, other researchers ruled out the effects of aflS on aflM and aflP’s expression.
- It is vital for LaeA to target a particular gene cluster. Furthermore, it is sensitive to temperature during incubation; henceforth, the expressions of aflS and aflR were increased by 24 times at 30 °C compared to 37 °C [46].
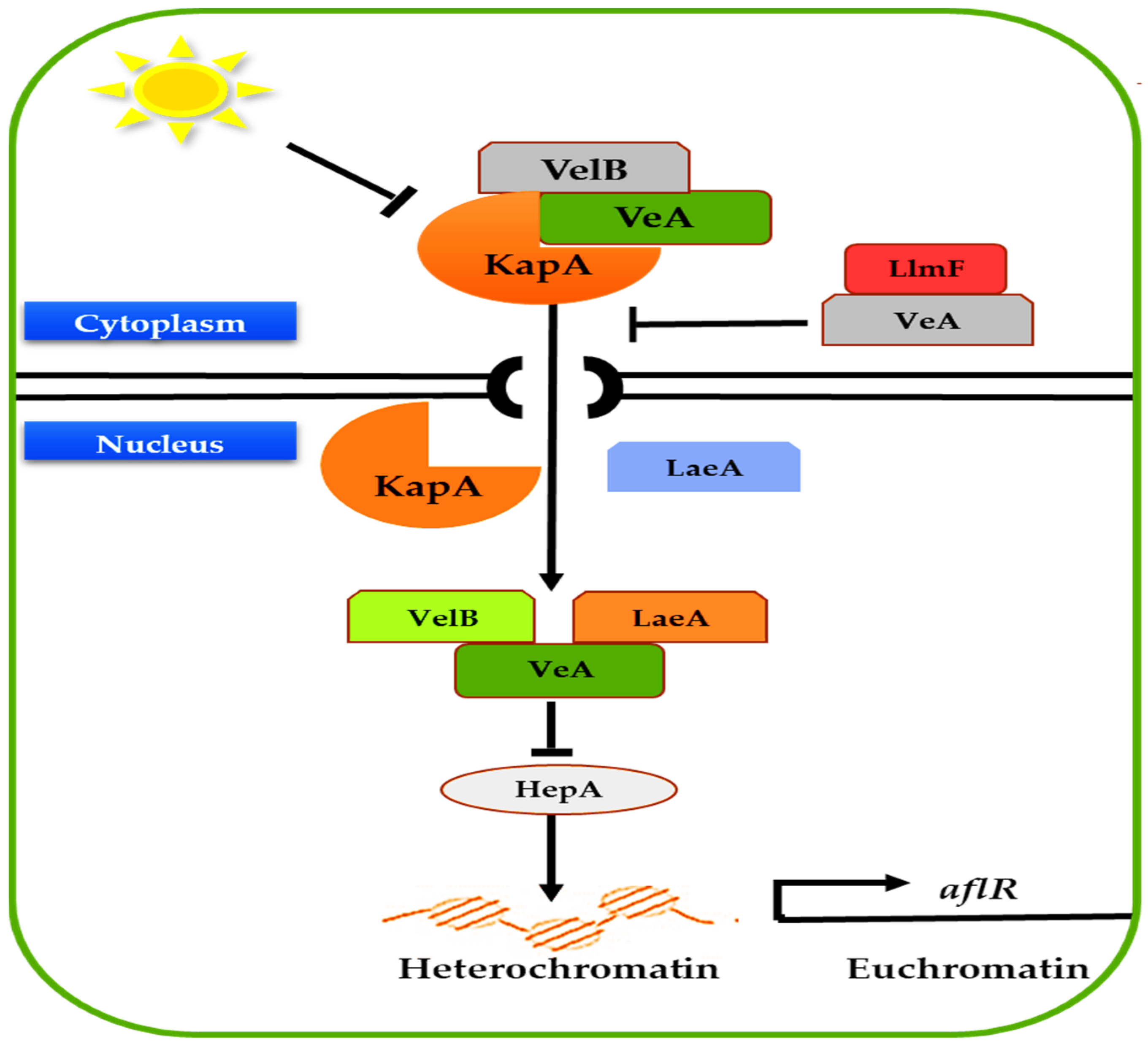
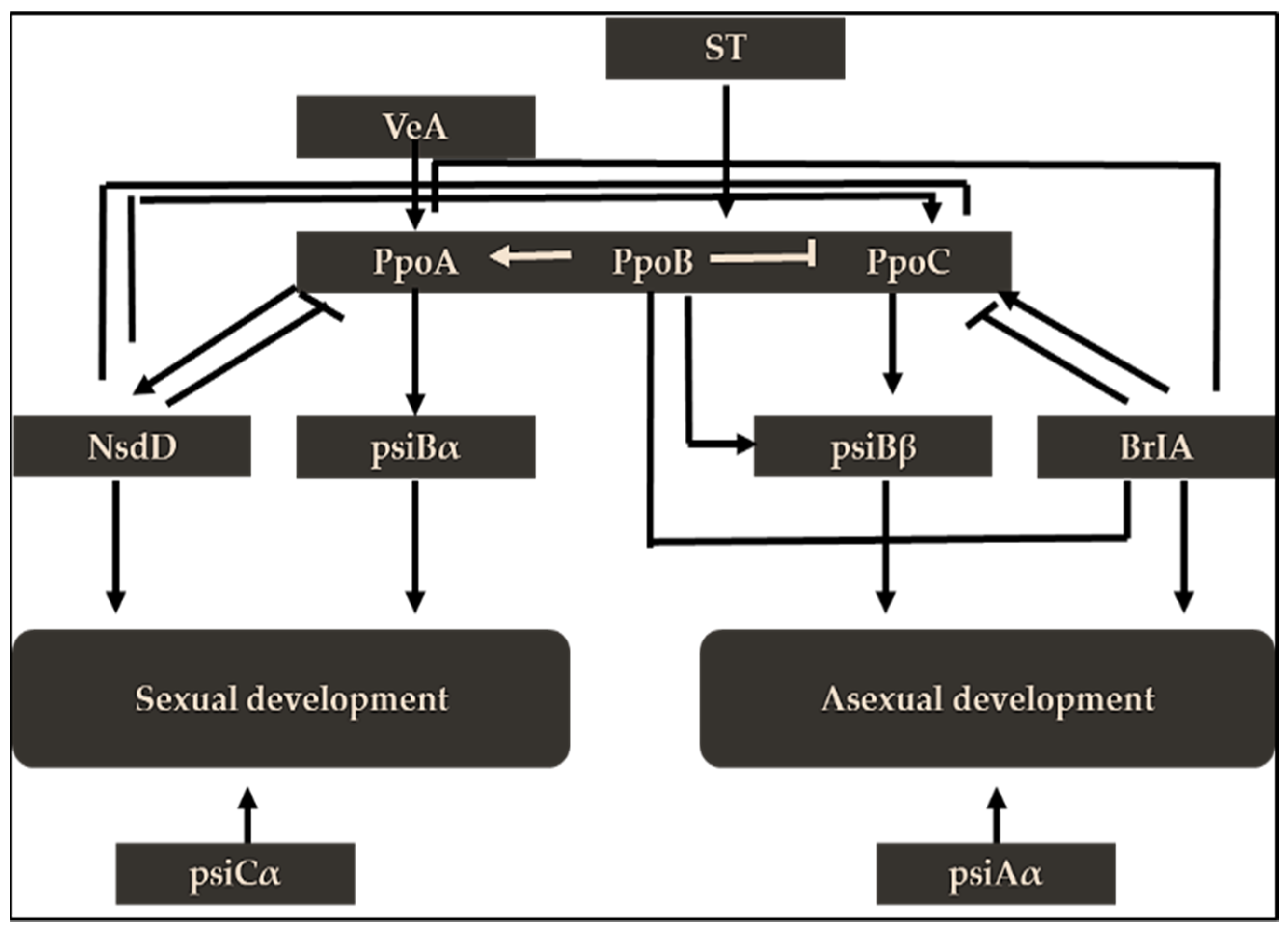
3.3. General Transcription Regulators
4. Aflatoxins Toxicity
4.1. Chronic Aflatoxicosis
4.1.1. Immunotoxicity
4.1.2. Innate Immunity
4.1.3. Adaptive Immunity
4.1.4. Teratogenicity
4.1.5. Malnutrition
4.1.6. Neurodegenerative Diseases
4.2. Acute Toxicity
5. Strategies for Aflatoxin Mitigation
5.1. Bt Corn
5.2. Biocontrol
5.3. Clay
5.4. Plants Volatiles
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Benkerroum, N. Chronic and acute toxicities of aflatoxins: Mechanisms of action. Int. J. Environ. Res. Public Health 2020, 17, 423. [Google Scholar] [CrossRef] [PubMed]
- Pickova, D.; Ostry, V.; Malir, F. A recent overview of producers and important dietary sources of aflatoxins. Toxins 2021, 13, 186. [Google Scholar] [CrossRef]
- Khan, R.; Ghazali, M.F.; Mahyudin, N.A.; Samsudin, N.I.P. Morphological characterization, and determination of aflatoxigenic and non-aflatoxigenic Aspergillus flavus isolated from sweet corn kernels and soil in Malaysia. Agriculture 2020, 10, 450. [Google Scholar] [CrossRef]
- Krulj, J.; Đisalov, J.; Bočarov-Stančić, A.; Pezo, L.; Kojić, J.; Vidaković, A.; Solarov, M.B. Occurrence of aflatoxin B1 in Triticum species inoculated with Aspergillus flavus. World Mycotoxin J. 2018, 11, 247–257. [Google Scholar] [CrossRef]
- Khan, R.; Ghazali, F.M.; Mahyudin, N.A.; Samsudin, N.I.P. Chromatographic analysis of aflatoxigenic Aspergillus flavus isolated from Malaysian sweet corn. Separations 2021, 8, 98. [Google Scholar] [CrossRef]
- Raiola, A.; Tenore, G.C.; Manyes, L.; Meca, G.; Ritieni, A. Risk analysis of main mycotoxins occurring in food for children: An overview. Food Chem. Toxicol. 2015, 84, 169–180. [Google Scholar] [CrossRef]
- Gong, Y.Y.; Watson, S.; Routledge, M.N. Aflatoxin exposure and associated human health effects, a review of epidemiological studies. Food Saf. 2016, 4, 14–27. [Google Scholar] [CrossRef]
- Lauer, J.M.; Natamba, B.K.; Ghosh, S.; Webb, P.; Wang, J.S.; Griffiths, J.K. Aflatoxin exposure in pregnant women of mixed-status of human immunodeficiency virus infection and rate of gestational weight gain: A Ugandan cohort study. Trop. Med. Int. Health 2020, 25, 1145–1154. [Google Scholar] [CrossRef]
- Mitchell, N.J.; Bowers, E.; Hurburgh, C.; Wu, F. Potential economic losses to the US corn industry from aflatoxin contamination. Food Addit. Contam. A 2016, 33, 540–550. [Google Scholar] [CrossRef]
- Matumba, L.; Kimanya, M.; Chunga-Sambo, W.; Munthali, M.; Ayalew, A. Probabilistic dietary based estimation of the burden of aflatoxin-induced hepatocellular carcinoma among adult Malawians. World Mycotoxin J. 2019, 12, 409–419. [Google Scholar] [CrossRef]
- Verheecke, C.; Liboz, T.; Anson, P.; Diaz, R.; Mathieu, F. Reduction of aflatoxin production by Aspergillus flavus and Aspergillus parasiticus in interaction with Streptomyces. Microbiology 2015, 161, 967–972. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Ryu, D. Worldwide occurrence of mycotoxins in cereals and cereal-derived food products: Public health perspectives of their co-occurrence. J. Agric. Food Chem. 2017, 65, 7034–7051. [Google Scholar] [CrossRef]
- Hao, S.; Hu, J.; Song, S.; Huang, D.; Xu, H.; Qian, G.; Huang, K. Selenium alleviates aflatoxin B1-induced immune toxicity through improving glutathione peroxidase 1 and selenoprotein S expression in primary porcine splenocytes. J. Agric. Food Chem. 2016, 64, 1385–1393. [Google Scholar] [CrossRef]
- Xu, Y.; Gong, Y.Y.; Routledge, M.N. Aflatoxin exposure assessed by aflatoxin albumin adduct biomarker in populations from six African countries. World Mycotoxin J. 2018, 11, 411–419. [Google Scholar] [CrossRef]
- Sadhasivam, S.; Britzi, M.; Zakin, V.; Kostyukovsky, M.; Trostanetsky, A.; Quinn, E.; Sionov, E. Rapid detection, and identification of mycotoxigenic fungi and mycotoxins in stored wheat grain. Toxins 2017, 9, 302. [Google Scholar] [CrossRef] [PubMed]
- Caceres, I.; Khoury, A.A.; Khoury, R.E.; Lorber, S.; Oswald, I.P.; Khoury, A.E.; Bailly, J.D. Aflatoxin biosynthesis and genetic regulation: A review. Toxins 2020, 12, 150. [Google Scholar] [CrossRef]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef]
- Benkerroum, N. Retrospective and prospective look at aflatoxin research and development from a practical standpoint. Int. J. Environ. Res. Public Health 2019, 16, 3633. [Google Scholar] [CrossRef]
- Zeng, H.; Cai, J.; Hatabayashi, H.; Nakagawa, H.; Nakajima, H.; Yabe, K. VerA gene is involved in the step to make the xanthone structure of demethylsterigmatocystin in aflatoxin biosynthesis. Int. J. Mol. Sci. 2020, 21, 6389. [Google Scholar] [CrossRef]
- Ehrlich, K.C.; Mack, B.M. Comparison of expression of secondary metabolite biosynthesis cluster genes in Aspergillus flavus, A. parasiticus, and A. oryzae. Toxins 2014, 6, 1916–1928. [Google Scholar] [CrossRef]
- Amare, M.G.; Keller, N.P. Molecular mechanisms of Aspergillus flavus secondary metabolism and development. Fungal Genet. Biol. 2014, 66, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Buitimea-Cantúa, G.V.; Buitimea-Cantúa, N.E.; del Refugio Rocha-Pizaña, M.; Rosas-Burgos, E.C.; Hernández-Morales, A.; Molina-Torres, J. Antifungal and anti-aflatoxigenic activity of Heliopsis longipes roots and affinin/spilanthol against Aspergillus parasiticus by downregulating the expression of aflD and aflR genes of the aflatoxins biosynthetic pathway. J. Environ. Sci. Health B 2020, 55, 210–219. [Google Scholar] [CrossRef]
- Conradt, D.; Schätzle, M.A.; Haas, J.; Townsend, C.A.; Müller, M. New insights into the conversion of versicolorin A in the biosynthesis of aflatoxin B1. J. Am. Chem. Soc. 2015, 137, 10867–10869. [Google Scholar] [CrossRef]
- Herbst, D.A.; Townsend, C.A.; Maier, T. The architectures of iterative type I PKS and FAS. Nat. Prod. Rep. 2018, 35, 1046–1069. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Hubka, V.; Ezekiel, C.N.; Hong, S.B.; Nováková, A.; Chen, A.J.; Houbraken, J. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins, and other mycotoxins. Stud. Mycol. 2019, 93, 1–63. [Google Scholar] [CrossRef]
- Moon, Y.S.; Kim, H.M.; Chun, H.S.; Lee, S.E. Organic acids suppress aflatoxin production via lowering expression of aflatoxin biosynthesis-related genes in Aspergillus flavus. Food Control 2018, 88, 207–216. [Google Scholar] [CrossRef]
- Wu, Y.Z.; Lu, F.P.; Jiang, H.L.; Tan, C.P.; Yao, D.S.; Xie, C.F.; Liu, D.L. The furofuran-ring selectivity, hydrogen peroxide-production, and low Km value are the three elements for highly effective detoxification of aflatoxin oxidase. Food Chem. Toxicol. 2015, 76, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Yabe, K.; Hatabayashi, H.; Ikehata, A.; Zheng, Y.; Kushiro, M. Development of the dichlorvos-ammonia (DV-AM) method for the visual detection of aflatoxigenic fungi. Appl. Microbiol. Biotechnol. 2015, 99, 10681–10694. [Google Scholar] [CrossRef] [PubMed]
- Jahanshiri, Z.; Shams-Ghahfarokhi, M.; Allameh, A.; Razzaghi-Abyaneh, M. Inhibitory effect of eugenol on aflatoxin B1 production in Aspergillus parasiticus by downregulating the expression of significant genes in the toxin biosynthetic pathway. World J. Microbiol. Biotechnol. 2015, 31, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Sakuno, E.; Wen, Y.; Hatabayashi, H.; Arai, H.; Aoki, C.; Yabe, K.; Nakajima, H. Aspergillus parasiticus cyclase catalyzes two dehydration steps in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2005, 71, 2999–3006. [Google Scholar] [CrossRef]
- Wang, B.; Han, X.; Bai, Y.; Lin, Z.; Qiu, M.; Nie, X.; Yuan, J. Effects of nitrogen metabolism on growth and aflatoxin biosynthesis in Aspergillus flavus. J. Hazard. Mater. 2017, 324, 691–700. [Google Scholar] [CrossRef]
- Li, S.; Muhammad, I.; Yu, H.; Sun, X.; Zhang, X. Detection of aflatoxin adducts as potential markers and the role of curcumin in alleviating AFB1-induced liver damage in chickens. Ecotox. Environ. Saf. 2019, 176, 137–145. [Google Scholar] [CrossRef]
- Kolawole, O.; Meneely, J.; Petchkongkaew, A.; Elliott, C. A review of mycotoxin biosynthetic pathways: Associated genes and their expressions under the influence of climatic factors. Fungal Biol. Rev. 2021, 37, 8–26. [Google Scholar] [CrossRef]
- Hosseini, H.M.; Pour, S.H.; Amani, J.; Jabbarzadeh, S.; Hosseinabadi, M.; Mirhosseini, S.A. The effect of Propolis on inhibition of Aspergillus parasiticus growth, aflatoxin production, and expression of aflatoxin biosynthesis pathway genes. J. Environ. Health Sci. Eng. 2020, 18, 297. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Xing, F.; Selvaraj, J.N.; Liu, X.; Wang, L.; Hua, H.; Liu, Y. Inhibitory effect of cinnamaldehyde, citral, and eugenol on aflatoxin biosynthetic gene expression and aflatoxin B1 biosynthesis in Aspergillus flavus. J. Food Sci. 2015, 80, 2917–2924. [Google Scholar] [CrossRef]
- Keller, N.P. Translating biosynthetic gene clusters into fungal armor and weaponry. Nat. Chem. Biol. 2015, 11, 671–677. [Google Scholar] [CrossRef]
- Gallo, A.; Knox, B.P.; Bruno, K.S.; Solfrizzo, M.; Baker, S.E.; Perrone, G. Identification and characterization of the polyketide synthase involved in ochratoxin A biosynthesis in Aspergillus carbonarius. Int. J. Food Microbiol. 2014, 179, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Hathout, A.S.; Aly, S.E. Biological detoxification of mycotoxins: A review. Ann. Microbiol. 2014, 64, 905–919. [Google Scholar] [CrossRef]
- Rao, K.R.; Vipin, A.V.; Venkateswaran, G. Mechanism of inhibition of aflatoxin synthesis by non-aflatoxigenic strains of Aspergillus flavus. Microb. Pathog. 2020, 147, 104280. [Google Scholar]
- Yu, J. Current understanding on aflatoxin biosynthesis and future perspective in reducing aflatoxin contamination. Toxins 2012, 4, 1024–1057. [Google Scholar] [CrossRef]
- Callicott, K.; Cotty, P. Method for monitoring deletions in the aflatoxin biosynthesis gene cluster of Aspergillus flavus with multiplex PCR. Lett. Appl. Microbiol. 2015, 60, 60–65. [Google Scholar] [CrossRef]
- Tang, M.C.; Zou, Y.; Watanabe, K.; Walsh, C.T.; Tang, Y. Oxidative cyclization in natural product biosynthesis. Chem. Rev. 2017, 117, 5226–5333. [Google Scholar] [CrossRef]
- Price, M.S.; Yu, J.; Nierman, W.C.; Kim, H.S.; Pritchard, B.; Jacobus, C.A.; Bhatnagar, D.; Cleveland, T.E.; Payne, G.A. The aflatoxin pathway regulator aflR induces gene transcription inside and outside of the aflatoxin biosynthetic cluster. FEMS Microbiol. Lett. 2006, 255, 275–279. [Google Scholar] [CrossRef]
- Faustinelli, P.C.; Palencia, E.R.; Sobolev, V.S.; Horn, B.W.; Sheppard, H.T.; Lamb, M.C.; Arias, R.S. Study of the genetic diversity of the aflatoxin biosynthesis cluster in Aspergillus section Flavi using insertion/deletion markers in peanut seeds from Georgia, USA. Mycologia 2017, 109, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Oliver, R.A.; Townsend, C.A. Identification and characterization of the sulfazecin monobactams biosynthetic gene cluster. Cell Chem. Biol. 2017, 24, 24–34. [Google Scholar] [CrossRef]
- Ojiambo, P.S.; Battilani, P.; Cary, J.W.; Blum, B.H.; Carbone, I. Cultural and genetic approaches to managing aflatoxin contamination: Recent insights provide opportunities for improved control. Phytopathology 2018, 108, 1024–1037. [Google Scholar] [CrossRef] [PubMed]
- Alkhayyat, F.; Yu, J.H. Upstream regulation of mycotoxin biosynthesis. Adv. Appl. Microbiol. 2014, 86, 251–278. [Google Scholar] [PubMed]
- Chan, P.; Han, X.; Zheng, B.; DeRan, M.; Yu, J.; Jarugumilli, G.K.; Wu, X. Autopalmitoylation of TEAD proteins regulates the transcriptional output of the Hippo pathway. Nat. Chem. Biol. 2016, 12, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Pfannenstiel, B.T.; Zhao, X.; Wortman, J.; Wiemann, P.; Throckmorton, K.; Spraker, J.E.; Lim, F.Y. Revitalization of a forward genetic screen identifies three new regulators of fungal secondary metabolism in the genus Aspergillus. MBio 2017, 8, e01246-17. [Google Scholar] [CrossRef] [PubMed]
- Scala, V.; Giorni, P.; Cirlini, M.; Ludovici, M.; Visentin, I.; Cardinale, F.; Battilani, P. LDS1-produced oxylipins are negative regulators of growth, conidiation, and fumonisin synthesis in the fungal maize pathogen Fusarium verticillioides. Front. Microbiol. 2014, 5, 669. [Google Scholar] [CrossRef]
- Hong, S.Y.; Roze, L.V.; Wee, J.; Linz, J.E. Evidence that a transcription factor regulatory network coordinates oxidative stress response and secondary metabolism in Aspergilli. Microbiologyopen 2013, 2, 144–160. [Google Scholar] [CrossRef]
- Cary, J.; Han, Z.; Yin, Y.; Lohmar, J.; Shantappa, S.; Harris-Coward, P.; Arroyo-Manzanares, N. Transcriptome analysis of Aspergillus flavus reveals the veA-dependent regulation of secondary metabolite gene clusters, including the novel aflavarin cluster. Eukaryot. Cell 2015, 14, 983–997. [Google Scholar] [CrossRef]
- Alvarez-Dominguez, J.R.; Bai, Z.; Xu, D.; Yuan, B.; Lo, K.A.; Yoon, M.J.; Chen, S. De novo reconstruction of adipose tissue transcriptomes reveals long non-coding RNA regulators of brown adipocyte development. Cell Metab. 2015, 21, 764–776. [Google Scholar] [CrossRef]
- Martín, J.F. Key role of LaeA and velvet complex proteins on expression of β-lactam and PR-toxin genes in Penicillium chrysogenum: Cross-talk regulation of secondary metabolite pathways. J. Ind. Microbiol. Biotechnol. 2017, 44, 525–535. [Google Scholar] [CrossRef]
- Reyes-Dominguez, Y.; Bok, J.W.; Berger, H.; Shwab, E.K.; Basheer, A.; Gallmetzer, A.; Scazzocchio, C.; Keller, N.; Strauss, J. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol. Microbiol. 2010, 76, 1376–1386. [Google Scholar] [CrossRef]
- Fischer, G.J.; Keller, N.P. Production of cross-kingdom oxylipins by pathogenic fungi: An update on their role in development and pathogenicity. J. Microbiol. 2016, 54, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Han, J.E.; Seo, M.J.; Shin, K.C.; Oh, D.K. Production of 10R-hydroxy unsaturated fatty acids from hemp seed oil hydrolyzate by recombinant Escherichia coli cells expressing PpoC from Aspergillus nidulans. Appl. Microbiol. Biotechnol. 2016, 100, 7933–7944. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.A.; Kamlangdee, N.; Kelly, J.M. The CreB deubiquitinating enzyme does not directly target the CreA repressor protein in Aspergillus nidulans. Curr. Genet. 2017, 63, 647–667. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Khosravi, C.; Lubbers, R.J.; Kun, R.S.; Pontes, M.V.A.; Battaglia, E.; de Vries, R.P. CreA-mediated repression of gene expression occurs at low monosaccharide levels during fungal plant biomass conversion in a time and substrate-dependent manner. Cell Surf. 2021, 7, 100050. [Google Scholar] [CrossRef]
- Mannaa, M.; Kim, K.D. Influence of temperature and water activity on deleterious fungi and mycotoxin production during grain storage. Mycobiology 2017, 45, 240–254. [Google Scholar] [CrossRef]
- Bills, G.F.; Gloer, J.B. Biologically active secondary metabolites from the fungi. Microbiol. Spectr. 2016, 4, 4–6. [Google Scholar] [CrossRef]
- Fouad, M.A.; Ruan, D.; El-Senousey, K.H.; Chen, W.; Jiang, S.; Zheng, C. Harmful effects and control strategies of aflatoxin B1 produced by Aspergillus flavus and Aspergillus parasiticus strains on poultry: Review. Toxins 2019, 11, 176. [Google Scholar] [CrossRef]
- Kourousekos, G.D.; Theodosiadou, E. Effects of aflatoxins on male reproductive system: A review. J. Hell. Vet. Med. Soc. 2015, 66, 201–210. [Google Scholar] [CrossRef][Green Version]
- McGlynn, K.A.; Petrick, J.L.; El-Serag, H.B. Epidemiology of hepatocellular carcinoma. Hepatology 2021, 73, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Alshannaq, A.F.; Gibbons, J.G.; Lee, M.K.; Han, K.H.; Hong, S.B.; Yu, J.H. Controlling aflatoxin contamination and propagation of Aspergillus flavus by a soy-fermenting Aspergillus oryzae strain. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Marchese, S.; Polo, A.; Ariano, A.; Velotto, S.; Costantini, S.; Severino, L. Aflatoxin B1, and M1: Biological properties and their involvement in cancer development. Toxins 2018, 10, 214. [Google Scholar] [CrossRef] [PubMed]
- Klvana, M.; Bren, U. Aflatoxin B1–formamidopyrimidine DNA adducts relationships between structures, free energies, and melting temperatures. Molecules 2019, 24, 150. [Google Scholar] [CrossRef]
- Turner, P.C. The molecular epidemiology of chronic aflatoxin driven impaired child growth. Scientifica 2013, 2013, 152879. [Google Scholar] [CrossRef]
- WHO (World Health Organization). Evaluation of Certain Contaminants in Food: Eighty-Third Report of the Joint FAO/WHO Expert Committee on Food Additives; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Klingelhöfer, D.; Zhu, Y.; Braun, M.; Bendels, M.H.; Brüggmann, D.; Groneberg, D.A. Aflatoxin–publication analysis of a global health threat. Food Control 2018, 89, 280–290. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, F. Global burden of aflatoxin-induced hepatocellular carcinoma: A risk assessment. Environ. Health Perspect. 2010, 118, 818–824. [Google Scholar] [CrossRef] [PubMed]
- Habibi, N.; Nassiri-Toosi, M.; Sharafi, H.; Alavian, S.M.; Shams-Ghahfarokhi, M.; Razzaghi-Abyaneh, M. Aflatoxin B1 exposure and the risk of hepatocellular carcinoma in Iranian carriers of viral hepatitis B and C. Toxin Rev. 2019, 38, 234–239. [Google Scholar] [CrossRef]
- Gibb, H.; Devleesschauwer, B.; Bolger, P.M.; Wu, F.; Ezendam, J.; Cliff, J.; Zeilmaker, M.; Verger, P.; Pitt, J.; Baines, J. World Health Organization estimates of the global and regional disease burden of four foodborne chemical toxins, 2010: A data synthesis. F1000Reserch 2015, 4, 1393. [Google Scholar] [CrossRef]
- Yao, J.G.; Huang, X.Y.; Long, X.D. Interaction of DNA repair gene polymorphisms and aflatoxin B1 in the risk of hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 6231–6244. [Google Scholar] [PubMed]
- Vartanian, V.; Minko, I.G.; Chawanthayatham, S.; Egner, P.A.; Lin, Y.C.; Earley, L.F.; Makar, R.; Eng, J.R.; Camp, M.T.; Li, L. NEIL1 protects against aflatoxin-induced hepatocellular carcinoma in mice. Proc. Natl. Acad. Sci. USA 2017, 114, 4207–4212. [Google Scholar] [CrossRef] [PubMed]
- Long, X.D.; Huang, H.D.; Xia, Q. The polymorphism of XRCC3 codon 241 and the hotspot mutation in the TP53 gene in hepatocellular carcinoma induced by aflatoxin B1. J. Tumor 2014, 2, 272–277. [Google Scholar]
- Long, X.D.; Zhao, D.; Wang, C.; Huang, X.Y.; Yao, J.G.; Ma, Y.; Wei, Z.H.; Liu, M.; Zeng, L.X.; Mo, X.Q.; et al. Genetic polymorphisms in DNA repair genes XRCC4 and XRCC5 and aflatoxin B1-related hepatocellular carcinoma. Epidemiology 2013, 24, 671–681. [Google Scholar] [CrossRef]
- Arafa, M.; Besheer, T.; El-Eraky, A.M.; Abo El-khair, S.M.; Elsamanoudy, A.Z. Genetic variants of XRCC1 and risk of hepatocellular carcinoma in chronic hepatitis C patients. Br. J. Biomed. Sci. 2019, 76, 64–69. [Google Scholar] [CrossRef]
- Coppock, R.W.; Christian, R.G.; Jacobsen, B.J. Aflatoxins. In Veterinary Toxicology, 3rd ed.; Gupta, R.C., Ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 983–994. [Google Scholar]
- Raafat, N.; Emam, W.A.; Gharib, A.F.; Nafea, O.E.; Zakaria, M. Assessment of serum aflatoxin B1 levels in neonatal jaundice with glucose-6-phosphate dehydrogenase deficiency: A preliminary study. Mycotoxin Res. 2021, 37, 109–116. [Google Scholar] [CrossRef]
- Mohsenzadeh, M.S.; Hedayati, N.; Riahi-Zanjani, B.; Karimi, G. Immunosuppression following dietary aflatoxin B1 exposure: A review of the existing evidence. Toxin Rev. 2016, 35, 121–127. [Google Scholar] [CrossRef]
- Barany, A.; Guilloto, M.; Cosano, J.; De Boevre, M.; Oliva, M.; De Saeger, S.; Mancera, J.M. Dietary aflatoxin B1 (AFB1) reduces growth performance, affecting growth axis, metabolism, and tissue integrity in juvenile gilthead sea bream (Sparus aurata). Aquaculture 2021, 533, 736189. [Google Scholar] [CrossRef]
- Gashaw, M. Review on Mycotoxins in Feeds: Implications to livestock and human health. J. Agric. Res. Dev. 2016, 5, 137–144. [Google Scholar]
- Mohammadi, A.; Mehrzad, J.; Mahmoudi, M.; Schneider, M. Environmentally appropriate level of aflatoxin B1 dysregulates human dendritic cells through signaling on key toll-like receptors. Int. J. Toxicol. 2014, 33, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Jolly, P.E. Aflatoxin: Does it contribute to an increase in HIV viral load? Future Microbiol. 2014, 9, 121–124. [Google Scholar] [CrossRef] [PubMed]
- Shirani, K.; Zanjani, B.R.; Mahmoudi, M.; Jafarian, A.H.; Hassani, F.V.; Giesy, J.P.; Karimi, G. Immunotoxicity of aflatoxin M1: As a potent suppressor of innate and acquired immune systems in a subacute study. J. Sci. Food Agric. 2018, 98, 5884–5892. [Google Scholar] [CrossRef] [PubMed]
- Bianco, G.; Russo, R.; Marzocco, S.; Velotto, S.; Autore, G.; Severino, L. Modulation of macrophage activity by aflatoxins B1 and B2 and their metabolites aflatoxins M1 and M2. Toxicon 2012, 59, 644–650. [Google Scholar] [CrossRef]
- Chaytor, A.C.; See, M.T.; Hansen, J.A.; de Souza, A.L.P.; Middleton, T.F.; Kim, S.W. Effects of chronic exposure of diets with reduced aflatoxin concentrations deoxynivalenol on growth and immune status of pigs1. J. Anim. Sci. 2011, 89, 124–135. [Google Scholar] [CrossRef]
- Adhikari, M.; Negi, B.; Kaushik, N.; Adhikari, A.; Al-Khedhairy, A.A.; Kaushik, N.K.; Choi, E.H. T-2 mycotoxin: Toxicological effects and decontamination strategies. Oncotarget 2017, 8, 33933–33952. [Google Scholar] [CrossRef] [PubMed]
- Giles, A.J.; Hutchinson, M.K.N.; Sonnemann, H.M.; Jung, J.; Fecci, P.E.; Ratnam, N.M.; Gilbert, M.R. Dexamethasone-induced immunosuppression: Mechanisms and implications for immunotherapy. J. Immunother. Cancer 2018, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Valtchev, I.; Koynarski, T.; Sotirov, L.; Nikolov, Y.; Petkov, P. Effect of aflatoxin B1 on moulard duck’s natural immunity. Pak. Vet. J. 2015, 35, 67–70. [Google Scholar]
- Yunus, A.W.; Razzazi-Fazeli, E.; Bohm, J. Aflatoxin B1 in affecting broiler’s performance, immunity, and gastrointestinal tract: A review of history and contemporary issues. Toxins 2011, 3, 566–590. [Google Scholar] [CrossRef]
- Doi, K.; Uetsuka, K. Mechanisms of mycotoxin-induced dermal toxicity and tumorigenesis through oxidative stress-related pathways. J. Toxicol. Pathol. 2014, 27, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wangia, R.N.; Tang, L.; Wang, J.S. Occupational exposure to aflatoxins and health outcomes: A review. J. Environ. Sci. Health C 2019, 37, 215–234. [Google Scholar] [CrossRef]
- Chaudhary, Z.; Rehman, K.; Akash, M.S.H. Mechanistic Insight of Mycotoxin-Induced Neurological Disorders and Treatment Strategies. Environ. Contam. Neurol. Disord. 2021, 125–146. [Google Scholar] [CrossRef]
- Yin, H.; Jiang, M.; Peng, X.; Cui, H.; Zhou, Y.; He, M.; Zuo, Z.; Ouyang, P.; Fan, J.; Fang, J. The molecular mechanism of G2M cell cycle arrest induced by AFB1 in the jejunum. Oncotarget 2016, 7, 35592–35606. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zuo, Z.; Chen, K.; Gao, C.; Yang, Z.; Zhao, S.; Li, J.; Song, H.; Peng, X.; Fang, J.; et al. Histopathological injuries, ultrastructural changes, and depressed TLR expression in the small intestine of broiler chickens with aflatoxin B1. Toxins 2018, 10, 131. [Google Scholar] [CrossRef]
- Chatterjee, D.; Ghosh, P. Sub-cytotoxic concentration of aflatoxin B2 prevents NO-mediated Increased mitochondrial membrane potential and intracellular killing of Candida albicans in macrophages. Adv. Life Sci. 2012, 2, 52–56. [Google Scholar] [CrossRef]
- Ma, J.; Liu, Y.; Guo, Y.; Ma, Q.; Ji, C.; Zhao, L. Transcriptional profiling of aflatoxin B1-induced oxidative stress and inflammatory response in macrophages. Toxins 2021, 13, 401. [Google Scholar] [CrossRef]
- Brown, R.; Priest, E.; Naglik, J.R.; Richardson, J.P. Fungal Toxins and Host Immune Responses. Front. Microbiol. 2021, 12, 697. [Google Scholar] [CrossRef]
- Lin, S.; Gao, P.; Li, Q.; Zhang, Y.; Hu, J.; Cai, M.; Zhou, P. Aflatoxin influences achalasia symptomatology. Mol. Med. Rep. 2020, 21, 1276–1284. [Google Scholar] [CrossRef]
- Malvandi, A.M.; Mehrzad, J.; Saleh-Moghaddam, M. Biologically relevant doses of mixed aflatoxins B and G up-regulate MyD88, TLR2, TLR4 and CD14 transcripts in human PBMCs. Immunopharmacol. Immunotoxicol. 2013, 35, 528–532. [Google Scholar] [CrossRef]
- Chen, X.; Horn, N.; Cotter, P.F.; Applegate, T.J. Growth, serum biochemistry, complement activity, and liver gene expression responses of Pekin ducklings to graded levels of cultured aflatoxin B1. Poult. Sci. 2014, 93, 2028–2036. [Google Scholar] [CrossRef]
- Monson, M.S.; Coulombe, R.A.; Reed, K.M. Aflatoxicosis: Lessons from toxicity and responses to aflatoxin B1 in poultry. Agriculture 2015, 5, 742–777. [Google Scholar] [CrossRef]
- Pierron, A.; Alassane-Kpembi, I.; Oswald, I.P. Impact of mycotoxin on immune response and consequences for pig health. Anim. Nutr. 2016, 2, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Shahabi-Ghahfarokhi, B.; Gholami-Ahangaran, M.; Dehkordi, M. Aflatoxin effect on humoral and mucosal immune responses against infectious bronchitis vaccine in broilers. Thai Vet. Med. 2016, 46, 149–153. [Google Scholar]
- Mikami, O.; Yamaguchi, H.; Murata, H.; Nakajima, Y.; Miyazaki, S. Induction of apoptotic lesions in the liver and lymphoid tissues and modulation of cytokine mRNA expression by acute exposure to deoxynivalenol in piglets. J. Vet. Sci. 2010, 11, 107–113. [Google Scholar] [CrossRef]
- Alpsoy, L.; Kotan, E.; Tatar, A.; Agar, G. Protective effects of selenium against sister chromatid exchange induced by AFG1 in human lymphocytes in vitro. Hum. Exp. Toxicol. 2011, 30, 515–519. [Google Scholar] [CrossRef] [PubMed]
- Mamo, F.T.; Abate, B.A.; Tesfaye, K.; Nie, C.; Wang, G.; Liu, Y. Mycotoxins in Ethiopia: A review on prevalence, economic and health impacts. Toxins 2020, 12, 648. [Google Scholar] [CrossRef]
- Germic, N.; Frangez, Z.; Yousefi, S.; Simon, H.U. Regulation of the innate immune system by autophagy: Monocytes, macrophages, dendritic cells, and antigen presentation. Cell Death Differ. 2019, 26, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, A.T.; Hirooka, E.Y.; Alvares, E.; Silva, P.L.; Bracarense, A.P.F.R.L.; Flaiban, K.K.M.D.C.; Akagi, C.Y.; Kawamura, O.; Costa, M.C.D.; Itano, E.N. Impact of a single oral acute dose of aflatoxin B1 on liver function/cytokines and the lymphoproliferative response in C57Bl/6 mice. Toxins 2017, 9, 374. [Google Scholar] [CrossRef]
- Smith, L.E.; Prendergast, A.J.; Turner, P.C.; Humphrey, J.H.; Stoltzfus, R.J. Aflatoxin exposure during pregnancy, maternal anemia, and adverse birth outcomes. Am. J. Trop. Med. Hyg. 2017, 96, 770–776. [Google Scholar] [CrossRef]
- Maleki, F.; Abdi, S.; Davodian, E.; Haghani, K.; Bakhtiyari, S. Exposure of infants to aflatoxin M1 from mother’s breast milk in Ilam, Western Iran. Osong Public Health Res. Perspect. 2015, 6, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Khlangwiset, P.; Shephard, G.S.; Wu, F. Aflatoxins and growth impairment: A review. Crit. Rev. Toxicol. 2011, 41, 740–755. [Google Scholar] [CrossRef]
- Watson, S.; Moore, S.E.; Darboe, M.K.; Chen, G.; Tu, Y.K.; Huang, Y.T.; Gong, Y.Y. Impaired growth in rural Gambian infants exposed to aflatoxin: A prospective cohort study. BMC Public Health 2018, 18, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Partanen, H.A.; El-Nezami, H.S.; Leppänen, J.M.; Myllynen, P.K.; Woodhouse, H.J.; Vähäkangas, K.H. Aflatoxin B1 transfer and metabolism in human placenta. Toxicol. Sci. 2010, 113, 216–225. [Google Scholar] [CrossRef]
- Kyei, N.N.; Boakye, D.; Gabrysch, S. Maternal mycotoxin exposure and adverse pregnancy outcomes: A systematic review. Mycotoxin Res. 2020, 36, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Shuaib, F.M.; Jolly, P.E.; Ehiri, J.E.; Yatich, N.; Jiang, Y.; Funkhouser, E.; Person, S.D.; Wilson, C.; Ellis, W.O.; Wang, J.S. Association between birth outcomes and aflatoxin B1 biomarker blood levels in pregnant women in Kumasi, Ghana. Trop. Med. Int. Health 2010, 15, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Ghazali, F.M.; Mahyudin, N.A.; Samsudin, N.I.P. Biocontrol of aflatoxins using non-aflatoxigenic Aspergillus flavus: A literature review. J. Fungi. 2021, 7, 381. [Google Scholar] [CrossRef]
- Hayashi, A.; Dorantes-Aranda, J.; Bowman, P.J.; Hallegraeff, G. Combined cytotoxicity of the phycotoxin okadaic acid and mycotoxins on intestinal and neuroblastoma human cell models. Toxins 2018, 10, 526. [Google Scholar] [CrossRef]
- El Khoury, D.; Fayjaloun, S.; Nassar, M.; Sahakian, J.; Aad, Y.P. Updates on the effect of mycotoxins on male reproductive efficiency in mammals. Toxins 2019, 11, 515. [Google Scholar] [CrossRef]
- Obuseh, F.A.; Jolly, P.E.; Kulczycki, A.; Ehiri, J.; Waterbor, J.; Desmond, R.A.; Preko, P.O.; Jiang, Y.; Piyathilake, C.J. Aflatoxin levels, plasma vitamins A and E concentrations, and their association with HIV and hepatitis B virus infections in Ghanaians: A cross-sectional study. J. Int. AIDS Soc. 2011, 14, 53. [Google Scholar] [CrossRef]
- Zhao, L.; Feng, Y.; Deng, J.; Zhang, N.Y.; Zhang, W.P.; Liu, X.L.; Rajput, S.A.; Qi, D.S.; Sun, L.H. Selenium deficiency aggravates aflatoxin B1-induced immunotoxicity in chick spleen by regulating six selenoprotein genes and redox/inflammation/apoptotic signaling. J. Nutr. 2019, 149, 894–901. [Google Scholar] [CrossRef]
- Knipstein, B.; Huang, J.; Barr, E.; Sossenheimer, P.; Dietzen, D.; Egner, P.A.; Rudnick, D.A. Dietary aflatoxin-induced stunting in a novel rat model: Evidence for toxin-induced liver injury and hepatic growth hormone resistance. Pediatr. Res. 2015, 78, 120–127. [Google Scholar] [CrossRef]
- Monyo, E.S.; Njoroge, S.M.C.; Coe, R.; Osiru, M.; Madinda, F.; Waliyar, F.; Thakur, R.P.; Chilunjika, T.; Anitha, S. Occurrence and distribution of aflatoxin contamination in groundnuts (Arachis hypogaea L.) and population density of aflatoxigenic Aspergilli in Malawi. Crop Prot. 2012, 42, 149–155. [Google Scholar] [CrossRef]
- Onyemelukwe, G.; Ogoina, D.; Ibiam, G.E.; Ogbadu, G.H. Aflatoxins in body fluids and Nigerian children with protein-energy malnutrition. Afr. J. Food Agric. Nutr. Dev. 2012, 12, 6553–6566. [Google Scholar]
- Ayelign, A.; Woldegiorgis, A.Z.; Adish, A.; De Boevre, M.; Heyndrickx, E.; De Saeger, S. Assessment of aflatoxin exposure among young children in Ethiopia using urinary biomarkers. Food Addit. Contam. A 2017, 34, 1606–1616. [Google Scholar] [CrossRef]
- Soriano, J.M.; Rubini, A.; Morales-Suarez, M.; Merino-Torres, J.F.; Silvestre, D. Aflatoxins in organs and biological samples from children affected by kwashiorkor, marasmus, and marasmic-kwashiorkor: A scoping review. Toxicon 2020, 185, 174–183. [Google Scholar] [CrossRef] [PubMed]
- McMillan, A.; Renaud, J.B.; Burgess, K.M.; Orimadegun, A.E.; Akinyinka, O.O.; Allen, S.J.; Sumarah, M.W. Aflatoxin exposure in Nigerian children with severe acute malnutrition. Food Chem. Toxicol. 2018, 111, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Liu, J.; Wang, Y.; Lian, H.; Wang, J.; Xing, L.; Zhang, X. Aflatoxin G1-induced oxidative stress causes DNA damage and triggers apoptosis through MAPK signaling pathway in A549 cells. Food Chem. Toxicol. 2013, 62, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Wahhab, M.A.; El-Nekeety, A.A.; Hathout, A.S.; Salman, A.S.; Abdel-Aziem, S.H.; Sabry, B.A.; Jaswir, I. Bioactive compounds from Aspergillus niger extract enhance the antioxidant activity and prevent the genotoxicity in aflatoxin B1-treated rats. Toxicon 2020, 181, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Rushing, B.R.; Selim, M.I. Structure and oxidation of pyrrole adducts formed between aflatoxin B2a and biological amines. Chem. Res. Toxicol. 2017, 30, 1275–1285. [Google Scholar] [CrossRef]
- Zamir-Nasta, T.; Razi, M.; Shapour, H.; Malekinejad, H. Roles of p21, p53, cyclin D1, CDK-4, estrogen receptor alpha in aflatoxin B1-induced cytotoxicity in testicular tissue of mice. Environ. Toxicol. 2018, 33, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Diaz, G.J.; Murcia, H.W. An unusually high production of hepatic aflatoxin B1-dihydrodiol, the possible explanation for the high susceptibility of ducks to aflatoxin B1. Sci. Rep. 2019, 9, 8010. [Google Scholar] [CrossRef] [PubMed]
- Colakoglu, F.; Donmez, H.H. Effects of aflatoxin on the liver and shielding effectiveness of esterified glucomannan in Merino rams. Sci. World J. 2012, 2012, 462925. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Liu, L.; Zhao, L.; Wang, X.; Wang, D.; Huang, C.; Zhang, J.; Ji, C.; Ma, Q. Influence of Bacillus subtilis ANSB060 on growth, digestive enzyme and aflatoxin residue in Yellow River carp fed diets contaminated with aflatoxin B1. Food Chem. Toxicol. 2018, 113, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Chen, L.; Zhu, Y.; Huang, Y.; Hu, X.; Wu, Q.; Nüssler, A.K.; Liu, L.; Yang, W. Current major degradation methods for aflatoxins: A review. Trends Food Sci. Technol. 2018, 80, 155–166. [Google Scholar] [CrossRef]
- Bediako, K.A.; Ofori, K.; Offei, S.K.; Dzidzienyo, D.; Asibuo, J.Y.; Amoah, R.A. Aflatoxin contamination of groundnut (Arachis hypogaea L.): Predisposing factors and management interventions. Food Control 2019, 98, 61–67. [Google Scholar] [CrossRef]
- Theumer, M.G.; Henneb, Y.; Khoury, L.; Snini, S.P.; Tadrist, S.; Canlet, C.; Audebert, M. Genotoxicity of aflatoxins and their precursors in human cells. Toxicol. Lett. 2018, 287, 100–107. [Google Scholar] [CrossRef]
- Rohlfs, M. Fungal secondary metabolite dynamics in fungus–grazer interactions: Novel insights and unanswered questions. Front. Microbiol. 2015, 5, 2–5. [Google Scholar] [CrossRef][Green Version]
- Moral, J.; Garcia-Lopez, M.T.; Camiletti, B.X.; Jaime, R.; Michailides, T.J.; Bandyopadhyay, R.; Ortega-Beltran, A. Present status, and perspective on the future use of aflatoxin biocontrol products. Agronomy 2020, 10, 491. [Google Scholar] [CrossRef]
- Valencia-Quintana, R.; Sánchez-Alarcón, J.; Tenorio, M.G.; Deng, Y.; Waliszewski, S.M.; Valera, M.Á. Preventive strategies aimed at reducing the health risks of aflatoxin B1. Toxicol. Environ. Health Sci. 2012, 4, 71–79. [Google Scholar] [CrossRef]
- Bandyopadhyay, R.; Ortega-Beltran, A.; Akande, A.; Mutegi, C.; Atehnkeng, J.; Kaptoge, L.; Cotty, P. Biological control of aflatoxins in Africa: Current status and potential challenges in the face of climate change. World Mycotoxin J. 2016, 9, 771–789. [Google Scholar] [CrossRef]
- Dorner, J.W. Biological control of aflatoxin contamination of crops. J. Toxicol. Toxin Rev. 2004, 23, 425–450. [Google Scholar] [CrossRef]
- Bhatnagar-Mathur, P.; Sunkara, S.; Bhatnagar-Panwar, M.; Waliyar, F.; Sharma, K.K. Biotechnological advances for combating Aspergillus flavus and aflatoxin contamination in crops. Plant Sci. 2015, 234, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Ghazali, F.M.; Mahyudin, N.A.; Samsudin, N.I.P. Co-Inoculation of aflatoxigenic and non-aflatoxigenic strains of Aspergillus flavus to assess the efficacy of non-aflatoxigenic strains in growth inhibition and aflatoxin B1 reduction. Agriculture 2021, 11, 198. [Google Scholar] [CrossRef]
- Zhou, L.; Wei, D.D.; Selvaraj, J.N.; Shang, B.; Zhang, C.S.; Xing, F.G.; Liu, Y. A strain of Aspergillus flavus from China shows potential as a biocontrol agent for aflatoxin contamination. Biocontrol Sci. Technol. 2015, 25, 583–592. [Google Scholar] [CrossRef]
- Cotty, P.J. Influence of field application of an aflatoxigenic strain of Aspergillus flavus on the populations of A. flavus infecting cotton bolls and on the aflatoxin content of cottonseed. Phytopathology 1994, 84, 1270–1277. [Google Scholar] [CrossRef]
- Abbas, H.K.; Zablotowicz, R.M.; Bruns, H.A.; Abel, C.A. Biocontrol of aflatoxin in corn by inoculation with non-aflatoxigenic Aspergillus flavus isolates. Biocontrol Sci. Technol. 2006, 16, 437–449. [Google Scholar] [CrossRef]
- Cardwell, K.F.; Henry, S.H. Risk of exposure to and mitigation of the effect of aflatoxin on human health: A West African example. J. Toxicol. Toxin Rev. 2004, 23, 217–247. [Google Scholar] [CrossRef]
- Roze, L.V.; Hong, S.Y.; Linz, J.E. Aflatoxin biosynthesis: Current frontiers. Annu. Rev. Food Sci. Technol. 2013, 4, 293–311. [Google Scholar] [CrossRef]
- Pitt, J.I.; Hocking, A.D. Mycotoxins in Australia: Biocontrol of aflatoxin in peanuts. Mycopathologia 2006, 162, 233–243. [Google Scholar] [CrossRef]
- Yin, Y.N.; Yan, L.Y.; Jiang, J.H.; Ma, Z.H. Biological control of aflatoxin contamination of crops. J. Zhejiang Univ. Sci. B 2008, 9, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Phillips, T.D.; Afriyie-Gyawu, E.; Williams, J.; Huebner, H.; Ankrah, N.A.; Ofori-Adjei, D.; Wang, J.S. Reducing human exposure to aflatoxin through the use of clay: A review. Food Addit. Contam. 2008, 25, 134–145. [Google Scholar] [CrossRef]
- Roze, L.V.; Chanda, A.; Wee, J.; Awad, D.; Linz, J.E. Stress-related transcription factor atfB integrates secondary metabolism with oxidative stress response in Aspergilli. J. Biol. Chem. 2011, 286, 35137–35148. [Google Scholar] [CrossRef] [PubMed]
- Wee, J.; Day, D.M.; Linz, J.E. Effects of zinc chelators on aflatoxin production in Aspergillus parasiticus. Toxins 2016, 8, 171. [Google Scholar] [CrossRef]
- Grintzalis, K.; Vernardis, S.I.; Klapa, M.I.; Georgiou, C.D. Role of oxidative stress in sclerotial differentiation and aflatoxin B1 biosynthesis in Aspergillus flavus. Appl. Environ. Microbiol. 2014, 80, 5561–5571. [Google Scholar] [CrossRef] [PubMed]
- Alinezhad, S.; Kamalzadeh, A.; Shams-Ghahfarokhi, M.; Rezaee, M.B.; Jaimand, K.; Kawachi, M.; Razzaghi-Abyaneh, M. Search for novel antifungals from 49 indigenous medicinal plants: Foeniculum vulgare and Platycladus orientalis as strong inhibitors of aflatoxin production by Aspergillus parasiticus. Ann. Microbiol. 2011, 61, 673–681. [Google Scholar] [CrossRef]
- Chang, P.K.; Scharfenstein, L.L.; Mack, B.; Yu, J.; Ehrlich, K.C. Transcriptomic profiles of Aspergillus flavus CA42, a strain that produces small sclerotia, by decanal treatment and after recovery. Fungal Genet. Biol. 2014, 68, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Gómez, J.V.; Tarazona, A.; Mateo-Castro, R.; Gimeno-Adelantado, J.V.; Jiménez, M.; Mateo, E.M. Selected plant essential oils and their main active components, a promising approach to inhibit aflatoxigenic fungi and aflatoxin production in food. Food Addit. Contam. A 2018, 35, 1581–1595. [Google Scholar] [CrossRef]
- Singh, G.; Bhattacharyya, R.; Das, T.K.; Sharma, A.R.; Ghosh, A.; Das, S.; Jha, P. Crop rotation and residue management effects on soil enzyme activities, glomalin, and aggregate stability under zero tillage in the Indo-Gangetic Plains. Soil Till. Res. 2018, 184, 291–300. [Google Scholar] [CrossRef]
- Kluge, E.R.; Mendes, M.C.; Faria, M.V.; Santos, L.A.; Santos, H.O.D.; Szeuczuk, K. Effect of foliar fungicide and plant spacing on the expression of lipoxygenase enzyme and grain rot in maize hybrids. Acta Sci. Agron. 2017, 39, 407–415. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, R.; Ghazali, F.M.; Mahyudin, N.A.; Samsudin, N.I.P. Aflatoxin Biosynthesis, Genetic Regulation, Toxicity, and Control Strategies: A Review. J. Fungi 2021, 7, 606. https://doi.org/10.3390/jof7080606
Khan R, Ghazali FM, Mahyudin NA, Samsudin NIP. Aflatoxin Biosynthesis, Genetic Regulation, Toxicity, and Control Strategies: A Review. Journal of Fungi. 2021; 7(8):606. https://doi.org/10.3390/jof7080606
Chicago/Turabian StyleKhan, Rahim, Farinazleen Mohamad Ghazali, Nor Ainy Mahyudin, and Nik Iskandar Putra Samsudin. 2021. "Aflatoxin Biosynthesis, Genetic Regulation, Toxicity, and Control Strategies: A Review" Journal of Fungi 7, no. 8: 606. https://doi.org/10.3390/jof7080606
APA StyleKhan, R., Ghazali, F. M., Mahyudin, N. A., & Samsudin, N. I. P. (2021). Aflatoxin Biosynthesis, Genetic Regulation, Toxicity, and Control Strategies: A Review. Journal of Fungi, 7(8), 606. https://doi.org/10.3390/jof7080606