Destruxin A Interacts with Aminoacyl tRNA Synthases in Bombyx mori
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Compounds
2.2. Preparation of Protein and Biolayer Interferometry (BLI)
2.3. Homology Modeling and Molecular Docking
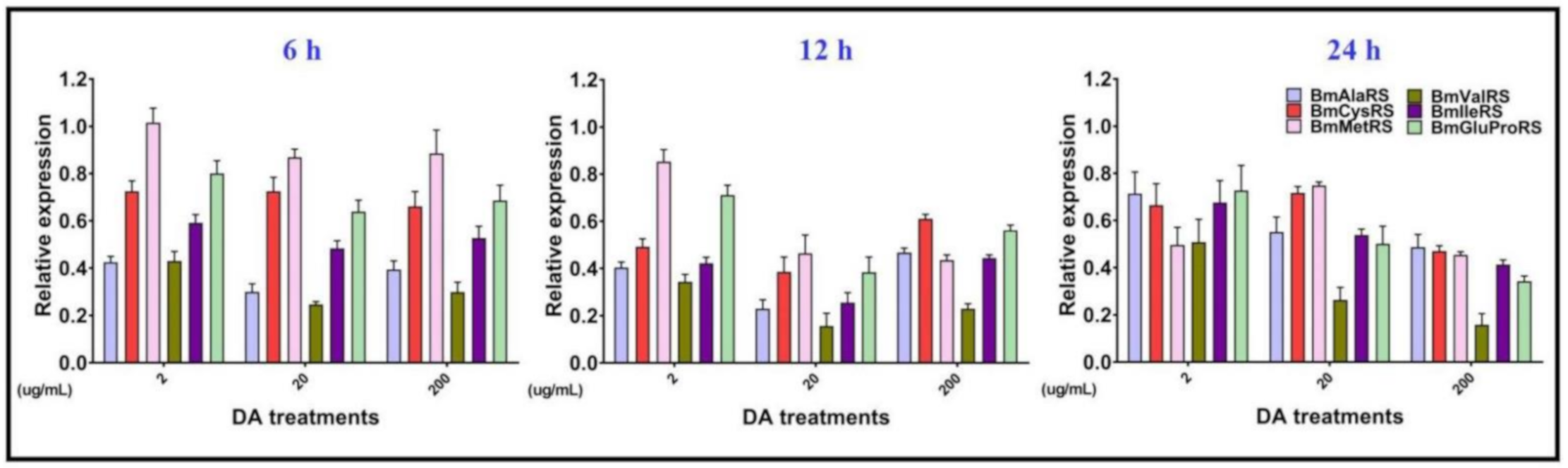
2.4. Assessment of Gene Expression
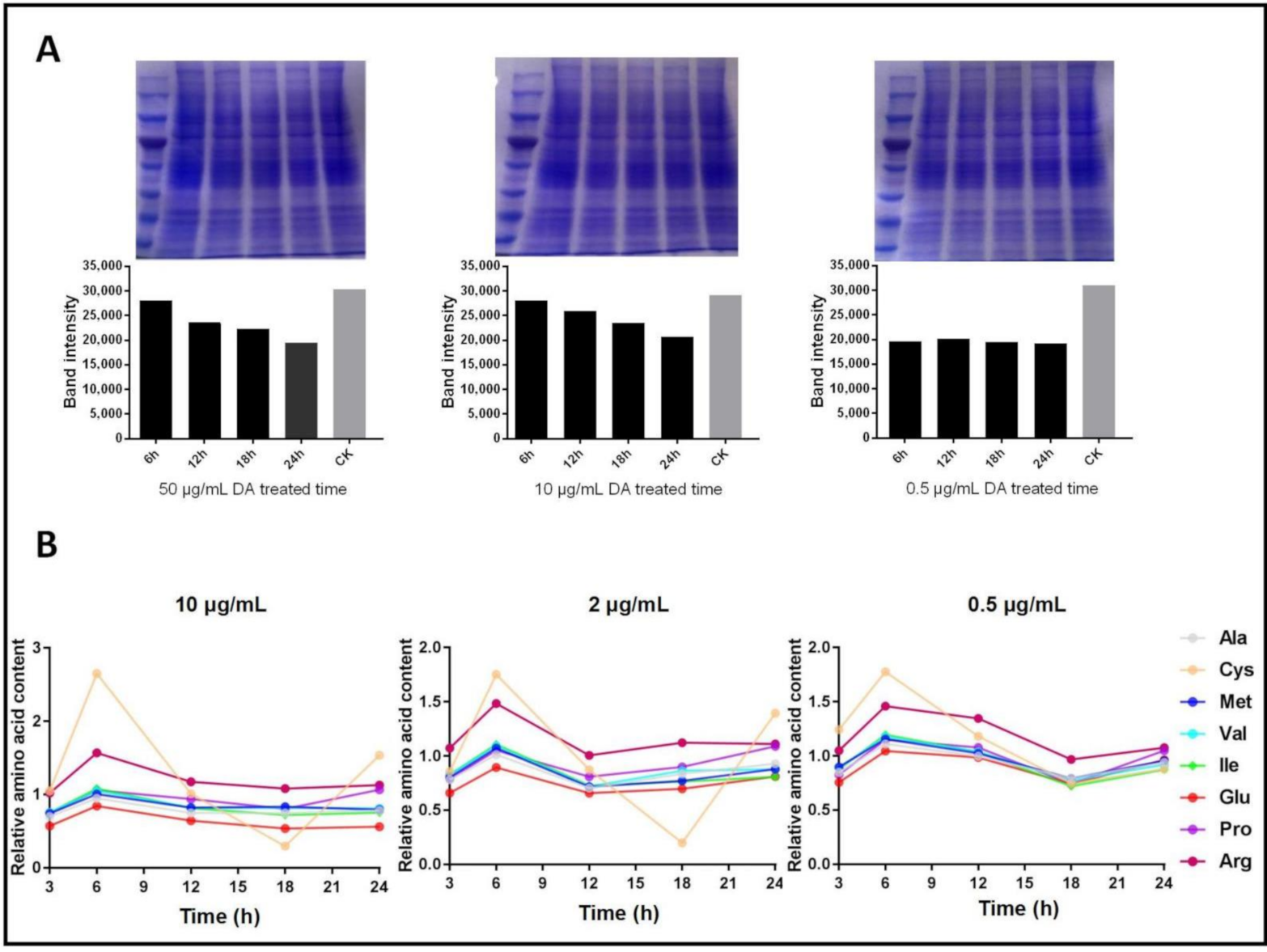
2.5. Detection of Soluble Protein and Free Amino-Acid Content
3. Results
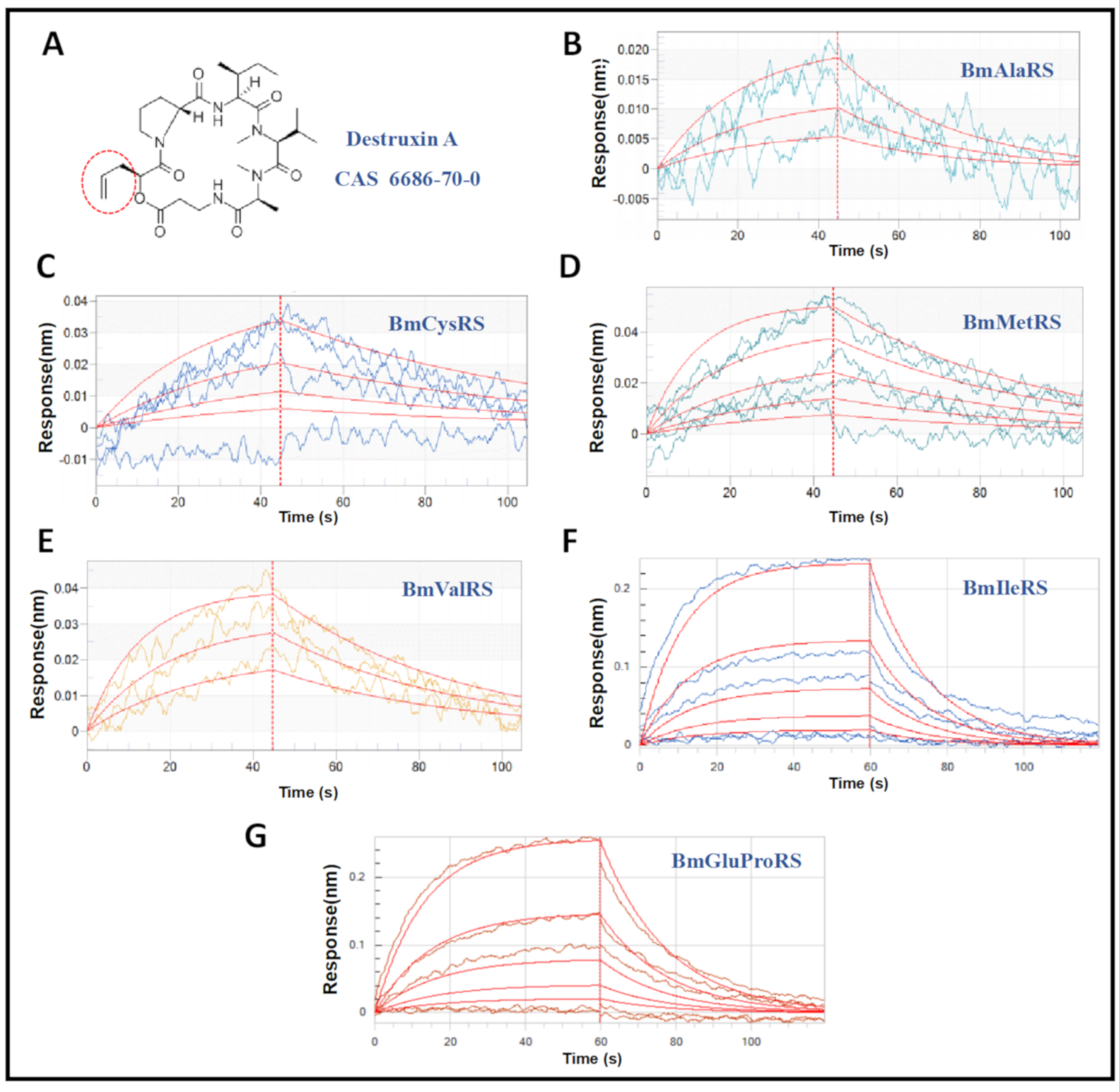
3.1. Interaction of DA with Six Silkworm ARSs
3.2. Analysis of Soluble Protein, Free Amino Acids, and Gene Expression Levels of ARSs in Bm12 Cells
3.3. Interaction Mode of DA with Silkworm ARSs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, B.L.; Tzeng, Y.M. Development and applications of destruxins: A review. Biotechnol. Adv. 2012, 30, 1242–1254. [Google Scholar] [CrossRef] [PubMed]
- Pedras, M.S.C.; Zaharia, L.I.; Ward, D.E. The destruxins: Synthesis, biosynthesis, biotransformation, and biological activity. Phytochemistry 2002, 59, 579–596. [Google Scholar] [CrossRef]
- Wang, B.; Kang, Q.; Lu, Y.; Bai, L.; Wang, C. Unveiling the biosynthetic puzzle of destruxins in Metarhizium species. PNAS 2012, 109, 1287–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiu-Run, C.; Qiong-Bo, H.; Xiao-Qiang, Y.; Shun-Xiang, R. Effects of destruxins on free calcium and hydrogen ions in insect hemocytes. Insect Sci. 2014, 21, 31–38. [Google Scholar]
- Pal, S.; St Leger, R.J.; Wu, L.P. Fungal Peptide Destruxin a Plays a Specific Role in Suppressing the Innate Immune Response in Drosophila melanogaster. J. Biol. Chem. 2007, 282, 8969–8977. [Google Scholar] [CrossRef] [Green Version]
- Kumar, K.K.; Sridhar, J.; Murali-Baskaran, R.K.; Senthil-Nathan, S.; Arthurs, S. Microbial biopesticides for insect pest management in India: Current status and future prospects. J. Invertebr. Pathol. 2018, 165, 74–81. [Google Scholar] [CrossRef]
- Senthil-Nathan, S. A Review of Biopesticides and Their Mode of Action Against Insect Pests; Springer: New Delhi, India, 2015. [Google Scholar]
- Wang, J.; Weng, Q.; Hu, Q. Effects of Destruxin A on Silkworm’s Immunophilins. Toxins 2019, 11, 349. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Hu, W.; Hu, Q. BmTudor-sn Is a Binding Protein of Destruxin A in Silkworm Bm12 Cells. Toxins 2019, 11, 67. [Google Scholar] [CrossRef] [Green Version]
- Rajendran, V.; Kalita, P.; Shukla, H.; Kumar, A.; Tripathi, T. Aminoacyl-tRNA synthetases: Structure, function, and drug discovery. Int. J. Biol. Macromol. 2018, 111, 400–414. [Google Scholar] [CrossRef]
- Vondenhoff, G.; Van Aerschot, A. Aminoacyl-tRNA synthetase inhibitors as potential antibiotics. Eur. J. Med. Chem. 2011, 46, 5227–5236. [Google Scholar] [CrossRef]
- Pang, Y.L.J.; Poruri, K.; Martinis, S.A. tRNA synthetase: tRNA aminoacylation and beyond. Wiley Interdiscip. Rev. RNA 2014, 5, 461–480. [Google Scholar] [CrossRef] [Green Version]
- Kwon, N.; Fox, P.; Kim, S. Aminoacyl-tRNA synthetases as therapeutic targets. Nat. Rev. Drug Discov. 2019, 18, 629–650. [Google Scholar] [CrossRef]
- Wang, J.; Weng, Q.; Yin, F.; Hu, Q. Interactions of Destruxin A with Silkworms’ Arginine tRNA Synthetase and Lamin-C Proteins. Toxins 2020, 12, 137. [Google Scholar] [CrossRef] [Green Version]
- Hu, Q.B.; Ren, S.X.; Wu, J.H.; Chang, J.M.; Musa, P.D. Investigation of destruxin A and B from 80 Metarhizium strains in China, and the optimization of cultural conditions for the strain MaQ10. Toxicon 2006, 48, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W. Aminoacyl-tRNA synthetase complexes: Beyond translation. J. Cell Sci. 2004, 117, 3725–3734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manickam, Y.; Chaturvedi, R.; Babbar, P.; Malhotra, N.; Jain, V.; Sharma, A. Drug targeting of one or more aminoacyl-tRNA synthetase in the malaria parasite Plasmodium falciparum. Drug Discov. Today 2018, 23, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, R.V.K.; Norquay, A.K.; Vederas, J.C. Natural products and their derivatives as tRNA synthetase inhibitors and antimicrobial agents. Medchemcomm 2016, 7, 1535–1545. [Google Scholar] [CrossRef]
- Dornetshuber-Fleiss, R.; Heffeter, P.; Mohr, T.; Hazemi, P.; Lemmens-Gruber, R. Destruxins: Fungal-derived Cyclohexadepsipeptides with Multifaceted Anticancer and Antiangiogenic Activities. Biochem. Pharmacol. 2013, 50, 361–377. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.H.; Chen, S.P.; Lu, T.M. Selective apoptotic cell death effects of oral cancer cells treated with destruxin B. BMC Complementary Altern. Med. 2014, 14, 207. [Google Scholar]
- Pavel, I.; Nancy, K.; Paul, A. Stress Granules and Processing Bodies in Translational Control. Cold Spring Harbor Perspectives in Biology. Cold Spring Harb. Perspect. Biol. 2019, 11, a032813. [Google Scholar]
- Mohler, K.; Ibba, M. Translational fidelity and mistranslation in the cellular response to stress. Nat. Microbiol. 2017, 2, 17117. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Gene | Primer (5′→3′) |
|---|---|
| BmAlaRS | F: ACATGGCGTATCGTGTCTTGGC |
| R: TCTGACGCATATCGCACAGCTC | |
| BmCysRS | F: ACGCCACACCGCAAGACAAC |
| R: CCATCCACCGCCGCAACC | |
| BmMetRS | F: TCTGGCTGACAGGTTCGTGGAG |
| R: CCGCACTTGTCGCACTGGTC | |
| BmValRS | F: ACCTGTGGCTCTACGACCTGTG |
| R: CGGTTACGAACGGCATGAAGGG | |
| BmIleRS | F: AGGGGCGCGTTTGAAAGGTG |
| R: CTGGTCACTGCCTGTAGCTTGG | |
| BmGluProRS | F: AGCAACCGCAACCTTTCCTGAG |
| R: CGCCGCCAGCAAACTTCTCC | |
| GAPDH | F: ATGTTTGTTGTGGGTGTTA |
| R: GTAGAGGCAGGAATGATT |
| Proteins | Kon (1/M·s) 1 | Kdis (1/s) 2 | KD (M) 3 |
|---|---|---|---|
| BmAlaRS | 1.15 × 102 | 3.66 × 10−2 | 3.18 × 10−4 |
| BmCysRS | 2.10 × 102 | 1.49 × 10−2 | 7.12 × 10−5 |
| BmMetRS | 3.07 × 102 | 2.03 × 10−2 | 6.61 × 10−5 |
| BmValRS | 5.72 × 102 | 2.32 × 10−2 | 4.39 × 10−5 |
| BmIleRS | 1.15 × 102 | 6.80 × 10−2 | 5.90 × 10−4 |
| BmGluProRS | 9.39 × 10 | 5.67 × 10−2 | 6.04 × 10−4 |
| Proteins | Docking Free Energy (kcal/mol) |
|---|---|
| BmAlaRS | −8.87 |
| BmGluProRS | −7.78 |
| BmCysRS | −8.97 |
| BmIleRS | −7.49 |
| BmMetRS | −8.25 |
| BmValRS | −9.23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Berestetskiy, A.; Hu, Q. Destruxin A Interacts with Aminoacyl tRNA Synthases in Bombyx mori. J. Fungi 2021, 7, 593. https://doi.org/10.3390/jof7080593
Wang J, Berestetskiy A, Hu Q. Destruxin A Interacts with Aminoacyl tRNA Synthases in Bombyx mori. Journal of Fungi. 2021; 7(8):593. https://doi.org/10.3390/jof7080593
Chicago/Turabian StyleWang, Jingjing, Alexander Berestetskiy, and Qiongbo Hu. 2021. "Destruxin A Interacts with Aminoacyl tRNA Synthases in Bombyx mori" Journal of Fungi 7, no. 8: 593. https://doi.org/10.3390/jof7080593
APA StyleWang, J., Berestetskiy, A., & Hu, Q. (2021). Destruxin A Interacts with Aminoacyl tRNA Synthases in Bombyx mori. Journal of Fungi, 7(8), 593. https://doi.org/10.3390/jof7080593







