Effects of Flurochloridone Application on Rhizosphere Soil Fungal Community and Composition in Potato Growing Areas of the Qinghai-Tibet Plateau
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Trials, Soil Sampling, and Preparation
2.2. Soil Properties Analysis
2.3. Next-Generation Sequencing
2.4. Sequence Analysis Processing
3. Results and Discussion
3.1. Physicochemical Properties
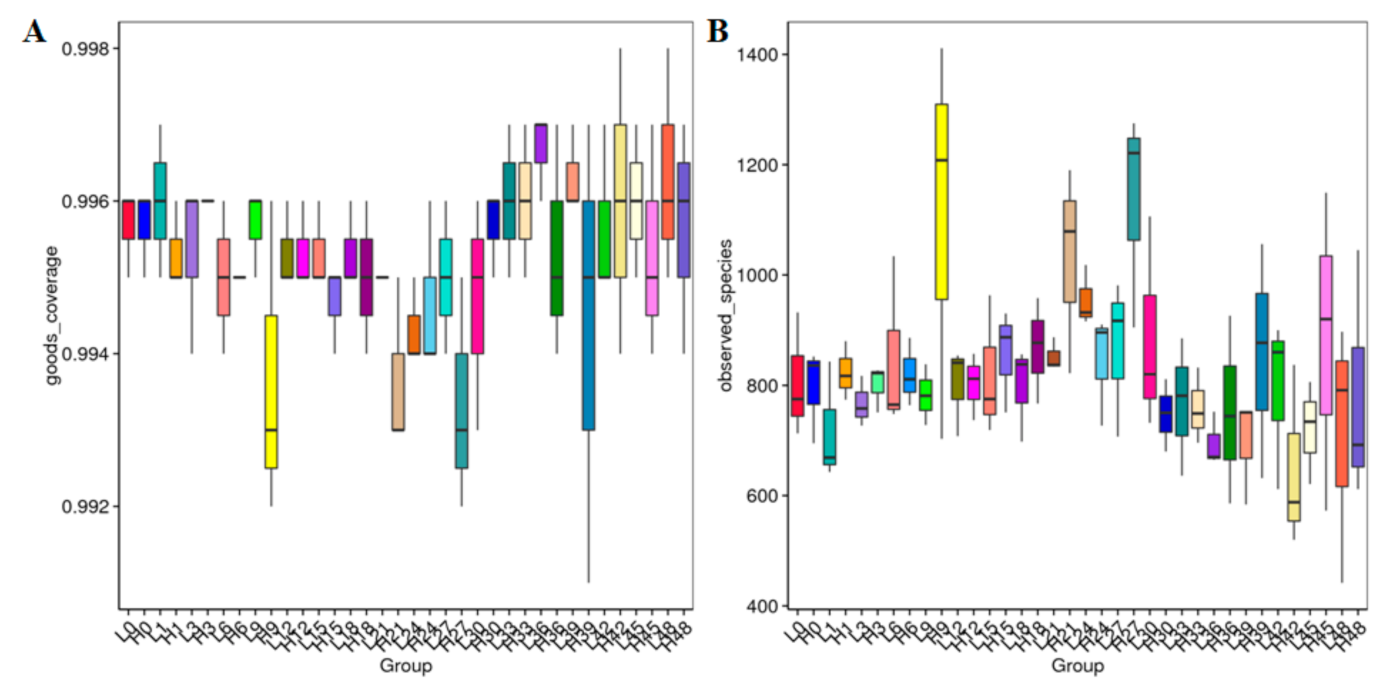
3.2. Phylogenetic Composition and Alpha-Diversity
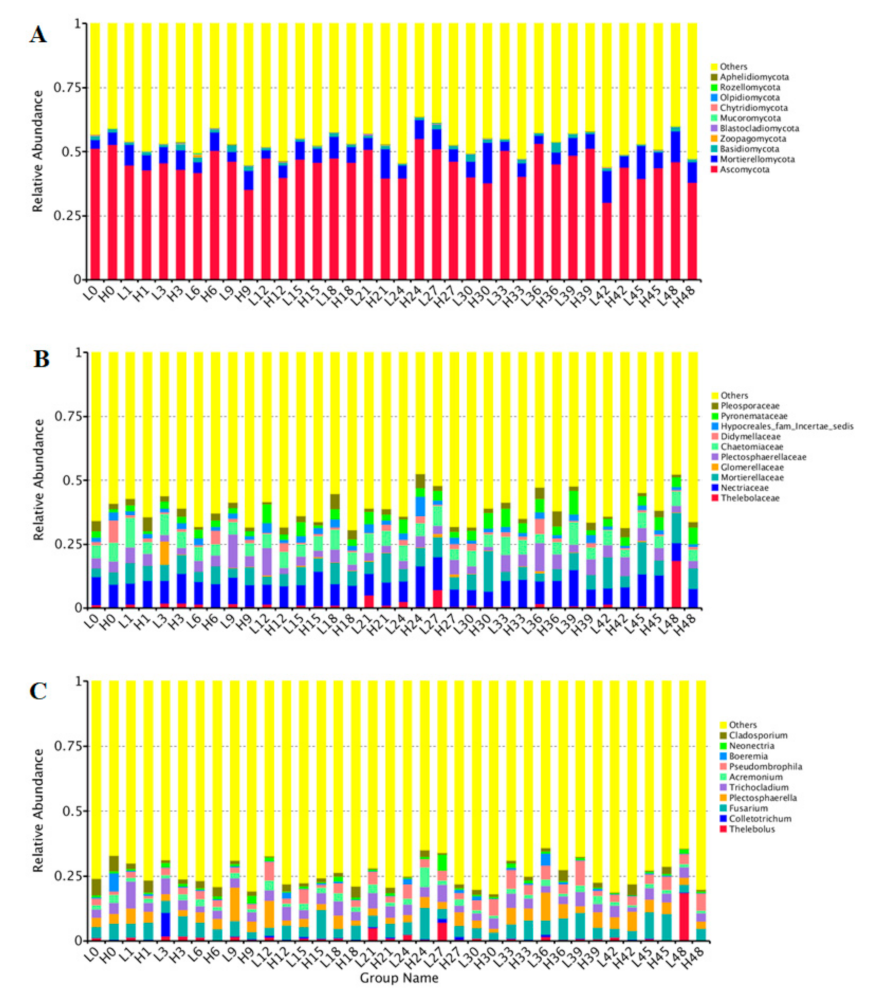
3.3. Rhizosphere Soil Fungal Community and Composition
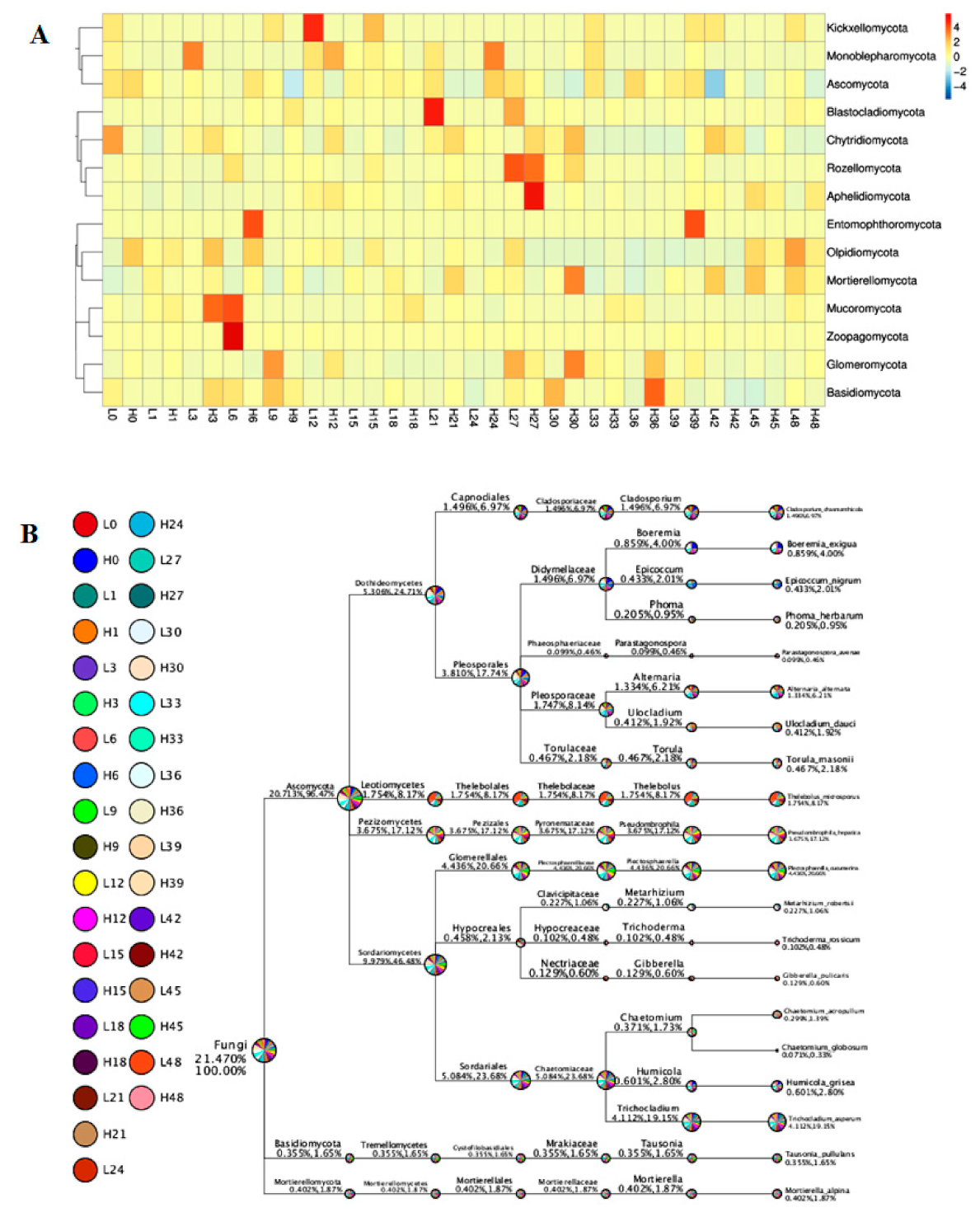
3.4. Comparison between LS and HS Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bolognesi, C. Genotoxicity of pesticides: A review of human biomonitoring studies. Mutat. Res. 2003, 543, 251–272. [Google Scholar] [CrossRef]
- Yüzbaşioğlu, D.; Ünal, F.; Sancak, C.; Kasap, R. Cytological effects of herbicide racer “fluorochloridone” on Allium cepa. Caryologia 2003, 56, 97–105. [Google Scholar] [CrossRef]
- Pareek, A.; Sopory, S.K.; Bohnert, H.J. Varietal Improvement for Abiotic Stress Tolerance in Crop Plants: Special Reference to Salinity in Rice. In Abiotic Stress Adaptation in Plants; Springer: Berlin, Germany, 2010; Volume 1, pp. 103–122. [Google Scholar] [CrossRef]
- Sondhia, S.; Choudhury, P.P.; Sharma, A.R. Environmental Fate of Herbicide Use in Central India. In Environmental Chemistry for a Sustainable World Herbicide Residue Research in India; Springer: Berlin, Germany, 2019; Volume 12, pp. 29–104, Chapter 2. [Google Scholar] [CrossRef]
- Rouchaud, J.; Neus, O.; Callens, D.; Bulcke, R. Herbicide flurochloridone soil biodegradation in potato crops. Toxicol. Environ. Chem. 1997, 61, 251–257. [Google Scholar] [CrossRef]
- Kaya, A.; Yigit, E. The physiological and biochemical effects of salicylic acid on sunflowers (Helianthus annuus) exposed to flurochloridone. Ecotoxicol. Environ. Saf. 2014, 106, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Jursik, M.; Soukup, J.; Holec, J.; Andr, J.; Hamouzova, K. Efficacy and selectivity of pre-emergent sunflower herbicides under different soil moisture conditions. Plant Prot. Sci. 2015, 51, 214–222. [Google Scholar] [CrossRef]
- Liu, S.; Shi, J.; Zhang, S.; Tang, L.; Zhou, Z. Research Status of Biotoxicity Caused by Flurochloridone. J. Environ. Occup. Med. 2015, 32, 987–989. [Google Scholar]
- Hengzhi, W.; Wenlei, G.; Zhaozhen, W.; Jinni, T.; Jinxin, W. Weed Control Efficacy and Crop Safety of Fluorochloridone and Its Mixtures in Cotton Fields. Chin. Agric. Sci. Bull. 2016, 32, 66–70. [Google Scholar]
- Shi, J.; Xie, C.; Liu, H.; Krausz, K.W.; Bewley, C.A.; Zhang, S.; Tang, L.; Zhou, Z.; Gonzalez, F.J. Metabolism and Bioactivation of Fluorochloridone, a Novel Selective Herbicide, in Vivo and in Vitro. Environ. Technol. 2016, 50, 9652–9660. [Google Scholar] [CrossRef]
- Ogawa, H.; Yamada, I.; Arai, K.; Hirase, K.; Wakabayashi, K. Mode of bleaching phytotoxicity of herbicidal diphenylpyrrolidinones. Pest Manag. Sci. 2015, 57, 33–40. [Google Scholar] [CrossRef]
- Celar, F.A.; Kos, K. Effects of selected herbicides and fungicides on growth, sporulation and conidial germination of entomopathogenic fungus Beauveria bassiana. Pest Manag. Sci. 2016, 72, 2110–2117. [Google Scholar] [CrossRef]
- Marschner, P.; Crowley, D.; Yang, C.H. Development of specific rhizosphere bacterial communities in relation to plant species, nutrition and soil type. Plant Soil 2004, 261, 199–208. [Google Scholar] [CrossRef]
- Saha, S.; Gopinath, K.A.; Mina, B.L.; Gupta, H.S. Influence of continuous application of inorganic nutrients to a Maize-Wheat rotation on soil enzyme activity and grain quality in a rainfed Indian soil. Eur. J. Soil Biol. 2008, 44, 521–531. [Google Scholar] [CrossRef]
- Jiang, L.; Yang, H. Prometryne-induced oxidative stress and impact on antioxidant enzymes in wheat. Ecotoxicol. Environ. Saf. 2009, 72, 1687–1693. [Google Scholar] [CrossRef]
- Waksman, S.A. Principles of Soil Microbiology; Williams and Wilkins Company: Baltimore, MD, USA, 1927. [Google Scholar]
- Fierer, N.; Strickland, M.S.; Liptzin, D.; Bradford, M.A.; Cleveland, C.C. Global patterns in belowground communities. Ecol. Lett. 2010, 12, 1238–1249. [Google Scholar] [CrossRef] [PubMed]
- Serna-Chavez, H.M.; Fierer, N.; Van Bodegom, P.M. Global drivers and patterns of microbial abundance in soil. Glob. Ecol. Biogeogr. 2013, 22, 1162–1172. [Google Scholar] [CrossRef]
- Liu, L.; Yang, J.; Yu, Z.; Wilkinson, D.M. The biogeography of abundant and rare bacterioplankton in the lakes and reservoirs of China. ISME J. 2015, 9, 2068–2077. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Chen, W.; Wei, G. Biogeography and ecological diversity patterns of rare and abundant bacteria in oil-contaminated soils. Mol. Ecol. 2017, 26, 5305–5317. [Google Scholar] [CrossRef]
- Yuanyuan, M.; Wenjing, Z.; Jun, Y.; Yuanshao, L.; Zheng, Y.; Senjie, L. Biogeographic patterns of abundant and rare bacterioplankton in three subtropical bays resulting from selective and neutral processes. ISME J. 2018, 12, 2198–2210. [Google Scholar]
- Torsvik, V.; Ovreas, L. Microbial diversity and function in soil: From genes to ecosystems. Curr. Opin. Microbiol. 2002, 5, 240–245. [Google Scholar] [CrossRef]
- Ramirez, K.S.; Leff, J.W.; Barberan, A.; Bates, S.T.; Betley, J.; Crowther, T.W.; Kelly, E.F.; Oldfield, E.E.; Shaw, E.A.; Steenbock, C. Biogeographic patterns in below-ground diversity in New York City’s Central Park are similar to those observed globally. Proc. Biol. Sci. 2014, 281, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [PubMed]
- Dupont, A.Ö.C.; Griffiths, R.I.; Bell, T.; Bass, D. Differences in soil micro-eukaryotic communities over soil pH gradients are strongly driven by parasites and saprotrophs. Environ. Microbiol. 2016, 18, 2010–2024. [Google Scholar] [CrossRef]
- Wang, K.B.; Zhang, Y.W.; Tang, Z.S.; Shangguan, Z.P.; Chang, F.; Jia, F.A.; Chen, Y.P.; He, X.H.; Shi, W.Y.; Deng, L. Effects of grassland afforestation on structure andfunction of soil bacterial and fungal communities. Sci. Total Environ. 2019, 676, 396–406. [Google Scholar] [CrossRef]
- Huaidong, H.E.; Waichin, L.I.; Riqing, Y.U.; Zhihong, Y.E. Illumina-Based Analysis of Bulk and Rhizosphere Soil Bacterial Communities in Paddy Fields Under Mixed Heavy Metal Contamination. Pedosphere 2017, 27, 569–578. [Google Scholar]
- Wang, R.; Xiao, Y.; Lv, F.; Hu, L.; Wei, L.; Yuan, Z.; Lin, H. Bacterial community structure and functional potential of rhizosphere soils as influenced by nitrogen addition and bacterial wilt disease under continuous sesame cropping. Appl. Soil Ecol. 2017, 125, 117–127. [Google Scholar] [CrossRef]
- Zhang, T.C.; Pang, H. Applications of Microelectrode Techniques to Measure pH and OxidationReduction Potential in Rhizosphere Soil. Environ. Sci. Technol. 1999, 33, 1293–1299. [Google Scholar] [CrossRef]
- Bowman, R.A. A reevaluation of the chromic acid colorimetric procedure for soil organic carbon. Commun. Soil Sci. Plant Anal. 1998, 29, 501–508. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes--application to the identification of mycorrhizae and rusts. Mol. Ecol. 2010, 2, 113–118. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Cheng, H.Y.; Chang, F.; She, X. Niche differentiation in the rhizosphere and endosphere fungal microbiome of wild Paris polyphylla Sm. PeerJ 2020, 8, e8510–e8530. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 16, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.C.; Mackenzie, A.F.; Schnitzer, M.; Monreal, C.M.; Voroney, P.R.; Beyaert, R.P. Management-induced change in labile soil organic matter under continuous corn in eastern Canadian soils. Biol. Fertil. Soils 1997, 26, 88–94. [Google Scholar] [CrossRef]
- Whalley, W.R.; Riseley, B.; Leeds-Harrison, P.B.; Bird, N.R.A.; Adderley, W.P. Structural differences between bulk and rhizosphere soil. Eur. J. Soil Sci. 2010, 56, 353–360. [Google Scholar] [CrossRef]
- Wang, B.; Liu, G.B.; Xue, S.; Zhu, B. Changes in soil physico-chemical and microbiological properties during natural succession on abandoned farmland in the Loess Plateau. Environ. Earth Sci. 2011, 62, 915–925. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Win, P.P.; De Waele, D.; Kyi, P.P.; Maung, Z.T.Z.; Myint, Y.Y. Effect of different water regimes on nematode reproduction, root galling, plant growth and yield of lowland and upland Asian rice varieties grown in two soil types infested by the rice root–knot nematode Meloidogyne graminicola. Field Crops Res. 2015, 165, 138–149. [Google Scholar]
- Mazzola, M.; Manici, L.M. Apple Replant Disease: Role of Microbial Ecology in Cause and Control. Annu. Rev. Phytopathol. 2012, 50, 45–65. [Google Scholar] [CrossRef]
- Zhou, S.M.; Zhang, M.; Zhang, K.K.; Yang, X.W.; Wang, C.Y. Effects of reduced nitrogen and suitable soil moisture on wheat (Triticum aestivum L.) rhizosphere soil microbiological, biochemical properties and yield in the Huanghuai Plain, China. J. Integr. Agric. 2020, 19, 234–250. [Google Scholar] [CrossRef]
- Schoch, C.L.; Spatafora, J.W. The Ascomycota Tree of Life: A Phylum. Syst. Biol. 2009, 58, 224–239. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef] [PubMed]
- Hacquard, S.; Garrido-Oter, R.; González, A.; Spaepen, S.; Ackermann, G.; Lebeis, S.; Mchardy, A.C.; Dangl, J.L.; Knight, R.; Ley, R. Microbiota and Host Nutrition across Plant and Animal Kingdoms. Cell Host Microbe 2015, 17, 603–616. [Google Scholar] [CrossRef]
- Kottke, I.; Haug, I.; Setaro, S.; Suárez, J.P.; Wei, M.; PreuIng, M.; Nebel, M.; Oberwinkler, F. Guilds of mycorrhizal fungi and their relation to trees, ericads, orchids and liverworts in a neotropical mountain rain forest. Basic Appl. Ecol. 2008, 9, 13–23. [Google Scholar] [CrossRef]
- Burke, D.J.; Lopez-Gutierrez, J.C.; Smemo, K.A.; Chan, C.R. Vegetation and soil environment influence the spatial distribution of root-associated fungi in a mature beech-maple forest. Appl. Environ. Microbiol. 2009, 75, 7639–7648. [Google Scholar] [CrossRef]
- Yu, L.; Nicolaisen, M.; Larsen, J.; Ravnskov, S. Molecular characterization of root-associated fungal communities in relation to health status of Pisum sativum using barcoded pyrosequencing. Plant Soil 2012, 357, 395–405. [Google Scholar] [CrossRef]
- Ladha, J.K.; Tirol-Padre, A.; Reddy, C.K.; Cassman, K.G.; Verma, S.; Powlson, D.S.; Van Kessel, C.; Daniel, D.B.R.; Chakraborty, D.; Pathak, H. Global nitrogen budgets in cereals: A 50-year assessment for maize, rice, and wheat production systems. Sci. Rep. 2016, 6, 19355–19364. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Shen, S.; Chen, H.; Zhang, Y.; Deng, L.; Liu, Y.; Shangguan, Z. Effects of Flurochloridone Application on Rhizosphere Soil Fungal Community and Composition in Potato Growing Areas of the Qinghai-Tibet Plateau. J. Fungi 2021, 7, 420. https://doi.org/10.3390/jof7060420
Li W, Shen S, Chen H, Zhang Y, Deng L, Liu Y, Shangguan Z. Effects of Flurochloridone Application on Rhizosphere Soil Fungal Community and Composition in Potato Growing Areas of the Qinghai-Tibet Plateau. Journal of Fungi. 2021; 7(6):420. https://doi.org/10.3390/jof7060420
Chicago/Turabian StyleLi, Wei, Shuo Shen, Hongyu Chen, Yang Zhang, Lei Deng, Yujiao Liu, and Zhouping Shangguan. 2021. "Effects of Flurochloridone Application on Rhizosphere Soil Fungal Community and Composition in Potato Growing Areas of the Qinghai-Tibet Plateau" Journal of Fungi 7, no. 6: 420. https://doi.org/10.3390/jof7060420
APA StyleLi, W., Shen, S., Chen, H., Zhang, Y., Deng, L., Liu, Y., & Shangguan, Z. (2021). Effects of Flurochloridone Application on Rhizosphere Soil Fungal Community and Composition in Potato Growing Areas of the Qinghai-Tibet Plateau. Journal of Fungi, 7(6), 420. https://doi.org/10.3390/jof7060420





