Transcription Activator Swi6 Interacts with Mbp1 in MluI Cell Cycle Box-Binding Complex and Regulates Hyphal Differentiation and Virulence in Beauveria bassiana
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Bioinformatic Identification and Functional Analyses of BbSwi6
2.3. Sub-Cellular Localization of BbMbp1 and BbSwi6
2.4. Quantification of Blastospore Production
2.5. Assay for Conidial Production
2.6. Fungal Growth
2.7. Insect Bioassay
2.8. Yeast Two Hybrid (Y2H) Assays
2.9. Analyzing the BbSwi6-Mediated Transcriptome during Sporulation
2.10. Data Analyses
3. Results
3.1. Bioinformatic Analysis of BbSwi6 and Generation of Its Gene Disruption and Complementation Strains
3.2. BbSwi6 Interacts with BbMbp1 in Nucleus
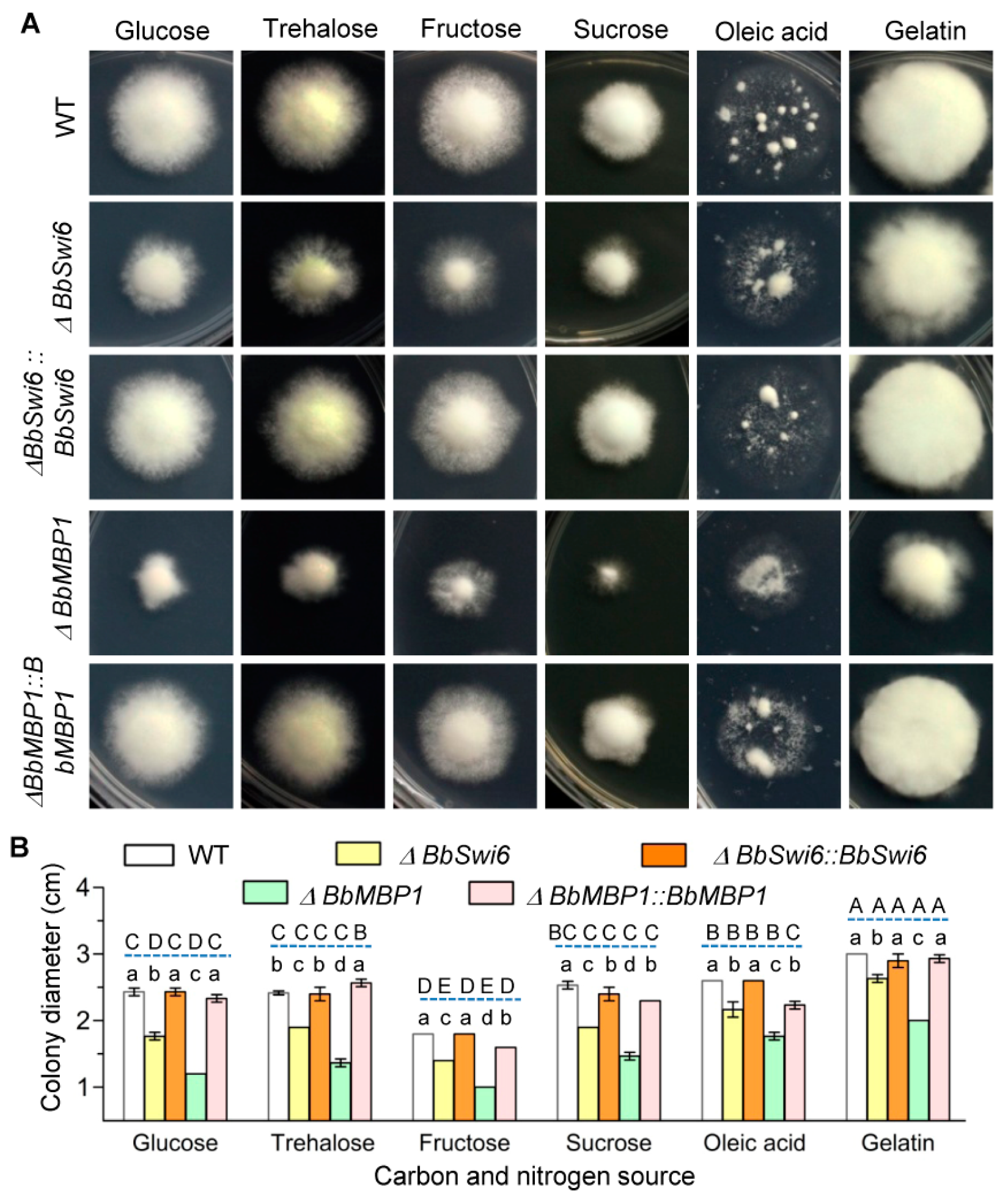
3.3. BbSwi6 Significantly Contributes to Vegetative Growth onNutrients
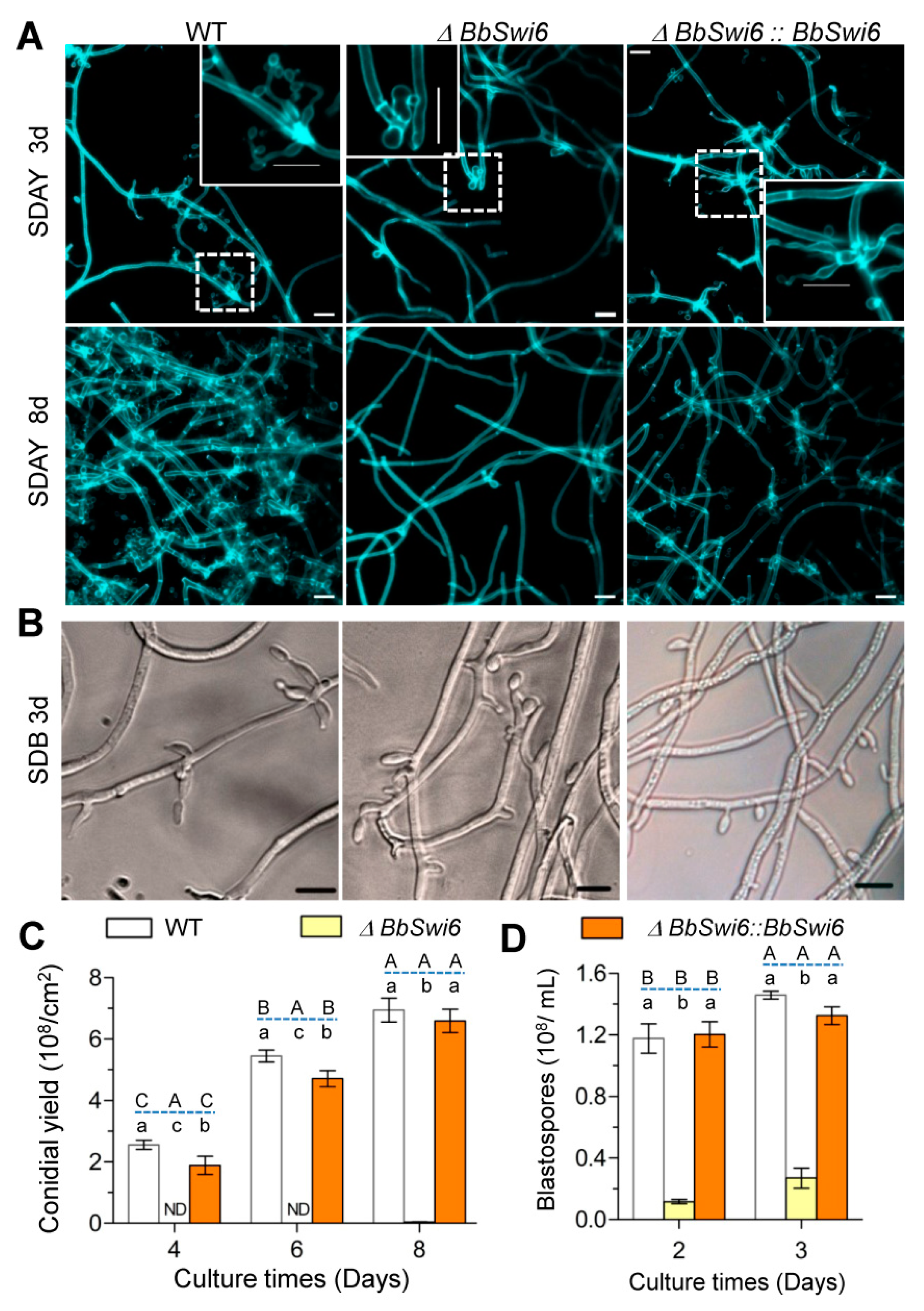
3.4. BbSwi6 Is Required for Asexual Sporulation
3.5. Disruption of BbSwi6Significantly Affected Fungal Pathogenicity
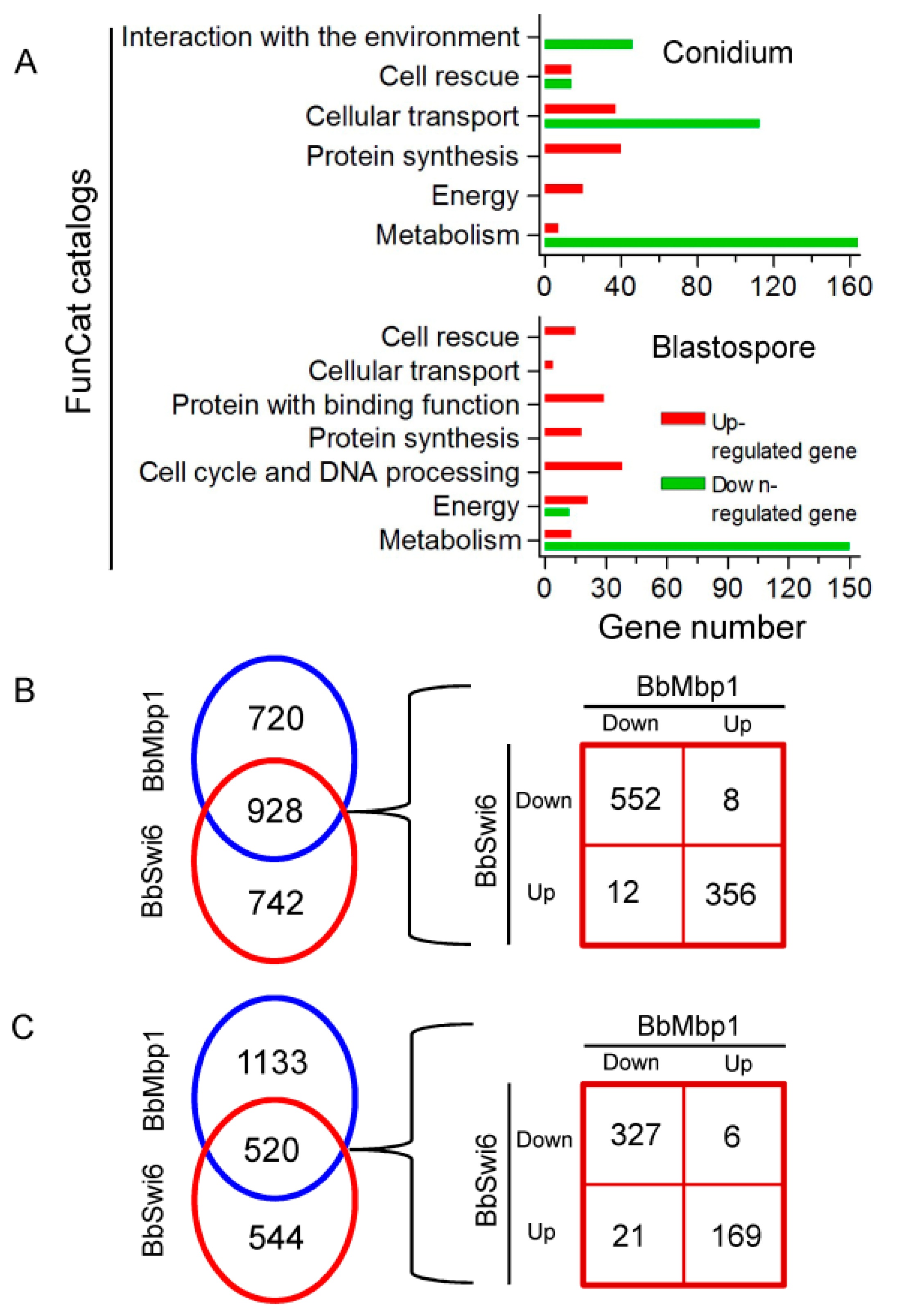
3.6. BbSwi6 Mediates the Genome-Wide Expression during Fungal Differentiation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Park, H.-S.; Yu, J.H. Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 2012, 15, 669–677. [Google Scholar] [CrossRef]
- Wang, C.S.; Wang, S.B. Insect pathogenic fungi: Genomics, molecular interactions, and genetic improvements. Annu. Rev. Entomol. 2017, 62, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, J.; Xie, X.Q.; Keyhani, N.O.; Feng, M.G.; Ying, S.H. The autophagy gene BbATG5, involved in the formation of the autophagosome, contributes to cell differentiation and growth but is dispensable for pathogenesis in the entomopathogenic fungus Beauveria bassiana. Microbiol. SGM 2013, 159, 243–252. [Google Scholar] [CrossRef]
- Peng, Y.J.; Wang, J.J.; Lin, H.Y.; Ding, J.L.; Feng, M.G.; Ying, S.H. HapX, an indispensable bZIP transcription factor for iron acquisition, regulates infection initiation by orchestrating conidial oleic acid homeostasis and cytomembrane functionality in mycopathogen Beauveria bassiana. mSystems 2020, 5, e00695-20. [Google Scholar] [CrossRef]
- Zhang, A.X.; Mouhoumed, A.Z.; Tong, S.M.; Ying, S.H.; Feng, M.G. BrlA and AbaA govern virulence-required dimorphic switch, conidiation, and pathogenicity in a fungal insect pathogen. mSystems 2019, 4, e00140-19. [Google Scholar] [CrossRef] [PubMed]
- He, P.H.; Wang, X.X.; Chu, X.L.; Feng, M.G.; Ying, S.H. RNA sequencing analysis identifies the metabolic and developmental genes regulated by BbSNF1 during conidiation of the entomopathogenic fungus Beauveria bassiana. Curr. Genet. 2015, 61, 143–152. [Google Scholar] [CrossRef]
- Hou, J.; Wang, J.J.; Lin, H.Y.; Feng, M.G.; Ying, S.H. Roles of autophagy-related genes in conidiogenesis and blastospore formation, virulence, and stress response of Beauveria bassiana. Fungal Biol. 2020, 124, 1052–1057. [Google Scholar] [CrossRef]
- Ah, F.A.; Judelson, H.S. Cell cycle regulator Cdc14 is expressed during sporulation but not hyphal growth in the fungus-like oomycete Phytophthora infestans. Mol. Microbiol. 2003, 50, 487–494. [Google Scholar]
- Wang, J.; Liu, J.; Hu, Y.; Ying, S.H.; Feng, M.G. Cytokinesis-required Cdc14 is a signaling hub of asexual development and multi-stress tolerance in Beauveria bassiana. Sci. Rep. 2013, 3, 3086. [Google Scholar] [CrossRef]
- Ding, J.L.; Lin, H.Y.; Feng, M.G.; Ying, S.H. Mbp1, a component of the MluI cell cycle box-binding complex, contributes to morphological transition and virulence in the filamentous entomopathogenic fungus Beauveria bassiana. Environ. Microbiol. 2020, 22, 584–597. [Google Scholar] [CrossRef] [PubMed]
- Bähler, J. Cell-cycle control of gene expression in budding and fission yeast. Annu. Rev. Genet. 2005, 39, 69–94. [Google Scholar] [CrossRef] [PubMed]
- Bean, J.M.; Siggia, E.D.; Cross, F.R. High functional overlap between MluI cell-cycle box binding factor and Swi4/6 cell-cycle box binding factor in the G1/S transcriptional program in Saccharomyces cerevisiae. Genetics 2005, 171, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Koch, C.; Moll, T.; Neuberg, M.; Ahorn, H.; Nasmyth, K. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science 1993, 261, 1551–1557. [Google Scholar] [CrossRef]
- Wittenberg, C.; Reed, S.I. Cell cycle-dependent transcription in yeast: Promoters, transcription factors, and transcriptomes. Oncogene 2005, 24, 2746–2755. [Google Scholar] [CrossRef]
- Hussein, B.; Huang, H.; Glory, A.; Osmani, A.; Kaminskyj, S.; Nantel, A.; Bachewich, C. G1/S transcription factor orthologues Swi4p and Swi6p are important but not essential for cell proliferation and influence hyphal development in the fungal pathogen Candida albicans. Eukaryot. Cell 2011, 10, 384–397. [Google Scholar] [CrossRef]
- Caligiuri, M.; Beach, D. Sct1 functions in partnership with Cdc10 in a transcription complex that activates cell cycle START and inhibits differentiation. Cell 1993, 72, 607–619. [Google Scholar] [CrossRef]
- Zhu, Y.; Takeda, T.; Whitehall, S.; Peat, N.; Jones, N. Functional characterization of the fission yeast Start-specific transcription factor Res2. EMBO J. 1997, 16, 1023–1034. [Google Scholar] [CrossRef]
- Qi, Z.; Wang, Q.; Dou, X.; Wang, W.; Zhao, Q.; Lv, R.; Zhang, H.; Zheng, X.; Wang, P.; Zhang, Z. MoSwi6, an APSES family transcription factor, interacts with MoMps1 and is required for hyphal and conidial morphogenesis, appressorial function and pathogenicity of Magnaportheoryzae. Mol. Plant Pathol. 2012, 13, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Fan, F.; Qiu, D.; Jiang, L. The transcription cofactor FgSwi6 plays a role in growth and development, carbendazim sensitivity, cellulose utilization, lithium tolerance, deoxynivalenol production and virulence in the filamentous fungus Fusarium graminearum. Fungal Genet. Biol. 2013, 58–59, 42–52. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, J.; Xin, C.; Xing, X.; Yin, Y.; Chen, L.; Song, Z. Regulation of conidiation, dimorphic transition, and microsclerotia formation by MrSwi6 transcription factor in dimorphic fungus Metarhiziumrileyi. World J. Microbiol. Biotechnol. 2019, 35, 46. [Google Scholar] [CrossRef]
- Xiao, G.; Ying, S.H.; Zheng, P.; Wang, Z.L.; Zhang, S.; Xie, X.Q.; Shang, Y.; Zheng, H.; Zhou, Y.; St. Leger, R.J.; et al. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci. Rep. 2012, 2, 483. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Cao, H.; Zhang, L.; Huang, P.; Lin, F. Systematic analysis of Zn2Cys6 transcription factors required for development and pathogenicity by high-throughput gene knockout in the rice blast fungus. PLoS Pathog. 2014, 10, e1004432. [Google Scholar] [CrossRef]
- Wang, J.J.; Peng, Y.J.; Ding, J.L.; Feng, M.G.; Ying, S.H. Mitochondrial fission is necessary for mitophagy, development and virulence of the insect pathogenic fungus Beauveria bassiana. J. Appl. Microbiol. 2020, 129, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.H.; Liu, J.; Chu, X.L.; Xie, X.Q.; Feng, M.G. The autophagy-related genes BbATG1 and BbATG8 have different functions in differentiation, stress resistance and virulence of mycopathogen Beauveria bassiana. Sci. Rep. 2016, 6, 26376. [Google Scholar] [CrossRef]
- El-Gohary, S.G.; Yousif-Khalil, S.I.; El-Maghraby, M.M.A.; Abd-Alla, S.M. Mass rearing of greater wax moth, Galleria mellonella L. Zagazig J. Agric. Res. 2018, 45, 495–503. [Google Scholar]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef]
- Priebe, S.; Kreisel, C.; Horn, F.; Guthke, R.; Linde, J. FungiFun2: A comprehensive online resource for systematic analysis of gene lists from fungal species. Bioinformatics 2015, 31, 445–446. [Google Scholar] [CrossRef]
- Dirick, L.; Moll, T.; Auer, H.; Nasmyth, K. A central role for SWI6 in modulating cell cycle Start-specific transcription in yeast. Nature 1992, 357, 508–513. [Google Scholar] [CrossRef]
- Iyer, V.R.; Horak, C.E.; Scafe, C.S.; Botstein, D.; Snyder, M.; Brown, P.O. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 2001, 409, 533–538. [Google Scholar] [CrossRef]
- Ortiz-Urquiza, A.; Keyhani, N.O. Molecular genetics of Beauveria bassiana infection of insects. Adv. Genet. 2016, 94, 165–249. [Google Scholar] [PubMed]
- He, P.H.; Dong, W.X.; Chu, X.L.; Feng, M.G.; Ying, S.H. The cellular proteome is affected by a gelsolin (BbGEL1) during morphological transitions in aerobic surface versus liquid growth in the entomopathogenic fungus Beauveria bassiana. Environ. Microbiol. 2016, 18, 4153–4169. [Google Scholar] [CrossRef]
- Hall, R.A.; Peterkin, D.D.; Ali, B.; Lopez, V.F. Influence of culture age on rate of conidiospore germination four deuteromycetousentomogenous fungi. Mycol. Res. 1994, 98, 763–768. [Google Scholar] [CrossRef]
- Gauthier, G.M. Dimorphism in fungal pathogens of mammals, plants, and insects. PLoS Pathog. 2015, 11, e1004608. [Google Scholar] [CrossRef]
- Wanchoo, A.; Lewis, M.W.; Keyhani, N.O. Lectin mapping reveals stage-specific display of surface carbohydrates in in vitro and haemolymph-derived cells of the entomopathogenic fungus Beauveria bassiana. Microbiol. SGM 2009, 155, 3121–3133. [Google Scholar] [CrossRef]
- Dorter, I.; Momany, M. Fungal cell cycle: A unicellular versus multicellular comparison. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Zhang, S.; Xia, Y.X.; Kim, B.; Keyhani, N.O. Two hydrophobinsare involved in fungal spore coat rodlet layer assembly and each play distinct roles in surface interactions, development and pathogenesis in the entomopathogenic fungus, Beauveria bassiana. Mol. Microbiol. 2011, 80, 811–826. [Google Scholar] [CrossRef] [PubMed]
- Moonjely, S.; Keyhani, N.O.; Bidochka, M.J. Hydrophobins contribute to root colonization and stress responses in the rhizosphere-competent insect pathogenic fungus Beauveria bassiana. Microbiol. SGM 2018, 164, 517–528. [Google Scholar] [CrossRef]
- Liu, X.H.; Lu, J.P.; Dong, B.; Gu, Y.; Lin, F.C. Disruption of MoCMK1, encoding a putative calcium/calmodulin-dependent kinase, in Magnaportheoryzae. Microbiol. Res. 2010, 165, 402–410. [Google Scholar] [CrossRef]
- Zhen, Z.; Zhang, G.; Yang, L.; Ma, N.; Li, Q.; Ma, Y.; Niu, X.; Zhang, K.Q.; Yang, J. Characterization and functional analysis of calcium/calmodulin-dependent protein kinases (CaMKs) in the nematode-trapping fungus Arthrobotrysoligospora. Appl. Microbiol. Biotechnol. 2019, 103, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Cen, J.; Chen, J.; Qiu, Y.; Zeng, W.; Zhang, J. Bioinformatic analysis of the pathogenic mechanism of Talaromycesmarneffei infection. Medicine 2020, 99, e23409. [Google Scholar] [CrossRef] [PubMed]
- Patiño-Medina, J.A.; Valle-Maldonado, M.I.; Vargas-Tejeda, D.; Chávez-Jacobo, V.M.; Corrales-Escobosa, A.R.; Ramirez-Emiliano, J.; Ruiz-Herrera, L.F.; Ramírez-Díaz, M.I.; Garre, V.; Meza-Carmen, V. Arf-like proteins (Arl1 and Arl2) are involved in mitochondrial homeostasis in Mucor circinelloides. Fungal Biol. 2020, 124, 619–628. [Google Scholar] [CrossRef]
- Chandler, J.M.; Treece, E.R.; Trenary, H.R.; Brenneman, J.L.; Flickner, T.J.; Frommelt, J.L.; Oo, Z.M.; Patterson, M.M.; Rundle, W.T.; Valle, O.V.; et al. Protein profiling of the dimorphic, pathogenic fungus, Penicillium marneffei. Proteome Sci. 2008, 6, 17. [Google Scholar] [CrossRef]
- Martinez-Soto, D.; Ruiz-Herrera, J. Transcriptomic analysis of the dimorphic transition of Ustilago maydis induced in vitro by a change in pH. Fungal Genet. Biol. 2013, 58–59, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ying, S.H.; Hu, Y.; Feng, M.G. Mas5, a homologue of bacterial DnaJ, is indispensable for the host infection and environmental adaptation of a filamentous fungal insect pathogen. Environ. Microbiol. 2016, 18, 1037–1047. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, J.; Fang, W.; Zhang, J.; Luo, Z.; Zhang, M.; Fan, Y.; Pei, Y. Mitogen-activated protein kinase hog1 in the entomopathogenic fungus Beauveria bassiana regulates environmental stress responses and virulence to insects. Appl. Environ. Microbiol. 2009, 75, 3787–3795. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, J.-L.; Hou, J.; Li, X.-H.; Feng, M.-G.; Ying, S.-H. Transcription Activator Swi6 Interacts with Mbp1 in MluI Cell Cycle Box-Binding Complex and Regulates Hyphal Differentiation and Virulence in Beauveria bassiana. J. Fungi 2021, 7, 411. https://doi.org/10.3390/jof7060411
Ding J-L, Hou J, Li X-H, Feng M-G, Ying S-H. Transcription Activator Swi6 Interacts with Mbp1 in MluI Cell Cycle Box-Binding Complex and Regulates Hyphal Differentiation and Virulence in Beauveria bassiana. Journal of Fungi. 2021; 7(6):411. https://doi.org/10.3390/jof7060411
Chicago/Turabian StyleDing, Jin-Li, Jia Hou, Xiu-Hui Li, Ming-Guang Feng, and Sheng-Hua Ying. 2021. "Transcription Activator Swi6 Interacts with Mbp1 in MluI Cell Cycle Box-Binding Complex and Regulates Hyphal Differentiation and Virulence in Beauveria bassiana" Journal of Fungi 7, no. 6: 411. https://doi.org/10.3390/jof7060411
APA StyleDing, J.-L., Hou, J., Li, X.-H., Feng, M.-G., & Ying, S.-H. (2021). Transcription Activator Swi6 Interacts with Mbp1 in MluI Cell Cycle Box-Binding Complex and Regulates Hyphal Differentiation and Virulence in Beauveria bassiana. Journal of Fungi, 7(6), 411. https://doi.org/10.3390/jof7060411







