Unfolded Protein Response and Scaffold Independent Pheromone MAP Kinase Signaling Control Verticillium dahliae Growth, Development, and Plant Pathogenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial and Fungal Cultivation Conditions
2.2. Verticillium Strain Construction
2.3. Quantification of Growth and Developmental Structures
2.4. Protein Extraction and Western Hybridization
2.5. Isolation of RNA, cDNA Synthesis, and Quantification of Gene Expressions
2.6. Confocal Microscopy
2.7. Arabidopsis thaliana Root Colonization Assays
2.8. Tomato Plant Infection Assays
2.9. Sequence Analyses
3. Results
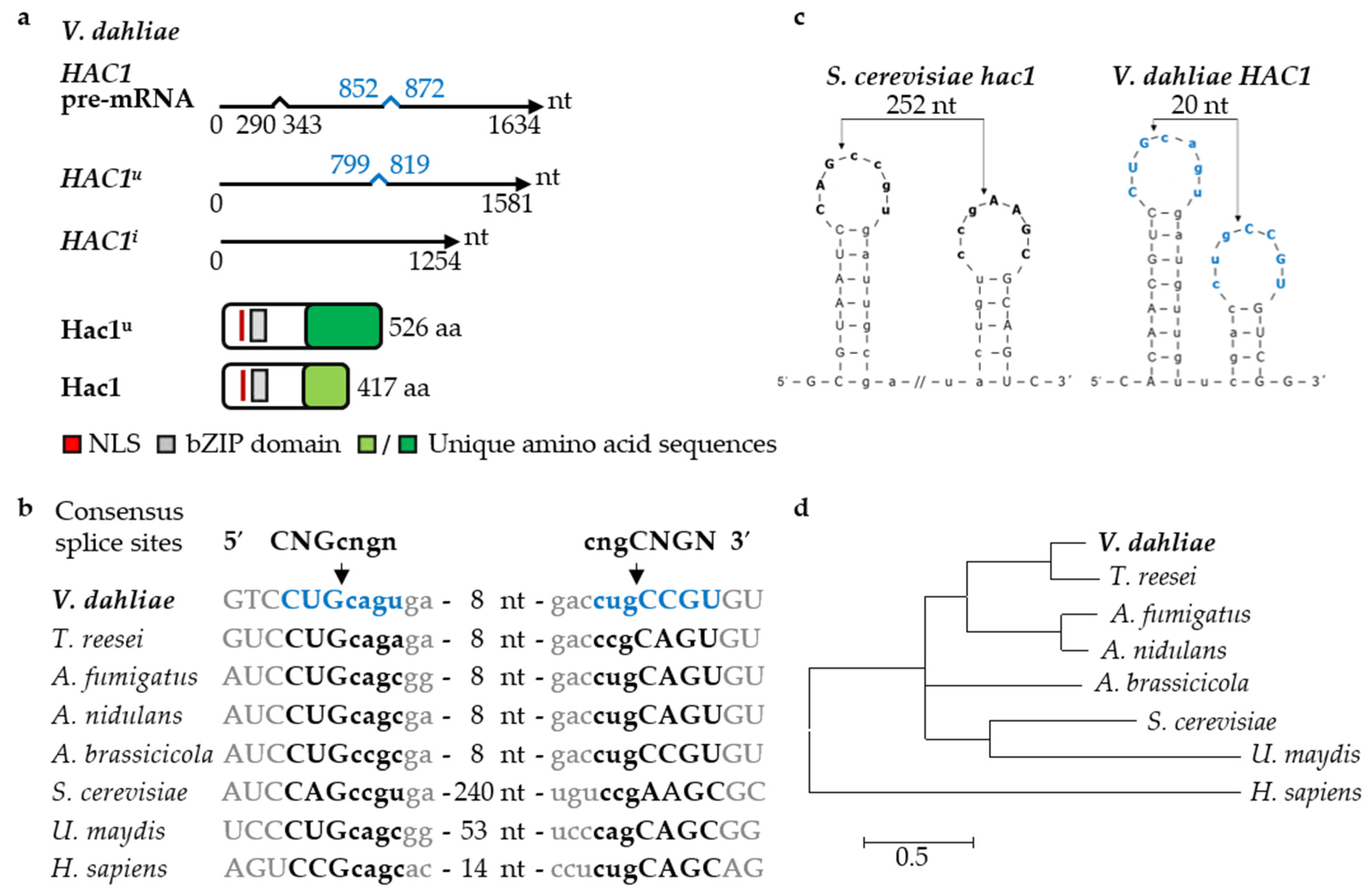
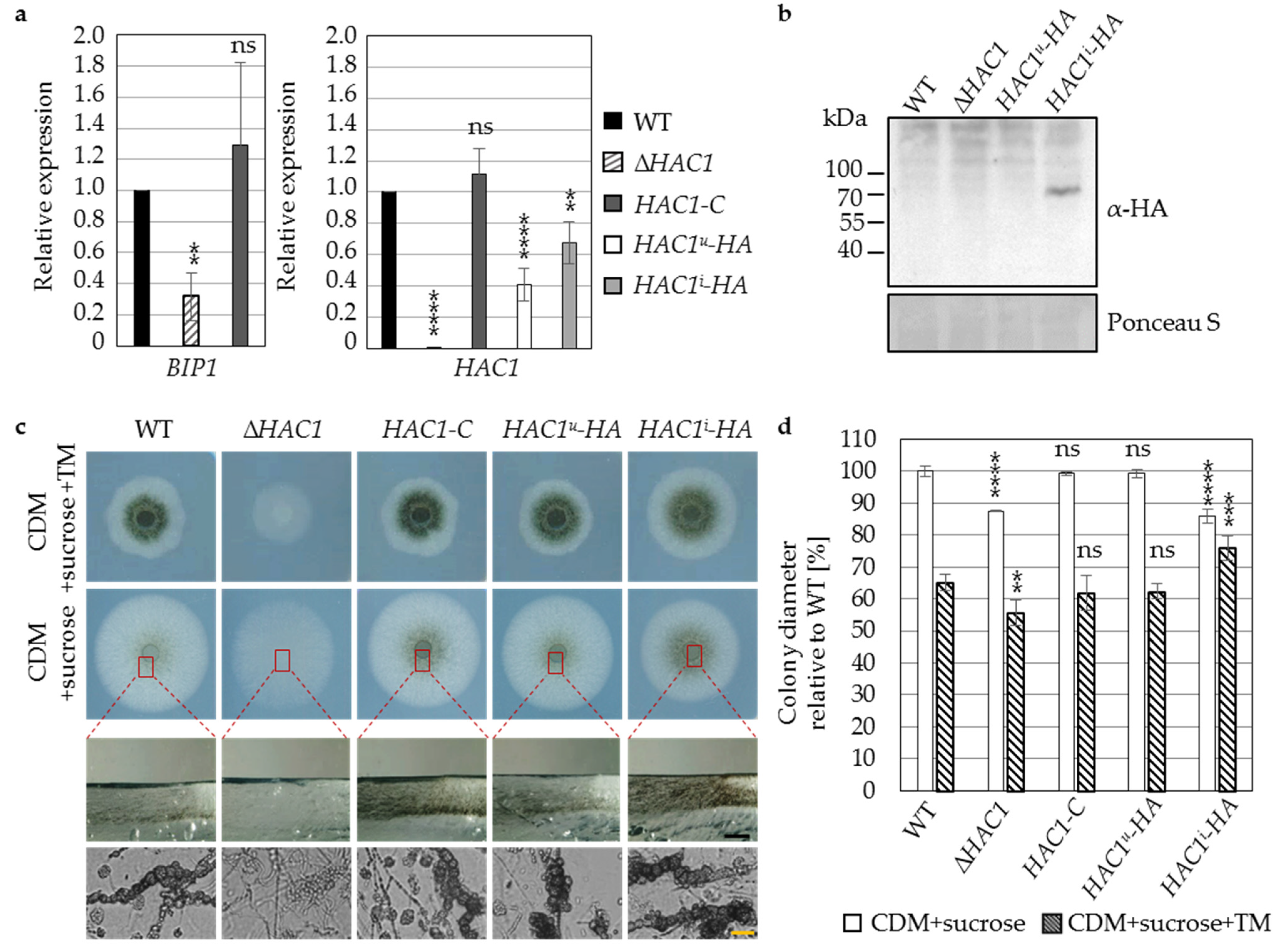
3.1. The Regulator of the Endoplasmic Reticulum-Associated Unfolded Protein Response Pathway Hac1 Supports Fungal Growth and Is Essential for Resting Structure Development in V. dahliae
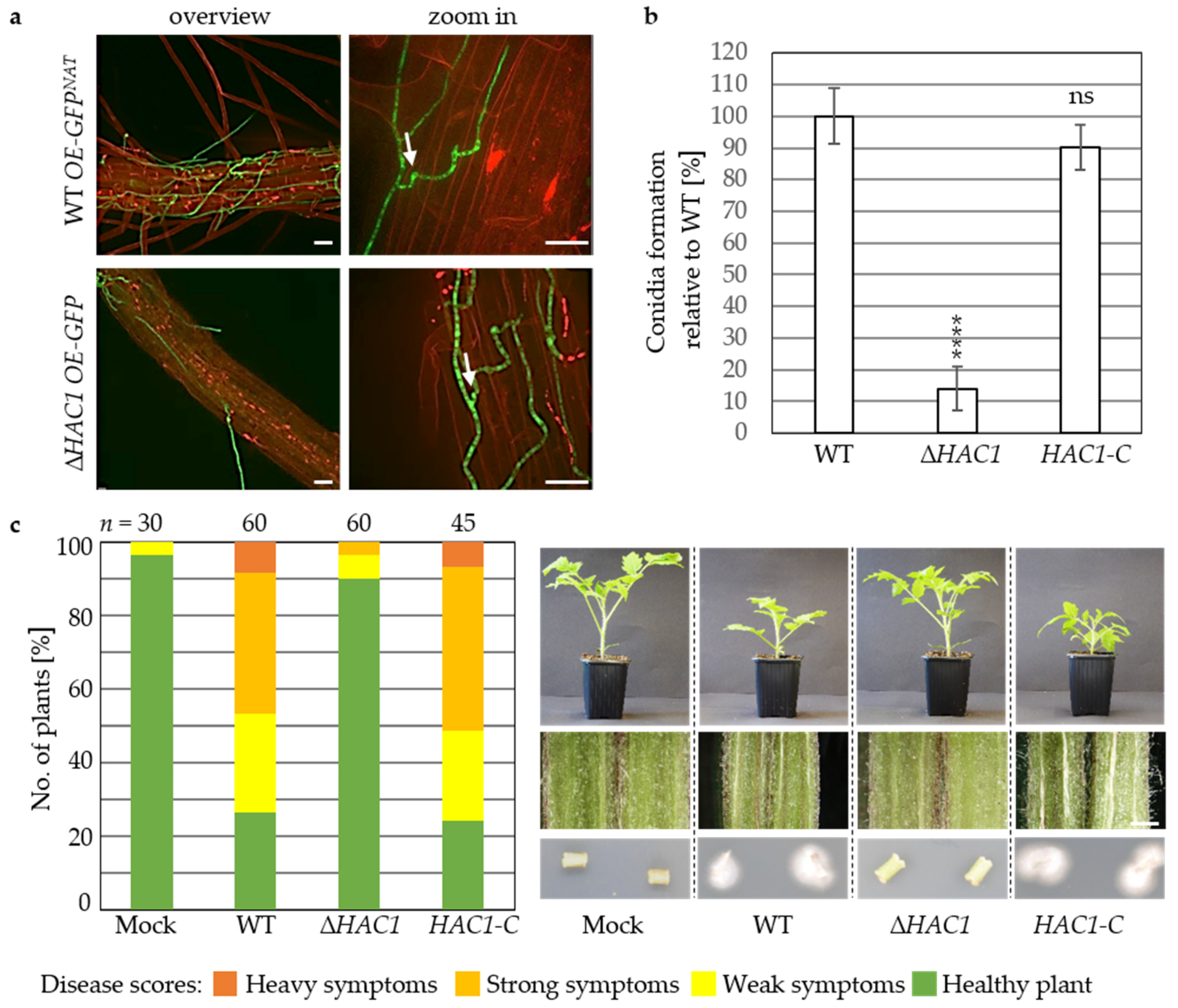
3.2. Virulence of V. dahliae Depends on the Unfolded Protein Response Transcription Factor Hac1
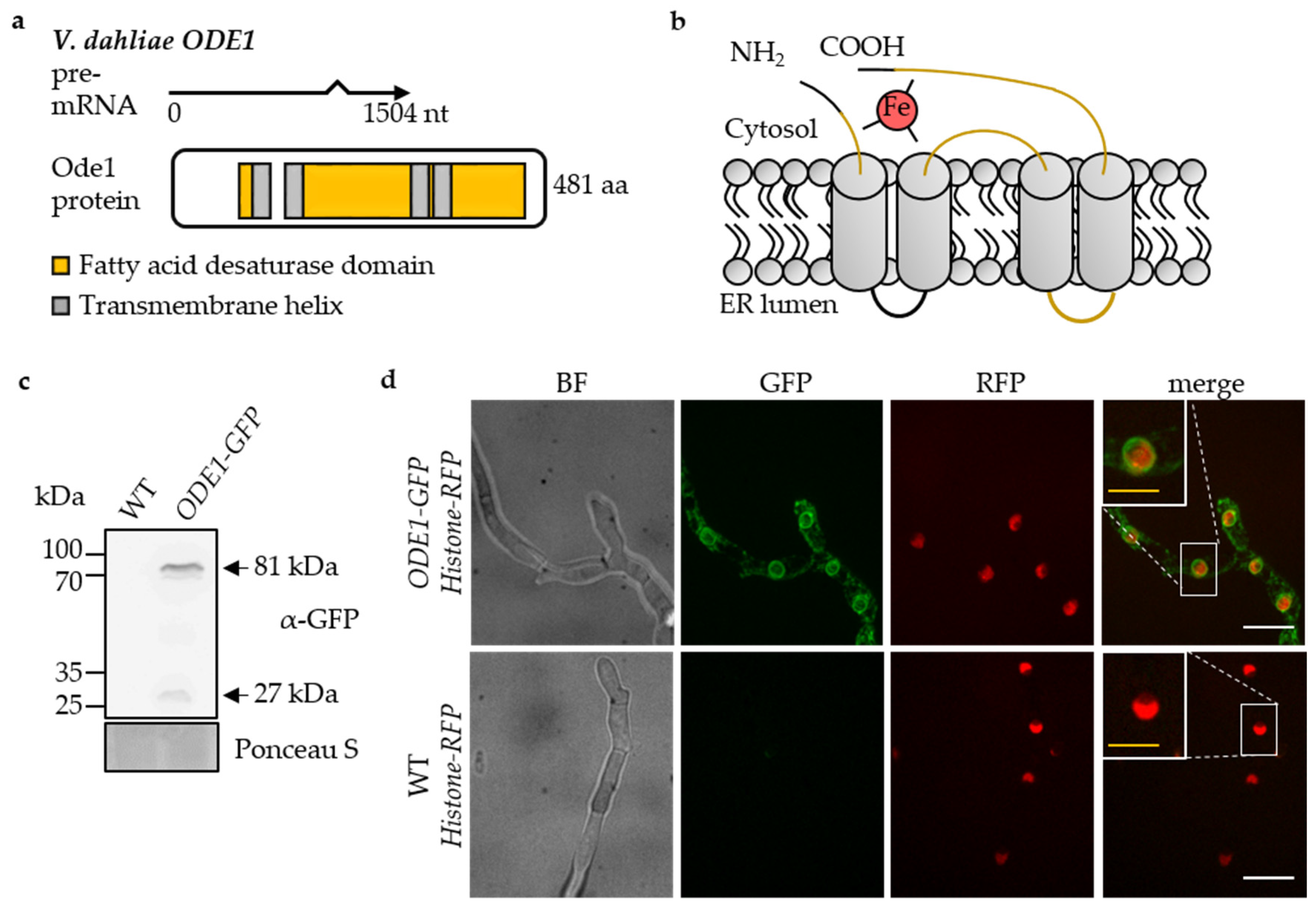
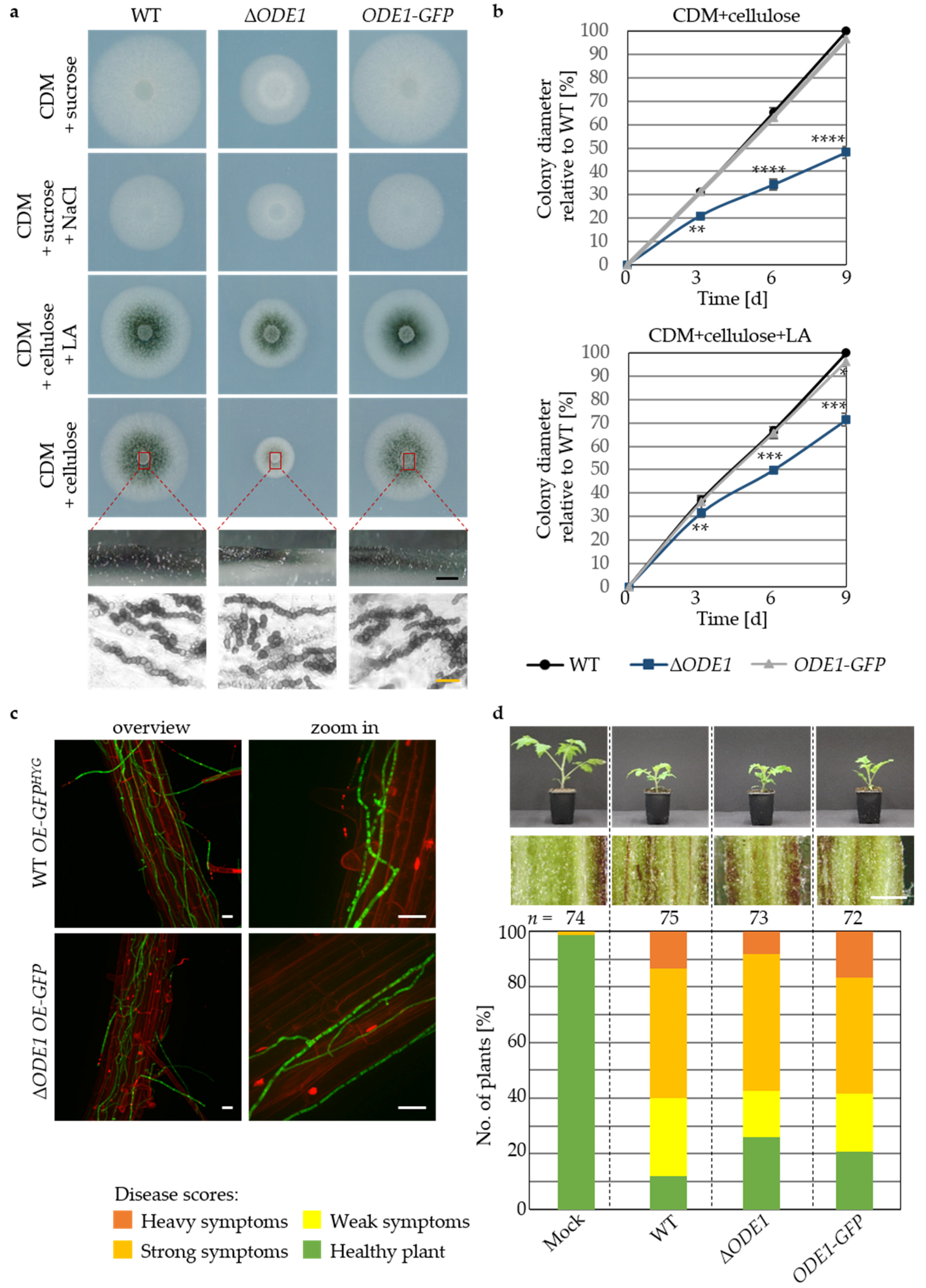
3.3. The Endoplasmic Reticulum-Associated V. dahliae Oleate ∆12-Fatty Acid Desaturase Ode1 Promotes Fungal Differentiation with Only a Minor Impact on Virulence
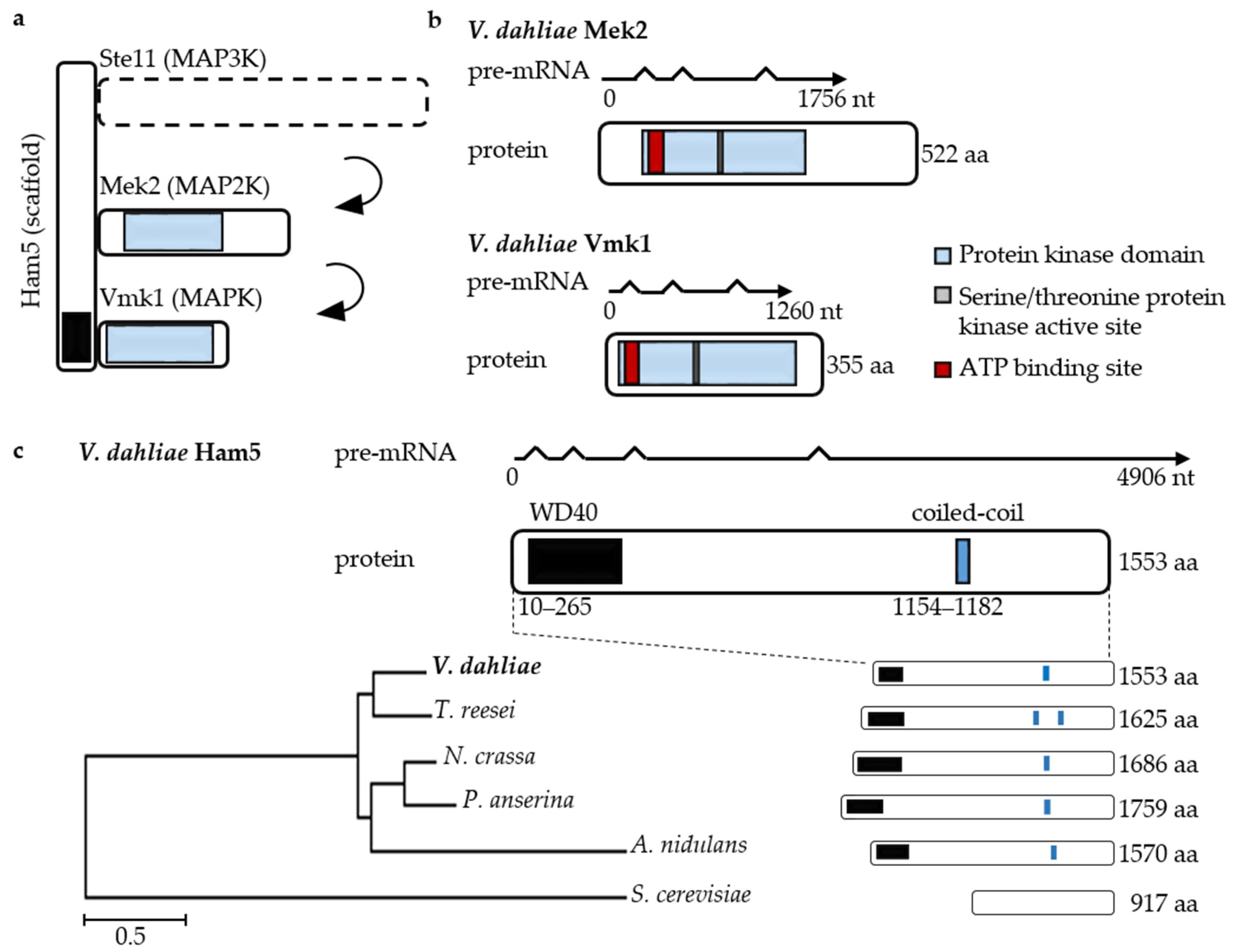
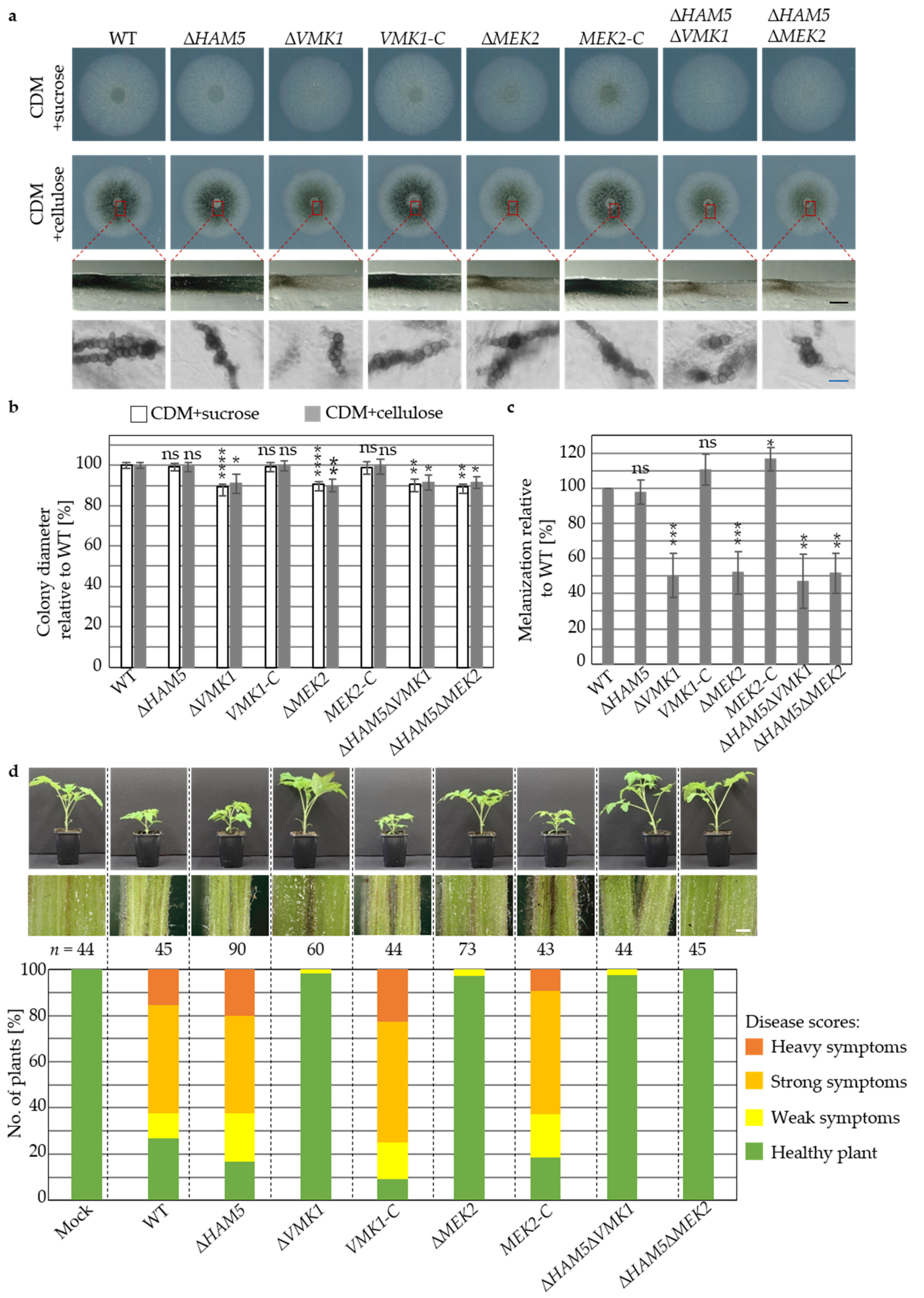
3.4. V. dahliae Development and Plant Disease Symptom Induction Require Pheromone Response MAP Kinase Activities Independently from the Ham5 Scaffold
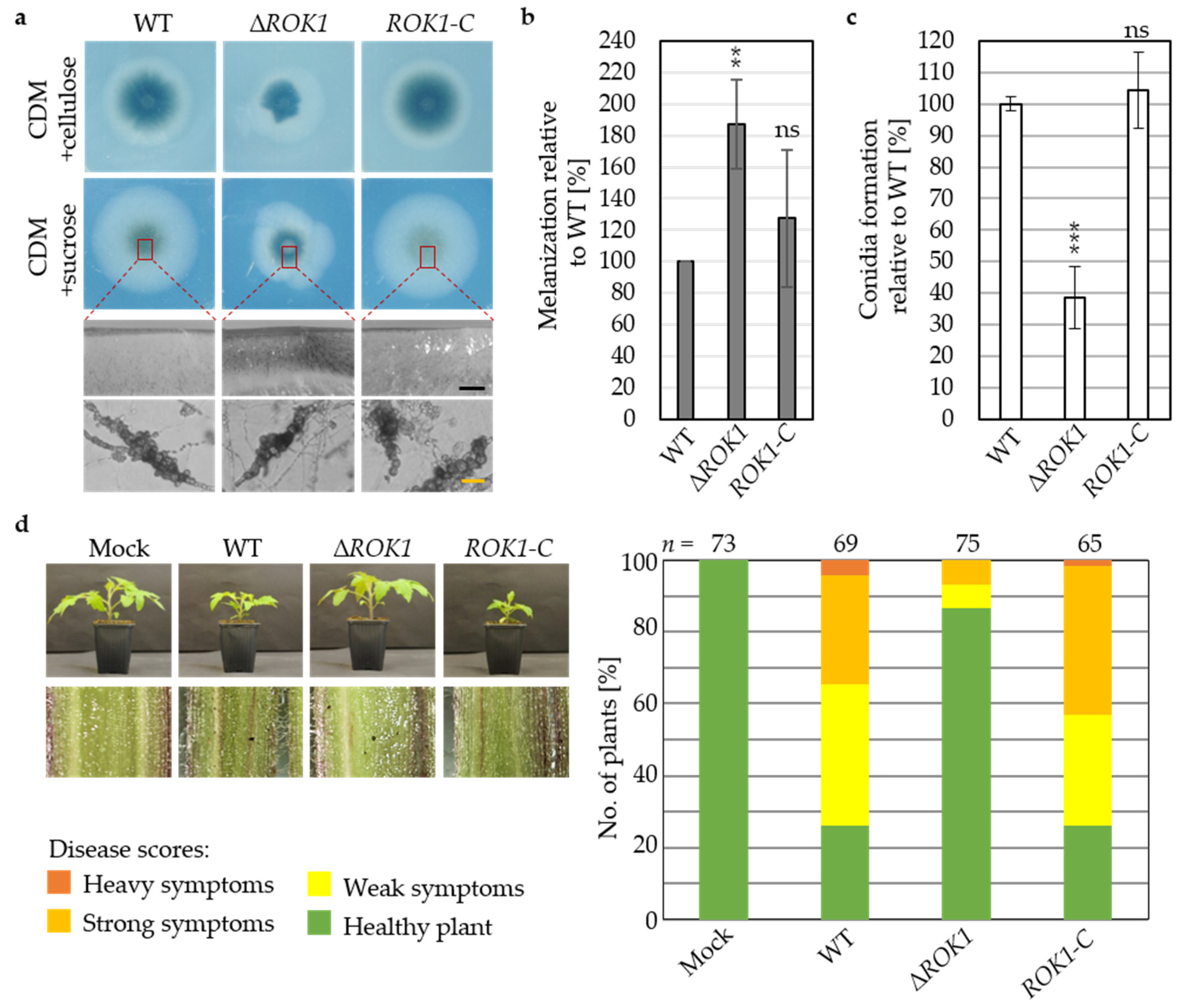
3.5. The Dual-Specificity Phosphatase Rok1 Limits V. dahliae Resting Structure Development and Promotes Growth and Conidiation
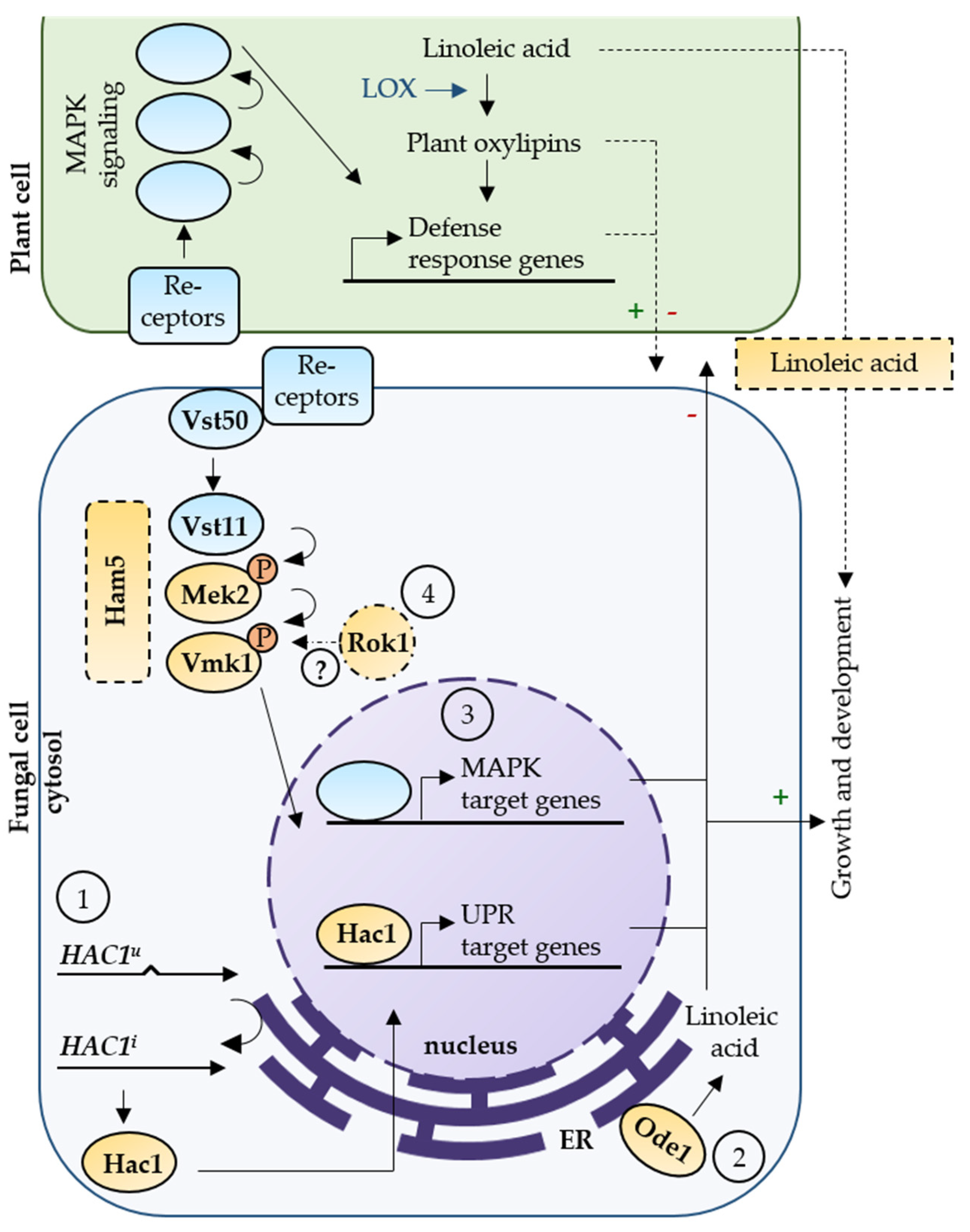
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pegg, G.F.; Brady, B.L. Verticillium Wilts; CABI Pub: Wallingford, UK, 2002; ISBN 9789679362664. [Google Scholar]
- Luo, X.; Xie, C.; Dong, J.; Yang, X.; Sui, A. Interactions between Verticillium dahliae and its host: Vegetative growth, pathogenicity, plant immunity. Appl. Microbiol. Biotechnol. 2014, 16, 6921–6932. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, S. Longevity of the Verticillium wilt fungus in the laboratory and field. Phytopathology 1955, 45, 180–181. [Google Scholar]
- Griffiths, D.A. The fine structure of developing microsclerotia of Verticillium dahliae Kleb. Arch. Mikrobiol. 1970, 74, 207–212. [Google Scholar] [CrossRef]
- Fitzell, R.; Evans, G.; Fahy, P. Studies on the colonization of plant roots by Verticillium dahliae Klebahn with use of immunofluorescent staining. Aust. J. Bot. 1980, 28, 357–368. [Google Scholar] [CrossRef]
- Eynck, C.; Koopmann, B.; Grunewaldt-Stoecker, G.; Karlovsky, P.; von Tiedemann, A. Differential interactions of Verticillium longisporum and V. dahliae with Brassica napus detected with molecular and histological techniques. Eur. J. Plant Pathol. 2007, 118, 259–274. [Google Scholar] [CrossRef]
- Vallad, G.E.; Subbarao, K.V. Colonization of resistant and susceptible lettuce cultivars by a green fluorescent protein-tagged isolate of Verticillium dahliae. Phytopathology 2008, 98, 871–885. [Google Scholar] [CrossRef]
- Su, X.; Lu, G.; Rehman, L.; Li, X.; Sun, L.; Guo, H.; Cheng, H. mCherry-labeled Verticillium dahliae could be utilized to investigate its pathogenicity process in Nicotiana benthamiana. Genes 2018, 9, 508. [Google Scholar] [CrossRef]
- Klosterman, S.J.; Atallah, Z.K.; Vallad, G.E.; Subbarao, K.V. Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 2009, 47, 39–62. [Google Scholar] [CrossRef] [PubMed]
- Fradin, E.F.; Thomma, B.P.H.J. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 2006, 7, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Gerke, J.; Köhler, A.M.; Meister, C.; Thieme, K.G.; Amedo, H.; Braus, G.H. Coordination of fungal secondary metabolism and development. In The Mycota; Benz, J.P., Schipper, K., Eds.; Springer: Cham, Switzerland, 2020; pp. 173–196. ISBN 978-3-030-49924-2. [Google Scholar]
- Kozutsumi, Y.; Segal, M.; Normington, K.; Gething, M.-J.; Sambrook, J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature 1988, 332, 462–464. [Google Scholar] [CrossRef]
- Kohno, K.; Normington, K.; Sambrook, J.; Gething, M.J.; Mori, K. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell. Biol. 1993, 13, 877–890. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C. The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012, 13, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Heimel, K. Unfolded protein response in filamentous fungi-implications in biotechnology. Appl. Microbiol. Biotechnol. 2015, 99, 121–132. [Google Scholar] [CrossRef]
- Okamura, K.; Kimata, Y.; Higashio, H.; Tsuru, A.; Kohno, K. Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem. Biophys. Res. Commun. 2000, 279, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Ma, W.; Gething, M.J.; Sambrook, J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 1993, 74, 743–756. [Google Scholar] [CrossRef]
- Harting, R.; Heimel, K. Genetics of the unfolded protein response in fungi. In The Mycota; Benz, J.P., Schipper, K., Eds.; Springer: Cham, Switzerland, 2020; pp. 49–76. ISBN 978-3-030-49924-2. [Google Scholar]
- Sidrauski, C.; Walter, P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 1997, 90, 1031–1039. [Google Scholar] [CrossRef]
- Gonzalez, T.N.; Sidrauski, C.; Dörfler, S.; Walter, P. Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J. 1999, 18, 3119–3132. [Google Scholar] [CrossRef]
- Mori, K.; Kawahara, T.; Yoshida, H.; Yanagi, H.; Yura, T. Signalling from endoplasmic reticulum to nucleus: Transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells 1996, 1, 803–817. [Google Scholar] [CrossRef]
- Mori, K.; Ogawa, N.; Kawahara, T.; Yanagi, H.; Yura, T. Palindrome with spacer of one nucleotide is characteristic of the cis -acting unfolded protein response element in Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 9912–9920. [Google Scholar] [CrossRef]
- Cox, J.S.; Shamu, C.E.; Walter, P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 1993, 73, 1197–1206. [Google Scholar] [CrossRef]
- Kaufman, R.J. Stress signaling from the lumen of the endoplasmic reticulum: Coordination of gene transcriptional and translational controls. Genes Dev. 1999, 13, 1211–1233. [Google Scholar] [CrossRef]
- Travers, K.J.; Patil, C.K.; Wodicka, L.; Lockhart, D.J.; Weissman, J.S.; Walter, P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 2000, 101, 249–258. [Google Scholar] [CrossRef]
- Conn, P.M. The unfolded protein response and cellular stress. In Methods in Enzymology; Conn, M.P., Ed.; Elsevier: Los Angeles, CA, USA, 2011; pp. 1–384. ISBN 9780123851154. [Google Scholar]
- Krishnan, K.; Askew, D.S. The fungal UPR: A regulatory hub for virulence traits in the mold pathogen Aspergillus fumigatus. Virulence 2014, 5, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, T.; Nakayama, H.; Nagayoshi, Y.; Kakeya, H.; Kohno, S. Dissection of Ire1 functions reveals stress response mechanisms uniquely evolved in Candida glabrata. PLoS Pathog. 2013, 9, e1003160. [Google Scholar] [CrossRef] [PubMed]
- Kimmig, P.; Diaz, M.; Zheng, J.; Williams, C.C.; Lang, A.; Aragón, T.; Li, H.; Walter, P. The unfolded protein response in fission yeast modulates stability of select mRNAs to maintain protein homeostasis. eLife 2012, 1, e00048. [Google Scholar] [CrossRef]
- Tang, W.; Ru, Y.; Hong, L.; Zhu, Q.; Zuo, R.; Guo, X.; Wang, J.; Zhang, H.; Zheng, X.; Wang, P.; et al. System-wide characterization of bZIP transcription factor proteins involved in infection-related morphogenesis of Magnaporthe oryzae. Environ. Microbiol. 2015, 17, 1377–1396. [Google Scholar] [CrossRef]
- Joubert, A.; Simoneau, P.; Campion, C.; Poupard, P.; François, J.M.; Georgeault, S.; Sellier, E.; Guillemette, T. Impact of the unfolded protein response on the pathogenicity of the necrotrophic fungus Alternaria brassicicola. Mol. Microbiol. 2011, 79, 1305–1324. [Google Scholar] [CrossRef] [PubMed]
- Guillemette, T.; Calmes, B.; Simoneau, P.; Index, F.H. Impact of the UPR on the virulence of the plant fungal pathogen A. brassicicola. Virulence 2014, 5, 357–364. [Google Scholar] [CrossRef]
- Heimel, K.; Scherer, M.; Schuler, D.; Kämper, J. The Ustilago maydis Clp1 protein orchestrates pheromone and b-dependent signaling pathways to coordinate the cell cycle and pathogenic development. Plant Cell 2010, 22, 2908–2922. [Google Scholar] [CrossRef]
- Hampel, M.; Jakobi, M.; Schmitz, L.; Meyer, U.; Finkernagel, F.; Doehlemann, G.; Heimel, K. Unfolded protein response (UPR) regulator Cib1 controls expression of genes encoding secreted virulence factors in Ustilago maydis. PLoS ONE 2016, 11, e0153861. [Google Scholar] [CrossRef]
- Pinter, N.; Hach, C.A.; Hampel, M.; Rekhter, D.; Zienkiewicz, K.; Feussner, I.; Poehlein, A.; Daniel, R.; Finkernagel, F.; Heimel, K. Signal peptide peptidase activity connects the unfolded protein response to plant defense suppression by Ustilago maydis. PLoS Pathog. 2019, 15, e1007734. [Google Scholar] [CrossRef]
- Cox, J.S.; Chapman, R.E.; Walter, P. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol. Biol. Cell 1997, 8, 1805–1814. [Google Scholar] [CrossRef]
- Kagiwada, S.; Hosaka, K.; Murata, M.; Nikawa, J.I.; Takatsuki, A. The Saccharomyces cerevisiae SCS2 gene product, a homolog of a synaptobrevin-associated protein, is an integral membrane protein of the endoplasmic reticulum and is required for inositol metabolism. J. Bacteriol. 1998, 180, 1700–1708. [Google Scholar] [CrossRef]
- Rambo, G.W.; Bean, G.A. Sterols and fatty acids of aflatoxin and non-aflatoxin producing isolates of Aspergillus. Phytochemistry 1974, 13, 195–198. [Google Scholar] [CrossRef]
- Evans, J.L.; Moclock, M.A.; Gealt, M.A. The fatty acid composition of the conidia and mycelia of the fungus Aspergillus nidulans. Can. J. Microbiol. 1986, 32, 179–181. [Google Scholar] [CrossRef] [PubMed]
- Castoria, R.; Fanelli, C.; Zoina, A.; Scala, F. Analysis of fatty acids in lipids of Verticillium dahliae and induction of lubimin accumulation in eggplant. Plant Pathol. 1995, 44, 791–795. [Google Scholar] [CrossRef]
- Goodrich-Tanrikulu, M.; Howe, K.; Stafford, A.; Nelson, M.A. Changes in fatty acid composition of Neurospora crassa accompany sexual development and ascospore germination. Microbiology 1998, 144, 1713–1720. [Google Scholar] [CrossRef]
- Gostinčar, C.; Turk, M.; Gunde-Cimerman, N. Environmental impacts on fatty acid composition of fungal membranes. In Fungi from Different Environments; Misra, J.K., Deshmukh, S.K., Eds.; Science Publishers: Enfield, NH, USA, 2009; pp. 278–325. ISBN 9781578085781. [Google Scholar]
- Uttaro, A.D. Biosynthesis of polyunsaturated fatty acids in lower eukaryotes. IUBMB Life 2006, 58, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Calvo, A.M.; Gardner, H.W.; Keller, N.P. Genetic connection between fatty acid metabolism and sporulation in Aspergillus nidulans. J. Biol. Chem. 2001, 276, 25766–25774. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.; Wilson, R.A.; Keller, N.P.; Cleveland, T.E. Deletion of the ∆12-oleic acid desaturase gene of a nonaflatoxigenic Aspergillus parasiticus field isolate affects conidiation and sclerotial development. J. Appl. Microbiol. 2004, 97, 1178–1184. [Google Scholar] [CrossRef]
- Wilson, R.A.; Calvo, A.M.; Chang, P.-K.; Keller, N.P. Characterization of the Aspergillus parasiticus 12-desaturase gene: A role for lipid metabolism in the Aspergillus-seed interaction. Microbiology 2004, 150, 2881–2888. [Google Scholar] [CrossRef]
- Fischer, G.J.; Keller, N.P. Production of cross-kingdom oxylipins by pathogenic fungi: An update on their role in development and pathogenicity. J. Microbiol. 2016, 54, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Brodhun, F.; Feussner, I. Oxylipins in fungi. FEBS J. 2011, 278, 1047–1063. [Google Scholar] [CrossRef] [PubMed]
- Alvaro, C.G.; Thorner, J. Heterotrimeric G protein-coupled receptor signaling in yeast mating pheromone response. J. Biol. Chem. 2016, 291, 7788–7795. [Google Scholar] [CrossRef]
- Cullen, P.J.; Sprague, G.F., Jr. The regulation of filamentous growth in yeast. Genetics 2012, 190, 23–49. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Zhang, X.; Liu, H.; Xu, J.-R. Mitogen-activated protein kinase signaling in plant pathogenic fungi. PLoS Pathog. 2018, 14, e1006875. [Google Scholar] [CrossRef]
- Rauyaree, P.; Ospina-Giraldo, M.D.; Kang, S.; Bhat, R.G.; Subbarao, K.V.; Grant, S.J.; Dobinson, K.F. Mutations in VMK1, a mitogen-activated protein kinase gene, affect microsclerotia formation and pathogenicity in Verticillium dahliae. Curr. Genet. 2005, 48, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Zhou, S.; Shang, X.; Wang, X. VdSho1 regulates growth, oxidant adaptation and virulence in Verticillium dahliae. J. Phytopathol. 2016, 164, 1064–1074. [Google Scholar] [CrossRef]
- Sarmiento-Villamil, J.L.; Prieto, P.; Klosterman, S.J.; García-Pedrajas, M.D. Characterization of two homeodomain transcription factors with critical but distinct roles in virulence in the vascular pathogen Verticillium dahliae. Mol. Plant Pathol. 2018, 19, 986–1004. [Google Scholar] [CrossRef]
- Li, J.-J.; Zhou, L.; Yin, C.-M.; Zhang, D.-D.; Klosterman, S.J.; Wang, B.-L.; Song, J.; Wang, D.; Hu, X.-P.; Subbarao, K.V.; et al. The Verticillium dahliae Sho1-MAPK pathway regulated melanin biosynthesis is required for cotton infection. Environ. Microbiol. 2019, 21, 4852–4874. [Google Scholar] [CrossRef]
- Yu, J.; Li, T.; Tang, C.; Klosterman, S.J.; Tian, C.; Wang, Y. Two Verticillium dahliae MAPKKKs, VdSsk2 and VdSte11, have distinct roles in pathogenicity, microsclerotial formation, and stress adaptation. mSphere 2019, 4, e00426-19. [Google Scholar] [CrossRef]
- Heimel, K.; Freitag, J.; Hampel, M.; Ast, J. Crosstalk between the unfolded protein response and pathways that regulate pathogenic development in Ustilago maydis. Plant Cell 2013, 25, 4262–4277. [Google Scholar] [CrossRef]
- Schmitz, L.; Schwier, M.A.; Heimel, K. The unfolded protein response regulates pathogenic development of Ustilago maydis by Rok1-dependent inhibition of mating-type signaling. mBio 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Dettmann, A.; Heilig, Y.; Valerius, O.; Ludwig, S.; Seiler, S. Fungal communication requires the MAK-2 pathway elements STE-20 and RAS-2, the NRC-1 adapter STE-50 and the MAP kinase scaffold HAM-5. PLoS Genet. 2014, 10, e1004762. [Google Scholar] [CrossRef]
- Jonkers, W.; Leeder, A.C.; Ansong, C.; Wang, Y.; Yang, F.; Starr, T.L.; Camp, D.G.; Smith, R.D.; Glass, N.L. HAM-5 functions as a MAP kinase scaffold during cell fusion in Neurospora crassa. PLoS Genet. 2014, 10, e1004783. [Google Scholar] [CrossRef]
- Frawley, D.; Karahoda, B.; Sarikaya Bayram, Ö.; Bayram, Ö. The HamE scaffold positively regulates MpkB phosphorylation to promote development and secondary metabolism in Aspergillus nidulans. Sci. Rep. 2018, 8, 16588. [Google Scholar] [CrossRef]
- Lazo, G.R.; Stein, P.A.; Ludwig, R.A. A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology 1991, 9, 963–967. [Google Scholar] [CrossRef] [PubMed]
- Bertani, G. Studies on lysogenesis I. The mode of phageliberation by lysogenic Eschericia coli. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Neumann, M.J.; Dobinson, K.F. Sequence tag analysis of gene expression during pathogenic growth and microsclerotia development in the vascular wilt pathogen Verticillium dahliae. Fungal Genet. Biol. 2003, 38, 54–62. [Google Scholar] [CrossRef]
- Hollensteiner, J.; Wemheuer, F.; Harting, R.; Kolarzyk, A.M.; Diaz Valerio, S.M.; Poehlein, A.; Brzuszkiewicz, E.B.; Nesemann, K.; Braus-Stromeyer, S.A.; Braus, G.H.; et al. Bacillus thuringiensis and Bacillus weihenstephanensis inhibit the growth of phytopathogenic Verticillium species. Front. Microbiol. 2017, 7, 2171. [Google Scholar] [CrossRef] [PubMed]
- Harting, R.; Höfer, A.; Tran, V.-T.; Weinhold, L.-M.; Barghahn, S.; Schlüter, R.; Braus, G.H. The Vta1 transcriptional regulator is required for microsclerotia melanization in Verticillium dahliae. Fungal Biol. 2020, 124, 490–500. [Google Scholar] [CrossRef]
- Fradin, E.F.; Zhang, Z.; Juarez Ayala, J.C.; Castroverde, C.D.M.; Nazar, R.N.; Robb, J.; Liu, C.-M.; Thomma, B.P.H.J. Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 2009, 150, 320–332. [Google Scholar] [CrossRef]
- Timpner, C.; Braus-Stromeyer, S.A.; Tran, V.-T.; Braus, G.H. The Cpc1 regulator of the cross-pathway control of amino acid biosynthesis is required for pathogenicity of the vascular pathogen Verticillium longisporum. Mol. Plant-Microbe Interact. 2013, 26, 1312–1324. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tran, V.-T.; Braus-Stromeyer, S.A.; Kusch, H.; Reusche, M.; Kaever, A.; Kühn, A.; Valerius, O.; Landesfeind, M.; Aßhauer, K.; Tech, M.; et al. Verticillium transcription activator of adhesion Vta2 suppresses microsclerotia formation and is required for systemic infection of plant roots. New Phytol. 2014, 202, 565–581. [Google Scholar] [CrossRef] [PubMed]
- Covert, S.F.; Kapoor, P.; Lee, M.; Briley, A.; Nairn, C.J. Agrobacterium tumefaciens-mediated transformation of Fusarium circinatum. Mycol. Res. 2001, 105, 259–264. [Google Scholar] [CrossRef]
- Leonard, M.; Kühn, A.; Harting, R.; Maurus, I.; Nagel, A.; Starke, J.; Kusch, H.; Valerius, O.; Feussner, K.; Feussner, I.; et al. Verticillium longisporum elicits media-dependent secretome responses with capacity to distinguish between plant-related environments. Front. Microbiol. 2020, 11, 1876. [Google Scholar] [CrossRef]
- Bayram, Ö.; Sarikaya Bayram, Ö.; Ahmed, Y.L.; Maruyama, J.-I.; Valerius, O.; Rizzoli, S.O.; Ficner, R.; Irniger, S.; Braus, G.H. The Aspergillus nidulans MAPK module AnSte11-Ste50-Ste7-Fus3 controls development and secondary metabolism. PLoS Genet. 2012, 8, e1002816. [Google Scholar] [CrossRef]
- Zhou, L.; Zhao, J.; Guo, W.; Zhang, T. Functional analysis of autophagy genes via Agrobacterium-mediated transformation in the vascular wilt fungus Verticillium dahliae. J. Genet. Genom. 2013, 40, 421–431. [Google Scholar] [CrossRef]
- Li, C.; Wen, A.; Shen, B.; Lu, J.; Huang, Y.; Chang, Y. FastCloning: A highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol. 2011, 11, 92. [Google Scholar] [CrossRef]
- Rasband ImageJ. Available online: https://imagej.nih.gov/ij/ (accessed on 30 November 2020).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the ∆∆CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Geer, L.Y.; Marchler-Bauer, A.; Geer, R.C.; Han, L.; He, J.; He, S.; Liu, C.; Shi, W.; Bryant, S.H. The NCBI BioSystems database. Nucleic Acids Res. 2010, 38, S492–S496. [Google Scholar] [CrossRef]
- Howe, K.L.; Contreras-Moreira, B.; De Silva, N.; Maslen, G.; Akanni, W.; Allen, J.; Alvarez-Jarreta, J.; Barba, M.; Bolser, D.M.; Cambell, L.; et al. Ensembl Genomes 2020-enabling non-vertebrate genomic research. Nucleic Acids Res. 2020, 48, D689–D695. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Hooks, K.B.; Griffiths-Jones, S. Conserved RNA structures in the non-canonical Hac1/Xbp1 intron. RNA Biol. 2011, 8, 552–556. [Google Scholar] [CrossRef] [PubMed]
- Brodhagen, M.; Tsitsigiannis, D.I.; Hornung, E.; Goebel, C.; Feussner, I.; Keller, N.P. Reciprocal oxylipin-mediated cross-talk in the Aspergillus-seed pathosystem. Mol. Microbiol. 2008, 67, 378–391. [Google Scholar] [CrossRef]
- Brodhun, F.; Go, C.; Hornung, E.; Feussner, I. Identification of PpoA from Aspergillus nidulans as a fusion protein of a fatty acid heme dioxygenase/ peroxidase and a cytochrome P450. J. Biol. Chem. 2009, 284, 11792–11805. [Google Scholar] [CrossRef]
- Reverberi, M.; Punelli, F.; Scarpari, M.; Camera, E.; Zjalic, S.; Ricelli, A.; Fanelli, C.; Fabbri, A.A. Lipoperoxidation affects ochratoxin A biosynthesis in Aspergillus ochraceus and its interaction with wheat seeds. Appl. Microbiol. Biotechnol. 2010, 85, 1935–1946. [Google Scholar] [CrossRef]
- Scala, V.; Giorni, P.; Cirlini, M.; Ludovici, M.; Visentin, I.; Cardinale, F.; Fabbri, A.A.; Fanelli, C.; Reverberi, M.; Battilani, P.; et al. LDS1-produced oxylipins are negative regulators of growth, conidiation and fumonisin synthesis in the fungal maize pathogen Fusarium verticillioides. Front. Microbiol. 2014, 5, 669. [Google Scholar] [CrossRef]
- Patkar, R.N.; Naqvi, N.I. Fungal manipulation of hormone-regulated plant defense. PLoS Pathog. 2017, 13, e1006334. [Google Scholar] [CrossRef] [PubMed]
- Los, D.A.; Murata, N. Structure and expression of fatty acid desaturases. Biochim. Biophys. Acta 1998, 1394, 3–15. [Google Scholar] [CrossRef]
- Doi, K.; Gartner, A.; Ammerer, G.; Errede, B.; Shinkawa, H.; Sugimoto, K.; Matsumoto, K. MSG5, a novel protein phosphatase promotes adaptation to pheromone response in S. cerevisiae. EMBO J. 1994, 13, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.L.; Deschenes, R.J.; Guan, K.L. Differential regulation of FUS3 MAP kinase by tyrosine-specific phosphatases PTP2/PTP3 and dual-specificity phosphatase MSG5 in Saccharomyces cerevisiae. Genes Dev. 1997, 11, 1690–1702. [Google Scholar] [CrossRef]
- Andersson, J.; Simpson, D.M.; Qi, M.; Wang, Y.; Elion, E.A. Differential input by Ste5 scaffold and Msg5 phosphatase route a MAPK cascade to multiple outcomes. EMBO J. 2004, 23, 2564–2576. [Google Scholar] [CrossRef]
- Di Stasio, M.; Brefort, T.; Mendoza-Mendoza, A.; Münch, K.; Kahmann, R. The dual specificity phosphatase Rok1 negatively regulates mating and pathogenicity in Ustilago maydis. Mol. Microbiol. 2009, 73, 73–88. [Google Scholar] [CrossRef]
- Xu, J.R.; Hamer, J.E. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 1996, 10, 2696–2706. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.R.; Staiger, C.J.; Hamer, J.E. Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc. Natl. Acad. Sci. USA 1998, 95, 12713–12718. [Google Scholar] [CrossRef] [PubMed]
- Saxena, M.; Mustelin, T. Extracellular signals and scores of phosphatases: All roads lead to MAP kinase. Semin. Immunol. 2000, 12, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Chapman, R.E.; Walter, P. Translational attenuation mediated by an mRNA intron. Curr. Biol. 1997, 7, 850–859. [Google Scholar] [CrossRef]
- Rüegsegger, U.; Leber, J.H.; Walter, P. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell 2001, 107, 103–114. [Google Scholar] [CrossRef]
- Di Santo, R.; Aboulhouda, S.; Weinberg, D.E. The fail-safe mechanism of post-transcriptional silencing of unspliced HAC1 mRNA. eLife 2016, 5, e20069. [Google Scholar] [CrossRef]
- Mulder, H.J.; Nikolaev, I. HacA-dependent transcriptional switch releases hacA mRNA from a translational block upon endoplasmic reticulum stress. Eukaryot. Cell 2009, 8, 665–675. [Google Scholar] [CrossRef]
- Saloheimo, M.; Valkonen, M.; Penttilä, M. Activation mechanisms of the HACI-mediated unfolded protein response in filamentous fungi. Mol. Microbiol. 2003, 47, 1149–1161. [Google Scholar] [CrossRef]
- Mulder, H.J.; Saloheimo, M.; Penttilä, M.; Madrid, S.M. The transcription factor HACA mediates the unfolded protein response in Aspergillus niger, and up-regulates its own transcription. Mol. Genet. Genom. 2004, 271, 130–140. [Google Scholar] [CrossRef]
- Nikawa, J.; Akiyoshi, M.; Hirata, S.; Fukuda, T. Saccharomyces cerevisiae IRE2/HAC1 is involved in IRE1-mediated KAR2 expression. Nucleic Acids Res. 1996, 24, 4222–4226. [Google Scholar] [CrossRef]
- Cheon, S.A.; Jung, K.W.; Chen, Y.L.; Heitman, J.; Bahn, Y.S.; Kang, H.A. Unique evolution of the UPR pathway with a novel bZIP transcription factor, HxL1, for controlling pathogenicity of Cryptococcus neoformans. PLoS Pathog. 2011, 7, e1002177. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, N.D.S.P.; Jørgensen, T.R.; Arentshorst, M.; Nitsche, B.M.; van den Hondel, C.A.; Archer, D.B.; Ram, A.F.J. Genome-wide expression analysis upon constitutive activation of the HacA bZIP transcription factor in Aspergillus niger reveals a coordinated cellular response to counteract ER stress. BMC Genom. 2012, 13, 350. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.; Harting, R.; Braus-Stromeyer, S.A.; Tran, V.-T.; Leonard, M.; Höfer, A.; Abelmann, A.; Bakti, F.; Valerius, O.; Schlüter, R.; et al. Verticillium dahliae transcription factors Som1 and Vta3 control microsclerotia formation and sequential steps of plant root penetration and colonisation to induce disease. New Phytol. 2019, 221, 2138–2159. [Google Scholar] [CrossRef]
- Herzog, B.; Popova, B.; Jakobshagen, A.; Shahpasandzadeh, H.; Braus, G.H. Mutual cross talk between the regulators Hac1 of the unfolded protein response and Gcn4 of the general amino acid control of Saccharomyces cerevisiae. Eukaryot. Cell 2013, 12, 1142–1154. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Scrimale, T.; Didone, L.; de Mesy Bentley, K.L.; Krysan, D.J. The unfolded protein response is induced by the cell wall integrity mitogen-activated protein kinase signaling cascade and is required for cell wall integrity in Saccharomyces cerevisiae. Mol. Biol. Cell 2009, 20, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.; Chi, M.-H.; Khang, C.H.; Park, S.-Y.; Kang, S.; Valent, B.; Lee, Y.-H. The ER chaperone LHS1 is involved in asexual development and rice infection by the blast fungus Magnaporthe oryzae. Plant Cell Online 2009, 21, 681–695. [Google Scholar] [CrossRef] [PubMed]
- Höfer, A.M.; Harting, R.; Aßmann, N.F.; Gerke, J.; Schmitt, K.; Starke, J.; Bayram, Ö.; Tran, V.-T.; Valerius, O.; Braus-Stromeyer, S.A.; et al. The velvet protein Vel1 controls initial plant root colonization and conidia formation for xylem distribution in Verticillium wilt. PLoS Genet. 2021, 17, e1009434. [Google Scholar] [CrossRef]
- Richie, D.L.; Feng, X.; Hartl, L.; Aimanianda, V.; Krishnan, K.; Powers-Fletcher, M.V.; Watson, D.S.; Galande, A.K.; White, S.M.; Willett, T.; et al. The virulence of the opportunistic fungal pathogen Aspergillus fumigatus requires cooperation between the endoplasmic reticulum-associated degradation pathway (ERAD) and the unfolded protein response (UPR). Virulence 2011, 2, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Burow, G.B.; Nesbitt, T.C.; Dunlap, J.; Keller, N.P. Seed lipoxygenase products modulate Aspergillus mycotoxin biosynthesis. Mol. Plant-Microbe Interact. 1997, 10, 380–387. [Google Scholar] [CrossRef]
- Calvo, A.M.; Hinze, L.L.; Gardner, H.W.; Keller, N.P. Sporogenic effect of polyunsaturated fatty acids on development of Aspergillus spp. Appl. Environ. Microbiol. 1999, 65, 3668–3673. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Kolomiets, M.V. Host-derived lipids and oxylipins are crucial signals in modulating mycotoxin production by fungi. Toxin Rev. 2009, 28, 79–88. [Google Scholar] [CrossRef]
- Horowitz Brown, S.; Scott, J.B.; Bhaheetharan, J.; Sharpee, W.C.; Milde, L.; Wilson, R.A.; Keller, N.P. Oxygenase coordination is required for morphological transition and the host-fungus interaction of Aspergillus flavus. Mol. Plant-Microbe Interact. 2009, 22, 882–894. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, H.J.; Weber, M.J. Mitogen-activated protein kinases: Specific messages from ubiquitous messengers. Mol. Cell. Biol. 1999, 19, 2435–2444. [Google Scholar] [CrossRef] [PubMed]
- Patterson, J.C.; Klimenko, E.S.; Thorner, J. Single-cell analysis reveals that insulation maintains signaling specificity between two yeast MAPK pathways with common components. Sci. Signal. 2010, 3, ra75. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Bobrowicz, P.; Wilkinson, H.H.; Ebbole, D.J. A mitogen-activated protein kinase pathway essential for mating and contributing to vegetative growth in Neurospora crassa. Genetics 2005, 170, 1091–1104. [Google Scholar] [CrossRef]
- Aldabbous, M.S.; Roca, M.G.; Stout, A.; Huang, I.-C.; Read, N.D.; Free, S.J. The ham-5, rcm-1 and rco-1 genes regulate hyphal fusion in Neurospora crassa. Microbiology 2010, 156, 2621–2629. [Google Scholar] [CrossRef]
- Wang, R.J.; Peng, J.; Li, Q.X.; Peng, Y.L. Phosphorylation-mediated regulatory networks in mycelia of Pyricularia oryzae revealed by phosphoproteomic analyses. Mol. Cell. Proteom. 2017, 16, 1669–1682. [Google Scholar] [CrossRef]
- Yun, Y.; Liu, Z.; Yin, Y.; Jiang, J.; Chen, Y.; Xu, J.R.; Ma, Z. Functional analysis of the Fusarium graminearum phosphatome. New Phytol. 2015, 207, 119–134. [Google Scholar] [CrossRef]
- Fernandes, T.R.; Sánchez Salvador, E.; Tapia, Á.G.; Di Pietro, A. Dual-specificity protein phosphatase Msg5 controls cell wall integrity and virulence in Fusarium oxysporum. Fungal Genet. Biol. 2021, 146, 103486. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, S.; Hou, R.; Zhao, Z.; Zheng, Q.; Xu, Q.; Zheng, D. Functional analysis of the kinome of the wheat scab fungus Fusarium graminearum. PLoS Pathog. 2011, 7, e1002460. [Google Scholar] [CrossRef]
- Xiong, D.; Wang, Y.; Ma, J.; Klosterman, S.J.; Xiao, S.; Tian, C. Deep mRNA sequencing reveals stage-specific transcriptome alterations during microsclerotia development in the smoke tree vascular wilt pathogen, Verticillium dahliae. BMC Genom. 2014, 15, 324. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Starke, J.; Harting, R.; Maurus, I.; Leonard, M.; Bremenkamp, R.; Heimel, K.; Kronstad, J.W.; Braus, G.H. Unfolded Protein Response and Scaffold Independent Pheromone MAP Kinase Signaling Control Verticillium dahliae Growth, Development, and Plant Pathogenesis. J. Fungi 2021, 7, 305. https://doi.org/10.3390/jof7040305
Starke J, Harting R, Maurus I, Leonard M, Bremenkamp R, Heimel K, Kronstad JW, Braus GH. Unfolded Protein Response and Scaffold Independent Pheromone MAP Kinase Signaling Control Verticillium dahliae Growth, Development, and Plant Pathogenesis. Journal of Fungi. 2021; 7(4):305. https://doi.org/10.3390/jof7040305
Chicago/Turabian StyleStarke, Jessica, Rebekka Harting, Isabel Maurus, Miriam Leonard, Rica Bremenkamp, Kai Heimel, James W. Kronstad, and Gerhard H. Braus. 2021. "Unfolded Protein Response and Scaffold Independent Pheromone MAP Kinase Signaling Control Verticillium dahliae Growth, Development, and Plant Pathogenesis" Journal of Fungi 7, no. 4: 305. https://doi.org/10.3390/jof7040305
APA StyleStarke, J., Harting, R., Maurus, I., Leonard, M., Bremenkamp, R., Heimel, K., Kronstad, J. W., & Braus, G. H. (2021). Unfolded Protein Response and Scaffold Independent Pheromone MAP Kinase Signaling Control Verticillium dahliae Growth, Development, and Plant Pathogenesis. Journal of Fungi, 7(4), 305. https://doi.org/10.3390/jof7040305





