Isolation and Molecular Characterization of the Romaine Lettuce Phylloplane Mycobiome
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Samples and Fungal Isolation
2.2. Identification of Fungal Isolates
2.3. Statistical Analyses
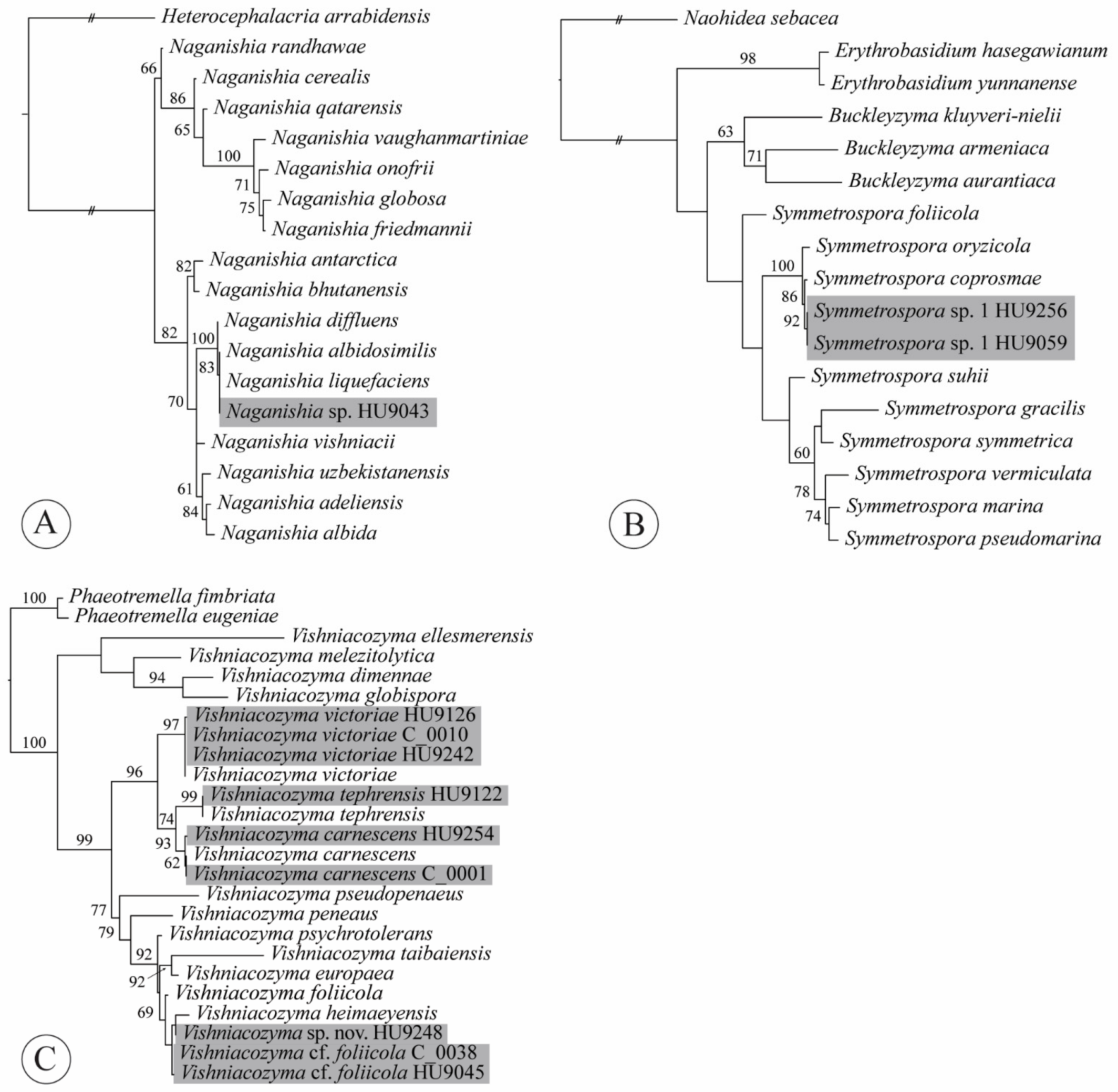
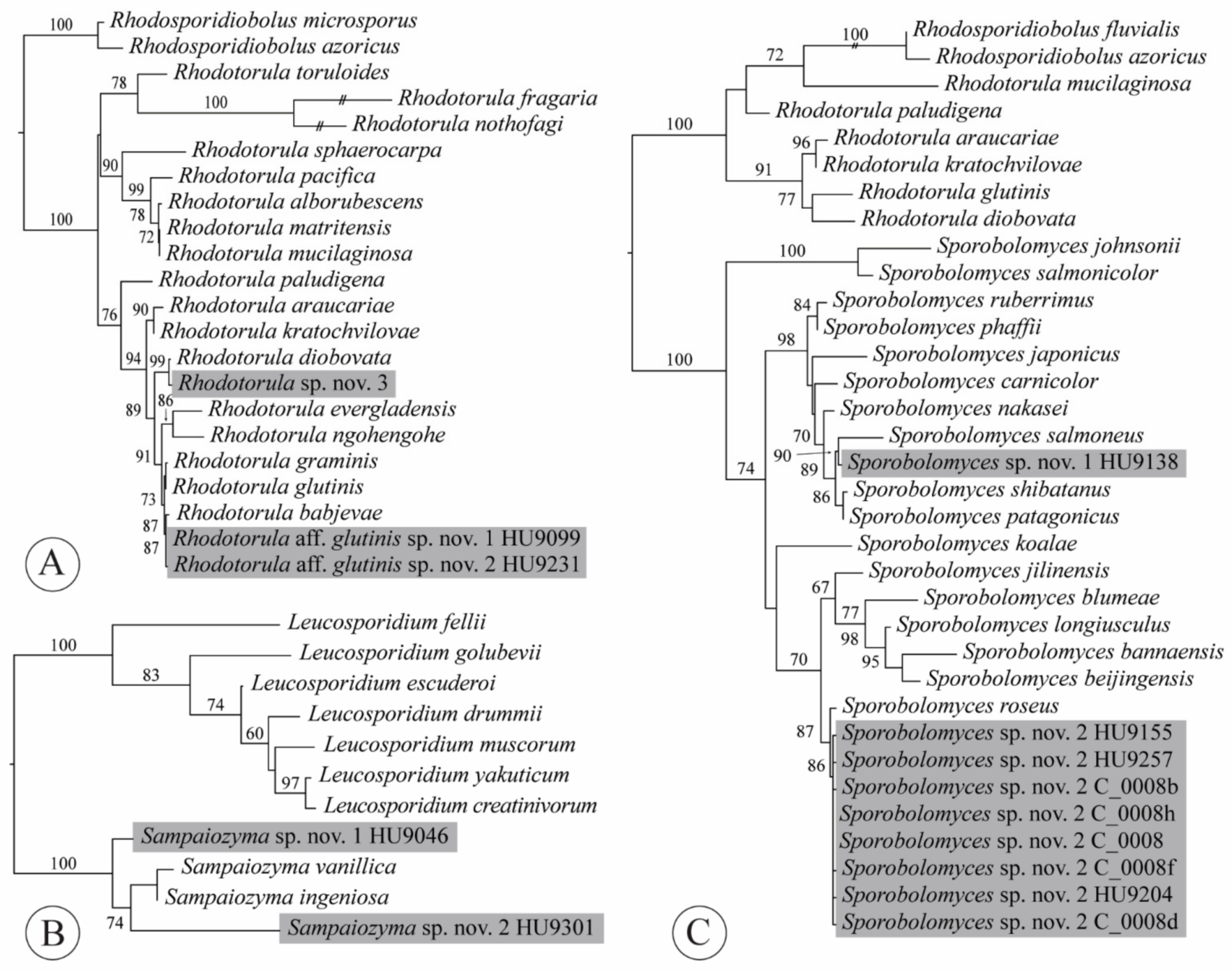
2.4. Phylogenetic Analyses
3. Results
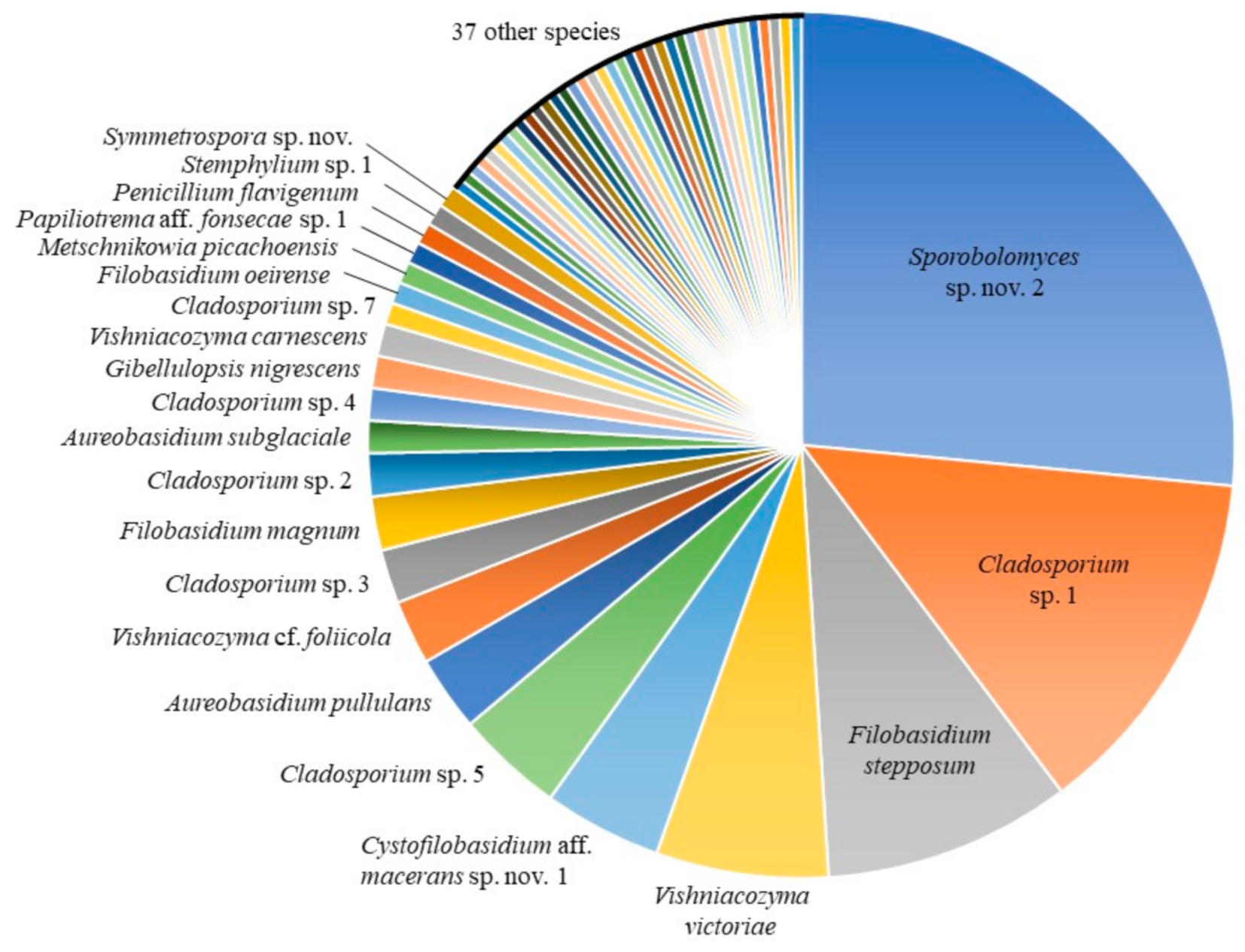
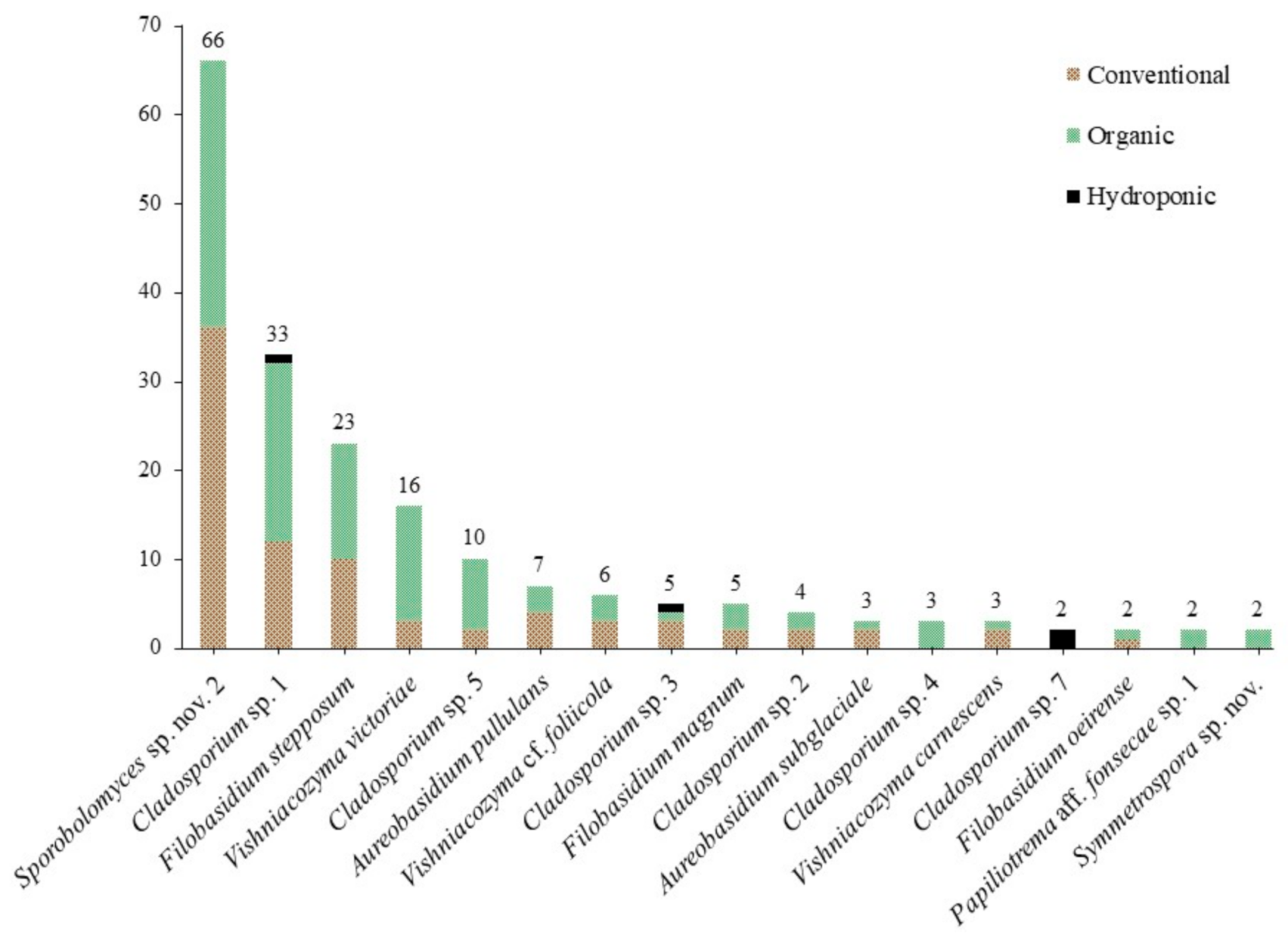
3.1. Culture-Based Community from Romaine Lettuce
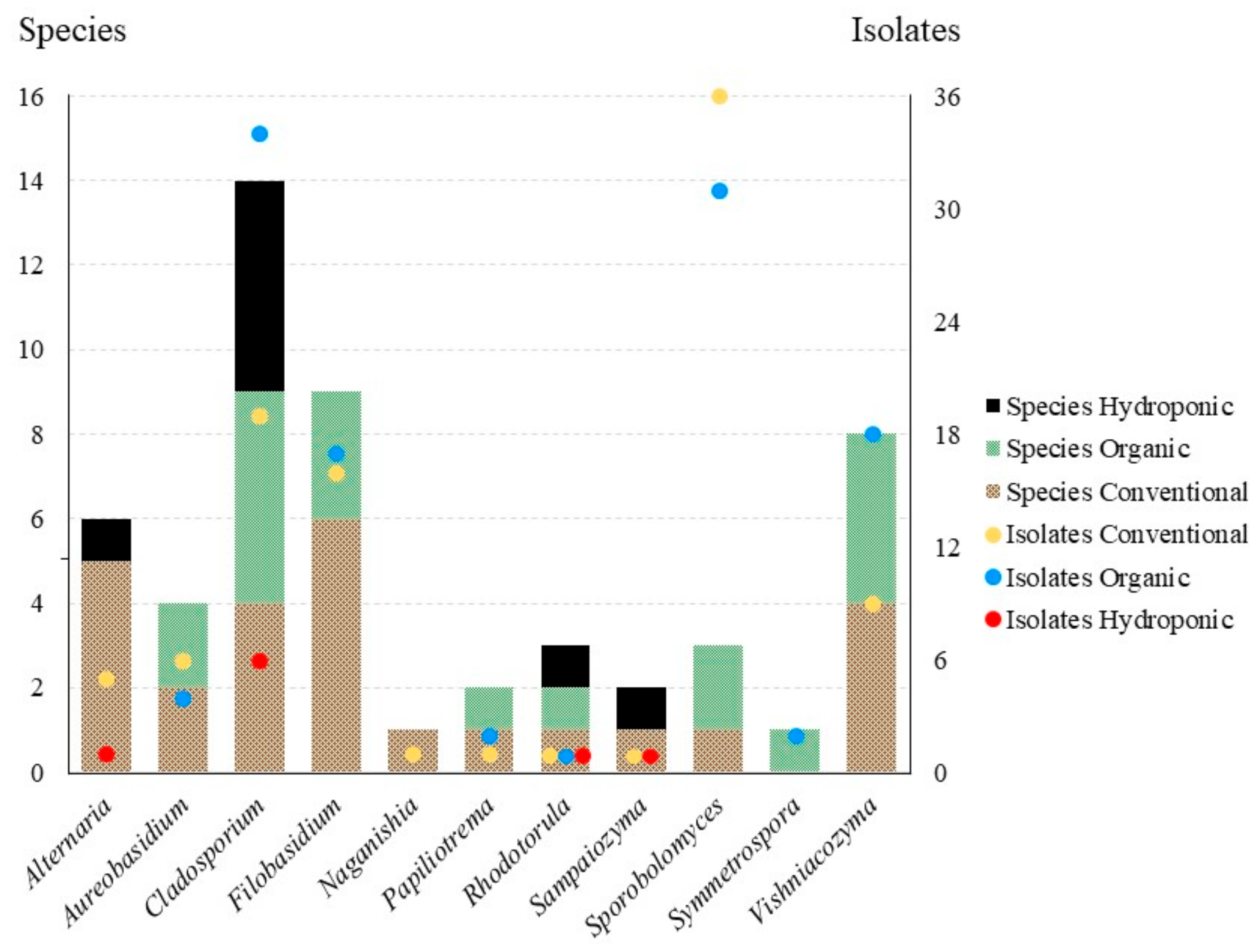
3.2. Statistical Analyses
3.3. Taxonomy
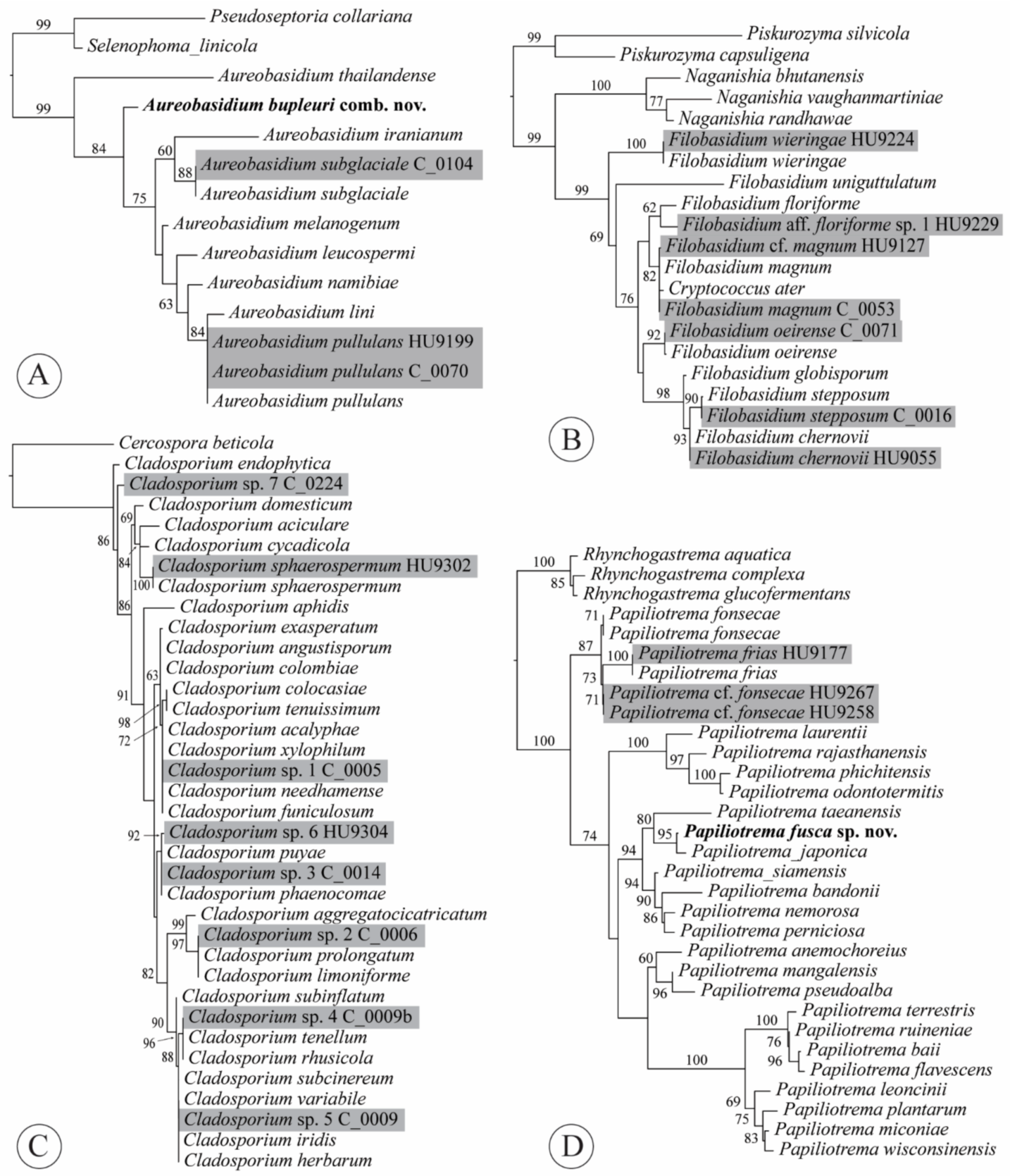
3.3.1. Aureobasidium bupleuri (Bills) Haelew. & Aime, comb. nov.
3.3.2. Curvibasidium nothofagi (C. Ramírez & A.E. González) Haelew. & Aime, comb. nov.
3.3.3. Filobasidium magnum (Lodder & Kreger-van Rij) Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, in Liu et al., Stud. Mycol. 81: 118 (2015)
3.3.4. Papiliotrema fusca J.P. Samp., J. Inácio, Fonseca and Fell ex Haelew., sp. nov.
4. Discussion
4.1. Alternaria
4.2. Aureobasidium
4.3. Cladosporium
4.4. Filobasidium
4.5. Naganishia
4.6. Papiliotrema
4.7. Rhodotorula
4.8. Sampaiozyma
4.9. Sporobolomyces
4.10. Symmetrospora
4.11. Vishniacozyma
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| Contig or Isolate | Species | Number of Isolates | Best Match | Strain | Sequence | Similarity [%] | Reference | |||
|---|---|---|---|---|---|---|---|---|---|---|
| All | C | O | H | |||||||
| C_0008 | Sporobolomyces sp. nov. 2 | 66 | 36 | 30 | Sporobolomyces roseus | CBS:7683T | KY105472 | 98.84 | [20] | |
| C_0005 | Cladosporium sp. 1 | 33 | 12 | 20 | 1 | Cladosporium xylophilum (a) | CBS:125997T | MH863875 | 100.00 | [20] |
| C_0016 | Filobasidium stepposum | 23 | 10 | 13 | Filobasidium stepposum | CBS:10265T | NR_111207 | 99.83 | [32] | |
| C_0010, HU9126, HU9242 | Vishniacozyma victoriae | 16 | 3 | 13 | Vishniacozyma victoriae | ATCC:MYA-305T | MH809977 | 99.81–100.00 | R.E. Rush et al. unpubl. | |
| C_0063 | Cystofilobasidium aff. macerans sp. nov. 1 | 11 | 7 | 4 | Cystofilobasidium macerans | CBS:10757T | KY103183 | 98.24 | [20] | |
| C_0009 | Cladosporium sp. 5 | 10 | 2 | 8 | Cladosporium iridis (b) | CBS:138.40T | EU167591 | 100.00 | [154] | |
| C_0070, HU9199 | Aureobasidium pullulans | 7 | 4 | 3 | Aureobasidium pullulans (c) | CBS 584.75T | KT693733 | 100.00 | [53] | |
| C_0038, HU9045 | Vishniacozyma cf. foliicola | 6 | 3 | 3 | Vishniacozyma foliicola | CBS:9920T | KY105821 | 98.67–98.86 | [20] | |
| C_0014 | Cladosporium sp. 3 | 5 | 3 | 1 | 1 | Cladosporium puyae (d) | CBS:274.80AT | NR_152298 | 100.00 | [69] |
| C_0053, HU9127 | Filobasidium magnum | 5 | 2 | 3 | Filobasidium magnum | CBS:140T | NR_130655 | 99.67–100.00 | [34] | |
| C_0006 | Cladosporium sp. 2 | 4 | 2 | 2 | Cladosporium limoniforme (d) | CBS:140484T | KT600397 | 100.00 | [69] | |
| C_0104 | Aureobasidium subglaciale | 3 | 2 | 1 | Aureobasidium subglaciale | CBS:123387T | KT693735 | 100.00 | [53] | |
| C_0009b | Cladosporium sp. 4 | 3 | 3 | Cladosporium rhusicola (d) | CBS:138.40T | NR_152299 | 100.00 | [69] | ||
| C_0003 | Gibellulopsis nigrescens | 3 | 2 | 1 | Gibellulopsis nigrescens | CBS:120949T | NR_149327 | 100.00 | [155] | |
| C_0001, HU9254 | Vishniacozyma carnescens | 3 | 2 | 1 | Vishniacozyma carnescens | CBS:973T | KY105817 | 99.84–100.00 | [20] | |
| C_0224 | Cladosporium sp. 7 | 2 | 2 | Cladosporium endophyticum | MFLUCC:17-0599T | MG646956 | 99.11 | [156] | ||
| C_0071 | Filobasidium oeirense | 2 | 1 | 1 | Filobasidium oeirense | CBS:8681T | KY103438 | 99.37 | [20] | |
| C_0215 | Metschnikowia picachoensis | 2 | 2 | Metschnikowia picachoensis | NRRL:Y-27607T | AY494780 | 100.00 | [157] | ||
| C_0205 | Papiliotrema aff. fonsecae sp. 1 | 2 | 2 | Papiliotrema fonsecae | EX:F-4087T | NR_119972 | 99.76 | [32] | ||
| C_0057 | Penicillium flavigenum | 2 | 1 | 1 | Penicillium flavigenum | CBS:419.89T | MH862182 | 100.00 | [158] | |
| C_0045 | Stemphylium sp. 1 | 2 | 2 | Stemphylium waikerieanum | VPRI:21969T | MK336832 | 100.00 | [70] | ||
| C_0048 | Symmetrospora sp. nov. | 2 | 2 | Symmetrospora coprosmae | CBS:7899T | NR_073317 | 99.48 | [101] | ||
| HU9265 | Acanthophysium sp. 1 | 1 | 1 | Acanthophysium bisporum | CBS:240.86T | MH861954 | 90.02 | [158] | ||
| HU9315 | Alternaria sp. 1 | 1 | 1 | Alternaria angustiovoidea (e) | CBS:195.86T | MH861939 | 100.00 | [158] | ||
| HU9181 | Alternaria sp. 2 | 1 | 1 | Alternaria ventricosa (e) | CBS:121546T | MH863116 | 100.00 | [158] | ||
| HU9087 | Alternaria sp. 3 | 1 | 1 | Alternaria dactylidicola | MFLUCC:15-0466T | NR_151852 | 99.01 | [159] | ||
| HU9178 | Alternaria sp. 4 | 1 | 1 | Alternaria conjuncta | CBS 196.86T | MH861940 | 99.67 | [158] | ||
| HU9194 | Alternaria sp. 5 | 1 | 1 | Alternaria eureka | CBS 193.86T | MH861937 | 98.78 | [158] | ||
| HU9226 | Alternaria sp. 6 | 1 | 1 | Alternaria arbusti (e) | CBS 596.93T | MH862447 | 100.00 | [158] | ||
| HU9303 | Beauveria cf. bassiana | 1 | 1 | Beauveria bassiana | ARSEF:1564T | NR_111594 | 98.91 | [32] | ||
| HU9220 | Botryotinia pelargonii | 1 | 1 | Botryotinia pelargonii | CBS:497.50T | AJ716290 | 100.00 | [160] | ||
| HU9201 | Bullera aff. unica sp. 1 | 1 | 1 | Bullera unica | CBS:8290T | NR_073256 | 99.62 | [32] | ||
| HU9228 | Bullera unica | 1 | 1 | Bullera unica | CBS:8290T | NR_073256 | 100.00 | [32] | ||
| HU9149 | Bulleromyces albus | 1 | 1 | Bulleromyces albus | CBS:500T | KY101811 | 100.00 | [20] | ||
| HU9304 | Cladosporium sp. 6 | 1 | 1 | Cladosporium puyae | CBS:274.80AT | NR_152298 | 99.82 | [69] | ||
| HU9302 | Cladosporium sphaerospermum | 1 | 1 | Cladosporium sphaerospermum | ATCC:11289T | AY361958 | 100.00 | [161] | ||
| HU9177 | Papiliotrema frias | 1 | 1 | Cryptococcus frias | CRUB 1250T | GU997162 | 100.00 | [89] | ||
| HU9106 | Cystofilobasidium infirmominiatum | 1 | 1 | Cystofilobasidium infirmominiatum | CBS:323T | NR_073232 | 100.00 | [32] | ||
| HU9025 | Epicoccum nigrum complex sp. 1 | 1 | 1 | Epicoccum layuense | CGMCC: 3.18362T | NR_158265 | 100.00 | [162] | ||
| HU9229 | Filobasidium aff. floriforme sp. 1 | 1 | 1 | Filobasidium floriforme | CBS:6241T | NR_119429 | 99.06 | [32] | ||
| HU9055 | Filobasidium chernovii | 1 | 1 | Filobasidium chernovii | CBS:8679T | KY103413 | 100.00 | [20] | ||
| HU9224 | Filobasidium wieringae | 1 | 1 | Filobasidium wieringae | CBS:1937T | NR_077105 | 100.00 | [20] | ||
| HU9215 | Holtermanniella sp. nov. 1 | 1 | 1 | Holtermanniella takashimae | CBS:11174T | NR_137721 | 99.42 | [163] | ||
| HU9066 | Leucosporidium yakuticum | 1 | 1 | Leucosporidium yakuticum | CBS:8621T | NR_155332 | 100.00 | [20] | ||
| HU9283 | Metschnikowia cf. rubicola | 1 | 1 | Metschnikowia rubicola | NRRL:Y-6064T | MG050888 | 99.64 | [164] | ||
| HU9309 | Moesziomyces aphidis | 1 | 1 | Moesziomyces aphidis | CBS:517.83T | NR_145336 | 100.00 | [165] | ||
| HU9011 | Mucor circinelloides f. circinelloides | 1 | 1 | Mucor circinelloides f. circinelloides | CBS:195.68T | HQ154604 | 99.12 | [166] | ||
| HU9043 | Naganishia sp. | 1 | 1 | Naganishia liquefaciens (f) | CBS:968T | NR_073220 | 100.00 | [32] | ||
| HU9061 | Ophiostoma sp. 1 | 1 | 1 | Ophiostoma rufum (g) | CBS:144871T | MH837040 | 100.00 | [167] | ||
| HU9060 | Penicillium sp. 1 | 1 | 1 | Penicillium hoeksii | CBS:137776T | NR_137913 | 99.63 | [168] | ||
| HU9099 | Rhodotorula aff. glutinis sp. nov. 1 | 1 | 1 | Rhodotorula babjevae | CBS:7808T | NR_077096 | 99.66 | [101] | ||
| HU9231 | Rhodotorula aff. glutinis sp. nov. 2 | 1 | 1 | Rhodotorula babjevae | CBS:7808T | NR_077096 | 99.49 | [101] | ||
| HU9305 | Rhodotorula sp. nov. 3 | 1 | 1 | Rhodotorula diobovata | CBS:6085T | KY104768 | 99.51 | [20] | ||
| HU9046 | Sampaiozyma sp. nov. 1 | 1 | 1 | Sampaiozyma ingeniosa | CBS:4240T | NR_111080 | 97.49 | [32] | ||
| HU9301 | Sampaiozyma sp. nov. 2 | 1 | 1 | Sampaiozyma vanillica | CBS:7404T | KY105310 | 93.94 | [20] | ||
| HU9138 | Sporobolomyces sp. nov. 1 | 1 | 1 | Sporobolomyces patagonicus | CBS:9657T | KY105521 | 98.82 | [20] | ||
| HU9240 | Tilletiopsis aff. pallescens sp. nov. 1 | 1 | 1 | Tilletiopsis pallescens | CBS 606.83T | NR_111216 | 98.49 | [32] | ||
| HU9248 | Vishniacozyma aff. heimaeyensis sp. nov. | 1 | 1 | Vishniacozyma heimaeyensis | CBS:8933T | NR_077070 | 98.63 | [32] | ||
| HU9122 | Vishniacozyma tephrensis | 1 | 1 | Vishniacozyma tephrensis | CBS:8935T | NR_144812 | 100.00 | [169] | ||
References
- Pitt, J.I.; Hocking, A.D. Fungi and Food Spoilage, 3rd ed.; Springer: New York, NY, USA, 2009. [Google Scholar] [CrossRef]
- Kibe, E.N. Occurrence of Mycotoxigenic Fungi in Maize from Food Commodity Markets in Kenya. Master’s Thesis, Ghent University, Ghent, Belgium, 2015. [Google Scholar]
- Marasas, W.F.O. Fumonisins: History, world-wide occurrence and impact. In Fumonisins in Food; Jackson, L.S., DeVries, J.W., Bullerman, L.B., Eds.; Springer: New York, NY, USA, 1996; pp. 1–17. [Google Scholar] [CrossRef]
- Shephard, G.S. Risk assessment of aflatoxins in food in Africa. Food Addit. Contam. 2008, 25, 1246–1256. [Google Scholar] [CrossRef]
- USDA (United States Department of Agriculture). 2017 Census of Agriculture. Summary and State Data. Vol. 1. Geogr. Area Ser. 2019, 51, 1–711. [Google Scholar]
- Sivapalasingam, S.; Friedman, C.R.; Cohen, L.; Tauxe, R.V. Fresh produce: A growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. J. Food Prot. 2004, 67, 2342–2353. [Google Scholar] [CrossRef] [PubMed]
- Brandl, M.T. Fitness of human enteric pathogens on plants and implications for food safety. Annu. Rev. Phytopathol. 2006, 44, 367–392. [Google Scholar] [CrossRef]
- Rangel, J.M.; Sparling, P.H.; Crowe, C.; Griffin, P.M.; Swerdlow, D.L. Epidemiology of Escherichia coli O157: H7 outbreaks, united states, 1982–2002. Emerg. Infect. Dis. 2005, 11, 603. [Google Scholar] [CrossRef] [PubMed]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Slayton, R.B.; Turabelidze, G.; Bennett, S.D.; Schwensohn, C.A.; Yaffee, A.Q.; Khan, F.; Butler, C.; Trees, E.; Ayers, T.L.; Davis, M.L.; et al. Outbreak of shiga toxin-producing Escherichia coli (STEC) O157:H7 associated with romaine lettuce consumption, 2011. PLoS ONE 2013, 8, e55300. [Google Scholar] [CrossRef]
- Jeamsripong, S.; Chase, J.A.; Jay-Russell, M.T.; Buchanan, R.L.; Atwill, E.R. Experimental in-field transfer and survival of Escherichia coli from animal feces to romaine lettuce in Salinas Valley, California. Microorganisms 2019, 7, 408. [Google Scholar] [CrossRef]
- Ma, J.; Mark Ibekwe, A.; Crowley, D.E.; Yang, C.-H. Persistence of Escherichia coli O157 and non-O157 strains in agricultural soils. Sci. Total Environ. 2014, 490, 822–829. [Google Scholar] [CrossRef]
- Sai, C.B.; Srinivasan, N.; Zachariah, J.K.; Dananjeyan, B. Experimentation on artificial inoculation studies for persistence of shiga-like toxin-producing Escherichia coli (E. coli O157) in agricultural soils and vegetables using real-time PCR. J. Food Biochem. 2019, 43, e13035. [Google Scholar] [CrossRef]
- CDC (Centers for Disease Control and Prevention). Multistate Outbreak of E. coli O157:H7 Infections Linked to Romaine Lettuce (Final Update). Available online: https://www.cdc.gov/ecoli/2018/o157h7-04-18/index.html (accessed on 7 March 2021).
- Albu, S.; Toome, M.; Aime, M.C. Violaceomyces palustris gen. et sp. nov. and a new monotypic lineage, Violaceomycetales ord. nov. in Ustilaginomycetes. Mycologia 2015, 107, 1193–1204. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Hunter, P.J.; Pink, D.A.; Bending, G.D. Cultivar-level genotype differences influence diversity and composition of lettuce (Lactuca sp.) phyllosphere fungal communities. Fungal Ecol. 2015, 17, 183–186. [Google Scholar] [CrossRef]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; Szöke, S.; Cardinali, G.; Eberhardt, U.; Stielow, B.; de Vries, M.; Verkleij, G.J.M.; Crous, P.W.; Boekhout, T.; et al. DNA barcoding analysis of more than 9000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud. Mycol. 2016, 85, 91–105. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; Institute of Electrical and Electronics Engineers: Piscataway, NJ, USA, 2010; pp. 1–8. [Google Scholar] [CrossRef]
- Baral, H.-O.; Weber, E.; Marson, G.; Quijada, L. A new connection between wood saprobism and beetle endosymbiosis: The rarely reported saprobic discomycete Tromeropsis is congeneric with the symbiotic yeast Symbiotaphrina (Symbiotaphrinales, Xylonomycetes) and two asexual morphs misplaced in Hyphozyma. Mycol. Prog. 2018, 17, 215–254. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Chernomor, O.; von Haeseler, A.; Minh, B.Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2017, 35, 518–522. [Google Scholar] [CrossRef]
- Bills, G.F.; González Menéndez, V.; Platas, G. Kabatiella bupleuri sp. nov. (Dothideales), a pleomorphic epiphyte and endophyte of the Mediterranean plant Bupleurum gibraltarium (Apiaceae). Mycologia 2012, 104, 962–973. [Google Scholar] [CrossRef] [PubMed]
- Humphries, Z.; Seifert, K.A.; Hirooka, Y.; Visagie, C.M. A new family and genus in Dothideales for Aureobasidium-like species isolated from house dust. IMA Fungus 2017, 8, 299–315. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T. The Yeasts: A Taxonomic Study, 5th ed.; Elsevier: London, UK, 2011; Volume 3. [Google Scholar]
- Wang, Q.-M.; Yurkov, A.M.; Göker, M.; Lumbsch, H.T.; Leavitt, S.D.; Groenewald, M.; Theelen, B.; Liu, X.-Z.; Boekhout, T.; Bai, F.-Y. Phylogenetic classification of yeasts and related taxa within Pucciniomycotina. Stud. Mycol. 2015, 81, 149–189. [Google Scholar] [CrossRef]
- Schoch, C.L.; Robbertse, B.; Robert, V.; Vu, D.; Cardinali, G.; Irinyi, L.; Meyer, W.; Nilsson, R.H.; Hughes, K.; Miller, A.N.; et al. Finding needles in haystacks: Linking scientific names, reference specimens and molecular data for Fungi. Database 2014, 2014, bau061. [Google Scholar] [CrossRef]
- Liu, X.-Z.; Wang, Q.-M.; Göker, M.; Groenewald, M.; Kachalkin, A.V.; Lumbsch, H.T.; Millanes, A.M.; Wedin, M.; Yurkov, A.M.; Boekhout, T.; et al. Towards an integrated phylogenetic classification of the Tremellomycetes. Stud. Mycol. 2015, 81, 85–147. [Google Scholar] [CrossRef] [PubMed]
- Fell, J.W.; Boekhout, T.; Fonseca, A.; Scorzetti, G.; Statzell-Tallman, A. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/ D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 2000, 50, 1351–1371. [Google Scholar] [CrossRef]
- Index Fungorum. Search Index Fungorum. Available online: http://www.indexfungorum.org/Names/Names.asp (accessed on 5 March 2021).
- Thompson, I.P.; Bailey, M.J.; Fenlon, J.S.; Fermor, T.R.; Lilley, A.K.; Lynch, J.M.; McCormack, P.J.; McQuilken, M.P.; Purdy, K.J.; Rainey, P.B.; et al. Quantitative and qualitative seasonal changes in the microbial community from the phyllosphere of sugar-beet (Beta vulgaris). Plant Soil 1993, 150, 177–191. [Google Scholar] [CrossRef]
- McCormack, P.J.; Wildman, H.G.; Jeffries, P. Production of antibacterial compounds by phylloplane-inhabiting yeasts and yeastlike fungi. Appl. Environ. Microbiol. 1994, 60, 927–931. [Google Scholar] [CrossRef]
- Andrews, J.H.; Harris, R.F. The ecology and biogeography of microorganisms on plant surfaces. Ann. Rev. Phytopathol. 2000, 38, 145–180. [Google Scholar] [CrossRef] [PubMed]
- Whipps, J.M.; Hand, P.; Pink, D.A.C.; Bending, G.D. Phyllosphere microbiology with special reference to diversity and plant genotype. J. Appl. Microbiol. 2008, 104, 1744–1755. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.T.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Zhao, R.L.; Aptroot, A.; Leontyev, D.V.; Saxena, R.K.; et al. Outline of Fungi and fungus-like taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Barth, M.; Hankinson, T.R.; Zhuang, H.; Breidt, F. Microbiological spoilage of fruits and vegetables. In Compendium of the Microbiological Spoilage of Foods and Beverages; Sperber, W., Doyle, M., Eds.; Springer: New York, NY, USA, 2009; pp. 135–183. [Google Scholar] [CrossRef]
- Pryor, B.; Michailides, T. Morphological, pathogenic, and molecular characterization of Alternaria isolates associated with alternaria late blight of pistachio. Phytopathology 2002, 92, 406–416. [Google Scholar] [CrossRef]
- Badotti, F.; de Oliveira, F.S.; Garcia, C.F.; Vaz, A.B.M.; Fonseca, P.L.C.; Nahum, L.A.; Oliveira, G.; Góes-Neto, A. Effectiveness of ITS and sub-regions as DNA barcode markers for the identification of Basidiomycota (Fungi). BMC Microbiol. 2017, 17, 42. [Google Scholar] [CrossRef]
- Lawrence, D.P.; Gannibal, P.B.; Peever, T.L.; Pryor, B.M. The sections of Alternaria: Formalizing species-group concepts. Mycologia 2013, 105, 530–546. [Google Scholar] [CrossRef]
- Pryor, B.M.; Bigelow, D.M. Molecular characterization of Embellisia and Nimbya species and their relationship to Alternaria, Ulocladium and Stemphylium. Mycologia 2003, 95, 1141–1154. [Google Scholar] [CrossRef]
- Joly, P. Le genre Alternaria: Recherches Physiologiques, Biologiques et Systématiques; Lechevalier: Paris, France, 1964. [Google Scholar]
- Strandberg, J.O. Alternaria species that attack vegetable crops: Biology and options for diseases management. In Alternaria Biology, Plant Diseases and Metabolites; Chelkowski, J., Visconti, A., Eds.; Elsevier Science Publishers: Amsterdam, The Netherlands, 1992; pp. 175–208. [Google Scholar]
- Boedo, C.; Benichou, S.; Berruyer, R.; Bersihand, S.; Dongo, A.; Simoneau, P.; Lecomte, M.; Briard, M.; Le Clerc, V.; Poupard, P. Evaluating aggressiveness and host range of Alternaria dauci in a controlled environment. Plant Pathol. 2012, 61, 63–75. [Google Scholar] [CrossRef]
- Anita, D.D.; Sridhar, K.R.; Bhat, R. Diversity of fungi associated with mangrove legume Sesbania bispinosa (Jacq.) W. Wight (Fabaceae). Livest. Res. Rural Dev. 2009, 21, 67. [Google Scholar]
- Varvas, T.; Kullman, B. First records of two ascomycetes on Phleum pratense in Estonia. Folia Cryptogam. Estonica 2012, 49, 73–76. [Google Scholar]
- Tymon, L.S.; Peever, T.L.; Johnson, D.A. Identification and enumeration of small-spored Alternaria species associated with potato in the US Northwest. Plant Dis. 2016, 100, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Poursafar, A.; Ghosta, Y.; Javan-Nikkhah, M. Identification of Alternaria species from the section Infectoriae associated with wheat and barley black (sooty) head mold in Iran. Taxon. Biosyst. 2017, 9, 13–30. [Google Scholar]
- van Nieuwenhuijzen, E.J.; Houbraken, J.A.; Meijer, M.; Adan, O.C.; Samson, R.A. Aureobasidium melanogenum: A native of dark biofinishes on oil treated wood. Ant. Van Leeuwenhoek 2016, 109, 661–683. [Google Scholar] [CrossRef] [PubMed]
- Species Fungorum. Search Species Fungorum. Available online: http://www.speciesfungorum.org/Names/Names.asp (accessed on 5 March 2021).
- Webb, T.A.; Mundt, J.O. Molds on vegetables at the time of harvest. Appl. Environ. Microbiol. 1978, 35, 655–658. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.R.; Singh, D.; Singh, R. Biological control of postharvest diseases of fruits and vegetables by microbial antagonists: A review. Biol. Control 2009, 50, 205–221. [Google Scholar] [CrossRef]
- Wilson, C.L.; Chalutz, E. Postharvest biocontrol of Penicillium rots of citrus with antagonistic yeasts and bacteria. Sci. Hortic. 1989, 40, 105–112. [Google Scholar] [CrossRef]
- Bencheqroun, S.K.; Baji, M.; Massart, S.; Labhilili, M.; El Jaafari, S.; Jijaki, M.H. In vitro and in situ study of postharvest apple blue mold biocontrol by Aureobasidium pullulans: Evidence for the involvement of competition for nutrients. Postharvest Biol. Technol. 2007, 46, 128–135. [Google Scholar] [CrossRef]
- Seibold, A.; Fried, A.; Kunz, S.; Moltmann, E.; Lange, E.; Jelkmann, W. Yeasts as antagonists against fireblight. EPPO Bull. 2004, 34, 389–390. [Google Scholar] [CrossRef]
- Gostinčar, C.; Ohm, R.A.; Kogej, T.; Sonjak, S.; Turk, M.; Zajc, J.; Zalar, P.; Grube, M.; Sun, H.; Han, J.; et al. Genome sequencing of four Aureobasidium pullulans varieties: Biotechnological potential, stress tolerance, and description of new species. BMC Genom. 2014, 15, 549. [Google Scholar] [CrossRef]
- Fradkin, A.; Tobin, R.S.; Tarlo, S.M.; Tucic-Porretta, M.; Malloch, D. Species identification of airborne molds and Its significance for the detection of indoor pollution. Int. J. Air Pollut. Control Hazard. Waste Manag. 1987, 37, 51–53. [Google Scholar] [CrossRef]
- Flannigan, B. Microorganisms in indoor air. In Microorganisms in Home and Indoor Work Environments: Diversity, Health Impacts, Investigation and Control, 2nd ed.; Flannigan, B., Samson, R., Miller, D., Eds.; CRC Press: Boca Raton, FL, USA, 2001; pp. 17–31. [Google Scholar] [CrossRef]
- Horner, W.E.; Worthan, A.G.; Morey, P.R. Air- and dustborne mycoflora in houses free of water damage and fungal growth. Appl. Environ. Microbiol. 2004, 70, 6394–6400. [Google Scholar] [CrossRef]
- Visagie, C.M.; Hirooka, Y.; Tanney, J.B.; Whitfield, E.; Mwange, K.; Meijer, M.; Amend, A.S.; Seifert, K.A.; Samson, R.A. Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Stud. Mycol. 2014, 78, 63–139. [Google Scholar] [CrossRef] [PubMed]
- Bensch, K.; Groenewald, J.Z.; Meijer, M.; Dijksterhuis, J.; Jurjević, Ž.; Andersen, B.; Houbraken, J.; Crous, P.W.; Samson, R.A. Cladosporium species in indoor environments. Stud. Mycol. 2018, 89, 177–301. [Google Scholar] [CrossRef] [PubMed]
- Sandoval-Denis, M.; Gené, J.; Sutton, D.A.; Wiederhold, N.P.; Cano-Lira, J.F.; Guarro, J. New species of Cladosporium associated with human and animal infections. Persoonia 2016, 36, 281–298. [Google Scholar] [CrossRef]
- Sandoval-Denis, M.; Sutton, D.A.; Martin-Vicente, A.; Cano-Lira, J.F.; Wiederhold, N.; Guarro, J.; Gené, J. Cladosporium species recovered from clinical samples in the United States. J. Clin. Microbiol. 2015, 53, 2990–3000. [Google Scholar] [CrossRef]
- Bensch, K.; Braun, U.; Groenewald, J.Z.; Crous, P.W. The genus Cladosporium. Stud. Mycol. 2012, 72, 1–401. [Google Scholar] [CrossRef] [PubMed]
- Bensch, K.; Groenewald, J.Z.; Braun, U.; Dijksterhuis, J.; de Jesús Yáñez-Morales, M.; Crous, P.W. Common but different: The expanding realm of Cladosporium. Stud. Mycol. 2015, 82, 23–74. [Google Scholar] [CrossRef]
- Marin-Felix, Y.; Hernández-Restrepo, M.; Iturrieta-González, I.; García, D.; Gené, J.; Groenewald, J.Z.; Cai, L.; Chen, Q.; Quaedvlieg, W.; Schumacher, R.K.; et al. Genera of phytopathogenic fungi: GOPHY 3. Stud. Mycol. 2019, 94, 1–124. [Google Scholar] [CrossRef]
- Bensch, K.; Groenewald, J.Z.; Dijksterhuis, J.; Starink-Willemse, M.; Andersen, B.; Summerell, B.A.; Shin, H.D.; Dugan, F.M.; Schroers, H.J.; Braun, U.; et al. Species and ecological diversity within the Cladosporium cladosporioides complex (Davidiellaceae, Capnodiales). Stud. Mycol. 2010, 67, 1–94. [Google Scholar] [CrossRef]
- Lavoie, K.; Touchette, M.; St-Gelais, D.; Labrie, S. Characterization of the fungal microflora in raw milk and specialty cheeses of province of Quebec. Dairy Sci. Technol. 2012, 92, 455–468. [Google Scholar] [CrossRef]
- Merín, M.G.; Mendoza, L.M.; Morata de Ambrosini, V.I. Pectinolytic yeasts from viticultural and enological environments: Novel finding of Filobasidium capsuligenum producing pectinases. J. Basic Microbiol. 2014, 54, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Šuranská, H.; Raspor, P.; Uroić, K.; Golić, N.; Kos, B.; Mihajlović, S.; Begović, J.; Šušković, J.; Topisirović, L.; Čadež, N. Characterisation of the yeast and mould biota in traditional white pickled cheeses by culture-dependent and independent molecular techniques. Folia Microbiol. 2016, 61, 455–463. [Google Scholar] [CrossRef]
- Lin, W.R.; PH, W.; Hsieh, S.Y.; Tsai, C.H.; Hsiao, S.C. Yeasts associated with Euploea butterflies. Mycosphere 2018, 9, 149–154. [Google Scholar] [CrossRef]
- Li, A.-H.; Yuan, F.-X.; Groenewald, M.; Bensch, K.; Yurkov, A.M.; Li, K.; Han, P.-J.; Guo, L.-D.; Aime, M.C.; Sampaio, J.P.; et al. Diversity and phylogeny of basidiomycetous yeasts from plant leaves and soil: Proposal of two new orders, three new families, eight new genera and one hundred and seven new species. Stud. Mycol. 2020, 96, 17–140. [Google Scholar] [CrossRef]
- Scupham, A.J.; Presley, L.L.; Wei, B.; Bent, E.; Griffith, N.; McPherson, M.; Zhu, F.; Oluwadara, O.; Rao, N.; Braun, J.; et al. Abundant and diverse fungal microbiota in the murine intestine. Appl. Environ. Microbiol. 2006, 72, 793–801. [Google Scholar] [CrossRef][Green Version]
- Gouba, N.; Raoult, D.; Drancourt, M. Eukaryote culturomics of the gut reveals new species. PLoS ONE 2014, 9, e106994. [Google Scholar] [CrossRef]
- Mattsson, R.; Haemig, P.D.; Olsen, B. Feral pigeons as carriers of Cryptococcus laurentii, Cryptococcus uniguttulatus and Debaryomyces hansenii. Med. Mycol. 1999, 37, 367–369. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Liao, W.; Hagen, F.; Theelen, B.; Shi, W.; Meis, J.F.; Boekhout, T. Meningitis caused by Filobasidium uniguttulatum: Case report and overview of the literature. Mycoses 2012, 55, 105–109. [Google Scholar] [CrossRef]
- Goto, S. On a new yeast genus Naganishia. J. Ferment. Tech. 1963, 41, 459–462. [Google Scholar]
- Fonseca, Á.; Scorzetti, G.; Fell, J.W. Diversity in the yeast Cryptococcus albidus and related species as revealed by ribosomal DNA sequence analysis. Can. J. Microbiol. 2000, 46, 7–27. [Google Scholar] [CrossRef]
- Fotedar, R.; Kolecka, A.; Boekhout, T.; Fell, J.W.; Anand, A.; Al Malaki, A.; Zeyara, A.; Al Marri, M. Naganishia qatarensis sp. nov., a novel basidiomycetous yeast species from a hypersaline marine environment in Qatar. Int. J. Syst. Evol. Microbiol. 2018, 68, 2924–2929. [Google Scholar] [CrossRef]
- Crous, P.W.; Carnegie, A.J.; Wingfield, M.J.; Sharma, R.; Mughini, G.; Noordeloos, M.E.; Santini, A.; Shouche, Y.S.; Bezerra, J.D.P.; Dima, B.; et al. Fungal Planet description sheets: 868–950. Persoonia 2019, 42, 291–473. [Google Scholar] [CrossRef] [PubMed]
- Kachalkin, A.V.; Turchetti, B.; Inácio, J.; Carvalho, C.; Mašínová, T.; Pontes, A.; Röhl, O.; Glushakova, A.M.; Akulov, A.; Baldrian, P.; et al. Rare and undersampled dimorphic basidiomycetes. Mycol. Prog. 2019, 18, 945–971. [Google Scholar] [CrossRef]
- Fonseca, Á.; Inácio, J. Phylloplane yeasts. In Biodiversity and Ecophysiology of Yeasts; Péter, G., Rosa, C., Eds.; Springer: Berlin, Germany, 2006; pp. 263–301. [Google Scholar] [CrossRef]
- Sampaio, J.P.; Weiß, M.; Gadanho, M.; Bauer, R. New taxa in the Tremellales: Bulleribasidium oberjochense gen. et sp. nov., Papiliotrema bandonii gen. et sp. nov. and Fibulobasidium murrhardtense sp. nov. Mycologia 2002, 94, 873–887. [Google Scholar] [CrossRef]
- Crestani, J.; Landell, M.F.; Faganello, J.; Vainstein, M.H.; Vishniac, H.S.; Valente, P. Cryptococcus terrestris sp. nov., a tremellaceous, anamorphic yeast phylogenetically related to Cryptococcus flavescens. Int. J. Syst. Evol. Microbiol. 2009, 59, 631–636. [Google Scholar] [CrossRef][Green Version]
- De Garcia, V.; Zalar, P.; Brizzio, S.; Gunde-Cimerman, N.; Van Broock, M. Cryptococcus species (Tremellales) from glacial biomes in the southern (Patagonia) and northern (Svalbard) hemispheres. FEMS Microbiol. Ecol. 2012, 82, 523–539. [Google Scholar] [CrossRef]
- Ferreira-Paim, K.; Ferreira, T.B.; Andrade-Silva, L.; Mora, D.J.; Springer, D.J.; Heitman, J.; Fonseca, F.M.; Matos, D.; Melhem, M.S.; Silva-Vergara, M.L. Phylogenetic analysis of phenotypically characterized Cryptococcus laurentii isolates reveals high frequency of cryptic species. PLoS ONE 2014, 9, e108633. [Google Scholar] [CrossRef]
- Surussawadee, J.; Khunnamwong, P.; Srisuk, N.; Limtong, S. Papiliotrema siamense f.a., sp. nov., a yeast species isolated from plant leaves. Int. J. Syst. Evol. Microbiol. 2014, 64, 3058–3062. [Google Scholar] [CrossRef] [PubMed]
- Handel, S.; Wang, T.; Yurkov, A.M.; König, H. Sugiyamaella mastotermitis sp. nov. and Papiliotrema odontotermitis f.a., sp. nov. from the gut of the termites Mastotermes darwiniensis and Odontotermes obesus. Int. J. Syst. Evol. Microbiol. 2016, 66, 4600–4608. [Google Scholar] [CrossRef]
- Pagani, D.M.; Brandao, L.R.; Santos, A.R.O.; Felix, C.R.; Ramos, J.P.; Broetto, L.; Scorzetti, G.; Fell, J.W.; Rosa, C.A.; Valente, P.; et al. Papiliotrema leoncinii sp. nov. and Papiliotrema miconiae sp. nov., two tremellaceous yeast species from Brazil. Int. J. Syst. Evol. Microbiol. 2016, 66, 1799–1806. [Google Scholar] [CrossRef][Green Version]
- Boekhout, T. A revision of ballistoconidia-forming yeasts and fungi. Stud. Mycol. 1991, 33, 1–194. [Google Scholar]
- Hamamoto, M.; Boekhout, T.; Nakase, T. Chapter 156. Sporobolomyces Kluyver & van Niel (1924). In The Yeasts, a Taxonomic Study, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; Volume 3, pp. 1929–1990. [Google Scholar] [CrossRef]
- Sampaio, J.P. Chapter 155. Rhodotorula Harrison (1928). In The Yeasts, a Taxonomic Study, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; Volume 3, pp. 1873–1927. [Google Scholar] [CrossRef]
- Aime, M.C.; Matheny, P.B.; Henk, D.A.; Frieders, E.M.; Nilsson, R.H.; Piepenbring, M.; McLaughlin, D.J.; Szabo, L.J.; Begerow, D.; Sampaio, J.P.; et al. An overview of the higher-level classification of Pucciniomycotina based on combined analyses of nuclear large and small subunit rDNA sequences. Mycologia 2006, 98, 895–905. [Google Scholar] [CrossRef]
- Bauer, R.; Begerow, D.; Sampaio, J.P.; Weiβ, M.; Oberwinkler, F. The simple-septate basidiomycetes: A synopsis. Mycol. Prog. 2006, 5, 41–66. [Google Scholar] [CrossRef]
- Wang, Q.-M.; Groenewald, M.; Takashima, M.; Theelen, B.; Han, P.-J.; Liu, X.-Z.; Boekhout, T.; Bai, F.-Y. Phylogeny of yeasts and related filamentous fungi within Pucciniomycotina determined from multigene sequence analyses. Stud. Mycol. 2015, 81, 27–53. [Google Scholar] [CrossRef] [PubMed]
- Gadanho, M.; Sampaio, J.P. Polyphasic taxonomy of the basidiomycetous yeast genus Rhodotorula: Rh. glutinis sensu stricto and Rh. dairenensis comb. nov. FEMS Yeast Res. 2002, 2, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Scorzetti, G.; Fell, J.W.; Fonseca, A.; Statzell-Tallman, A. Systematics of basidiomycetous yeasts: A comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res. 2002, 2, 495–517. [Google Scholar] [CrossRef]
- Wirth, F.; Goldani, L.Z. Epidemiology of Rhodotorula: An emerging pathogen. Interdiscip. Perspect. Infect. Dis. 2012, 2012, 465–717. [Google Scholar] [CrossRef]
- Gildemacher, P.; Heijne, B.; Silvestri, M.; Houbraken, J.; Hoekstra, E.; Theelen, B.; Boekhout, T. Interactions between yeasts, fungicides and apple fruit russeting. FEMS Yeast Res. 2006, 6, 1149–1156. [Google Scholar] [CrossRef]
- Magnuson, J.A.; King, A.D.; Török, T. Microflora of partially processed lettuce. Appl. Environ. Microbiol. 1990, 56, 3851–3854. [Google Scholar] [CrossRef] [PubMed]
- Török, T.; King, A.D. Comparative study on the identification of food-borne yeasts. Appl. Environ. Microbiol. 1991, 57, 1207–1212. [Google Scholar] [CrossRef] [PubMed]
- De Corato, U. Fungal population dynamics in ready-to-eat salads during a shelf-life in Italy. J. Agric. Sci. Technol. A 2012, 2, 569–576. [Google Scholar]
- Yafetto, L.; Ekloh, E.; Sarsah, B.; Amenumey, E.K.; Adator, E.H. Microbiological contamination of some fresh leafy vegetables sold in Cape Coast, Ghana. Ghana J. Sci. 2019, 60, 11–23. [Google Scholar] [CrossRef]
- Khodadad, C.L.; Hummerick, M.E.; Spencer, L.E.; Dixit, A.R.; Richards, J.T.; Romeyn, M.W.; Smith, T.M.; Wheeler, R.M.; Massa, G.D. Microbiological and nutritional analysis of lettuce crops grown on the International Space Station. Front. Plant Sci. 2020, 11, 199. [Google Scholar] [CrossRef] [PubMed]
- Urbina, H.; Aime, M.C. A closer look at Sporidiobolales: Ubiquitous microbial community members of plant and food biospheres. Mycologia 2018, 110, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Tuon, F.F.; Costa, S.F. Rhodotorula infection. A systematic review of 128 cases from literature. Rev. Iberoam. Micol. 2008, 25, 135–140. [Google Scholar] [CrossRef]
- Alvarez-Perez, S.; Mateos, A.; Dominguez, L.; Martinez-Nevado, E.; Blanco, J.L.; Garcia, M.E. Isolation of Rhodotorula mucilaginosa from skin lesions in a Southern sea lion (Otaria flavescens): A case report. Vet. Med. 2010, 55, 297–301. [Google Scholar] [CrossRef]
- Ioannou, P.; Vamvoukaki, R.; Samonis, G. Rhodotorula species infections in humans: A systematic review. Mycoses 2019, 62, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Dì Menna, M.E. Torulopsis ingeniosa n. sp., from grass leaves. J. Gen. Microbiol. 1958, 19, 581–583. [Google Scholar] [CrossRef]
- Sampaio, J.P. Utilization of low molecular weight lignin-related aromatic compounds for the selective isolation of yeasts: Rhodotorula vanillica, a new basidiomycetous yeast species. Syst. Appl. Microbiol. 1995, 17, 613–619. [Google Scholar] [CrossRef]
- Liu, X.-Z.; Groenewald, M.; Boekhout, T.; Bai, F.-Y. Heitmania gen. nov., a new yeast genus in Microbotryomycetes, and description of three novel species: Heitmania litseae sp. nov., Heitmania castanopsis sp. nov. and Heitmania elacocarpi sp. nov. Int. J. Syst. Evol. Microbiol. 2017, 67, 4534–4540. [Google Scholar] [CrossRef] [PubMed]
- Haelewaters, D.; Peterson, R.A.; Nevalainen, H.; Aime, M.C. Inopinatum lactosum gen. & comb. nov., the first pink-pigmented yeast in Leotiomycetes. Int. J. Syst. Evol. Microbiol. 2021. in review. [Google Scholar]
- Lorenzini, M.; Zapparoli, G.; Azzolini, M.; Carvalho, C.; Sampaio, J.P. Sporobolomyces agrorum sp. nov. and Sporobolomyces sucorum sp. nov., two novel basidiomycetous yeast species isolated from grape and apple must in Italy. Int. J. Syst. Evol. Microbiol. 2019, 69, 3385–3391. [Google Scholar] [CrossRef]
- Valadon, L.R.G. Carotenoids as additional taxonomic characters in fungi: A review. Trans. Brit. Mycol. Soc. 1976, 67, 1–15. [Google Scholar] [CrossRef]
- Ratledge, C. Microorganisms for lipids. Acta Biotechnol. 1991, 11, 429–438. [Google Scholar] [CrossRef]
- Davoli, P.; Mierau, V.; Weber, R.W. Carotenoids and fatty acids in red yeasts Sporobolomyces roseus and Rhodotorula glutinis. Appl. Biochem. Microbiol. 2004, 40, 460–465. [Google Scholar] [CrossRef]
- Manimala, M.R.A.; Murugesan, R. In vitro antioxidant and antimicrobial activity of carotenoid pigment extracted from Sporobolomyces sp. isolated from natural source. J. Appl. Nat. Sci. 2014, 6, 649–653. [Google Scholar] [CrossRef]
- Li, C.J.; Zhao, D.; Li, B.X.; Zhang, N.; Yan, J.Y.; Zou, H.T. Whole genome sequencing and comparative genomic analysis of oleaginous red yeast Sporobolomyces pararoseus NGR identifies candidate genes for biotechnological potential and ballistospores-shooting. BMC Genom. 2020, 21, 181. [Google Scholar] [CrossRef]
- Frengova, G.; Beshkova, M. Carotenoids from Rhodotorula and Phaffia: Yeasts of biotechnological importance. J. Ind. Microbiol. Biotechnol. 2009, 36, 163–180. [Google Scholar] [CrossRef] [PubMed]
- Mannazzu, I.; Landolfo, S.; da Silva, T.L.; Buzzini, P. Red yeasts and carotenoid production: Outlining a future for non-conventional yeasts of biotechnological interest. World J. Microbiol. Biotechnol. 2015, 31, 1665–1673. [Google Scholar] [CrossRef]
- Derx, H.G. Etude sur les Sporobolomycetes. Ann. Mycol. 1930, 28, 1–23. [Google Scholar]
- Tubaki, K. Studies on the Sporobolomycetaceae in Japan. III. On Sporobolomyces and Bullera. Nagaoa 1953, 3, 12–21. [Google Scholar]
- Last, F.T. Seasonal incidence of Sporobolomyces on cereal leaves. Trans. Brit. Mycol. Soc. 1955, 38, 221–239. [Google Scholar] [CrossRef]
- Hamamoto, M.; Nakase, T. Ballistosporous yeasts found on the surface of plant materials collected in New Zealand. 1. Six new species in the genus Sporobolomyces. Ant. Van Leeuwenhoek 1995, 67, 151–171. [Google Scholar] [CrossRef]
- Nakase, T. Expanding world of ballistosporous yeasts: Distribution in the phyllosphere, systematics and phylogeny. J. Gen. Appl. Microbiol. 2000, 46, 189–216. [Google Scholar] [CrossRef]
- Molnár, O.; Wuczkowski, M.; Prillinger, H. Yeast biodiversity in the guts of several pests on maize; comparison of three methods: Classical isolation, cloning and DGGE. Mycol. Prog. 2008, 7, 111–123. [Google Scholar] [CrossRef]
- Debode, J.; Van Hemelrijck, W.; Creemers, P.; Maes, M. Effect of fungicides on epiphytic yeasts associated with strawberry. Microbiol. Open 2013, 2, 482–491. [Google Scholar] [CrossRef]
- Janisiewicz, W.J.; Peterson, D.L.; Bors, R.; Sheu, J.J.; Jan, S.P.; Lee, H.T.; Yu, S.M.; Seastedt, T.R.; Coxwell, C.C.; Ojima, D.S.; et al. Control of storage decay of apples with Sporobolomyces roseus. Plant Dis. 1994, 78, 466–470. [Google Scholar] [CrossRef]
- Bashi, E.; Fokkema, N.J. Environmental factors limiting growth of Sporobolomyces roseus, an antagonist of Cochliobolus sativus, on wheat leaves. Trans. Brit. Mycol. Soc. 1977, 68, 17–25. [Google Scholar] [CrossRef]
- Bai, F.-Y.; Zhao, J.-H.; Takashima, M.; Jia, J.-H.; Boekhout, T.; Nakase, T. Reclassification of the Sporobolomyces roseus and Sporidiobolus pararoseus complexes, with the description of Sporobolomyces phaffii sp. nov. Int. J. Syst. Evol. Microbiol. 2002, 52, 2309–2314. [Google Scholar] [CrossRef]
- Serradilla, M.J.; del Carmen Villalobos, M.; Hernández, A.; Martín, A.; Lozano, M.; de Guía Córdoba, M. Study of microbiological quality of controlled atmosphere packaged ‘Ambrunés’ sweet cherries and subsequent shelf-life. Int. J. Food Microbiol. 2013, 166, 85–92. [Google Scholar] [CrossRef]
- Bourret, T.B.; Grove, G.G.; Vandemark, G.J.; Henick-Kling, T.; Glawe, D.A. Diversity and molecular determination of wild yeasts in a central Washington State vineyard. N. Am. Fungi 2013, 8, 1–32. [Google Scholar] [CrossRef]
- Barahona, S.; Yuivar, Y.; Socias, G.; Alcaíno, J.; Cifuentes, V.; Baeza, M. Identification and characterization of yeasts isolated from sedimentary rocks of Union Glacier at the Antarctica. Extremophiles 2016, 20, 479–491. [Google Scholar] [CrossRef]
- Nakase, T.; Takematsu, A.; Yamada, Y. Molecular approaches to the taxonomy of ballistosporous yeasts based on the analysis of the partial nucleotide sequences of 18S ribosomal ribonucleic acids. J. Gen. Appl. Microbiol. 1993, 39, 107–134. [Google Scholar] [CrossRef][Green Version]
- Boekhout, T.; Fonseca, A.; Sampaio, J.P.; Bandoni, R.J.; Fell, J.W.; Kwon-Chung, K.J. Chapter 100. Discussion of teleomorphic and anamorphic basidiomycetous yeasts. In The Yeasts, a Taxonomic Study, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; Volume 3, pp. 1339–1372. [Google Scholar] [CrossRef]
- Haelewaters, D.; Toome-Heller, M.; Albu, S.; Aime, M.C. Red yeasts from leaf surfaces and other habitats: Three new species and a new combination of Symmetrospora (Pucciniomycotina, Cystobasidiomycetes). Fungal Syst. Evol. 2020, 5, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Phaff, H.J.; Mrak, E.M.; Williams, O.B. Yeasts isolated from shrimp. Mycologia 1952, 44, 431–451. [Google Scholar] [CrossRef]
- Shivas, R.G.; de Miranda, L.R. Two new species of the genus Sporobolomyces and a new Rhodotorula species from leaf surfaces. Ant. Van Leeuwenhoek 1983, 49, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Nakase, T.; Suzuki, M. Bullera intermedia sp. nov. and Sporobolomyces oryzicola sp. nov. isolated from dead leaves of Oryza sativa. J. Gen. Appl. Microbiol. 1986, 32, 149–155. [Google Scholar] [CrossRef]
- Takashima, M.; Nakase, T. Four new species of the genus Sporobolomyces isolated from leaves in Thailand. Mycoscience 2000, 41, 357–369. [Google Scholar] [CrossRef]
- Wang, Q.-M.; Bai, F.-Y. Four new yeast species of the genus Sporobolomyces from plant leaves. FEMS Yeast Res. 2004, 4, 579–586. [Google Scholar] [CrossRef]
- Suh, S.-O.; Zhang, N.; Nguyen, N.; Gross, S.; Blackwell, M. Lab Manual for Yeast Study; Louisiana State University: Baton Rouge, LA, USA, 2008. [Google Scholar]
- Tsuji, M.; Tanabe, Y.; Vincent, W.F.; Uchida, M. Vishniacozyma ellesmerensis sp. nov., a psychrophilic yeast isolated from a retreating glacier in the Canadian High Arctic. Int. J. Syst. Evol. Microbiol. 2019, 69, 696–700. [Google Scholar] [CrossRef]
- Félix, C.R.; Andrade, D.A.; Almeida, J.H.; Navarro, H.M.; Fell, J.W.; Landell, M.F. Vishniacozyma alagoana sp. nov. a tremellomycetes yeast associated with plants from dry and rainfall tropical forests. Int. J. Syst. Evol. Microbiol. 2020, 70, 3449–3454. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Fungal Barcoding Consortium. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [PubMed]
- Péter, G.; Takashima, M.; Čadež, N. Yeast habitats: Different but global. In Yeasts in Natural Ecosystems: Ecology; Buzzini, P., Lachance, M.A., Yurkov, A., Eds.; Springer: Cham, Switzerland, 2017; pp. 39–71. [Google Scholar] [CrossRef]
- Morris, C.E. Phyllosphere. In Encyclopedia of Life Sciences; Nature Publishing Group: London, UK, 2001. [Google Scholar] [CrossRef]
- Morris, C.E.; Kinkel, L.L. Fifty years of phyllosphere microbiology: Significant contributions to research in related fields. In Phyllosphere Microbiology; Lindow, S.E., Hecht-Poinar, E.I., Elliott, V., Eds.; APS Press: St. Paul, MN, USA, 2002; pp. 365–375. [Google Scholar]
- Lindow, S.E.; Brandl, M.T. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 2003, 69, 1875–1883. [Google Scholar] [CrossRef]
- Simon, U.K.; Groenewald, J.Z.; Crous, P.W. Cymadothea trifolii, an obligate biotrophic leaf parasite of Trifolium, belongs to Mycosphaerellaceae as shown by nuclear ribosomal DNA analyses. Persoonia 2009, 22, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Zare, R.; Gams, W.; Starink-Willemse, M.; Summerbell, R.C. Gibellulopsis, a suitable genus for Verticillium nigrescens, and Musicillium, a new genus for V. theobromae. Nova Hedwig. 2007, 85, 463–489. [Google Scholar] [CrossRef]
- Tibpromma, S.; Hyde, K.D.; Bhat, J.D.; Mortimer, P.E.; Xu, J.; Promputtha, I.; Doilom, M.; Yang, J.B.; Tang, A.M.; Karunarathna, S.C. Identification of endophytic fungi from leaves of Pandanaceae based on their morphotypes and DNA sequence data from southern Thailand. MycoKeys 2018, 33, 25–67. [Google Scholar] [CrossRef]
- Suh, S.-O.; Gibson, C.M.; Blackwell, M. Metschnikowia chrysoperlae sp. nov., Candida picachoensis sp. nov. and Candida pimensis sp. nov., isolated from the green lacewings Chrysoperla comanche and Chrysoperla carnea (Neuroptera: Chrysopidae). Int. J. Syst. Evol. Microbiol. 2004, 54, 1883–1890. [Google Scholar] [CrossRef] [PubMed]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef]
- Thambugala, K.M.; Wanasinghe, D.N.; Phillips, A.J.L.; Camporesi, E.; Bulgakov, T.S.; Phukhamsakda, C.; Ariyawansa, H.A.; Goonasekara, I.D.; Phookamsak, R.; Dissanayake, A.; et al. Mycosphere notes 1–50: Grass (Poaceae) inhabiting Dothideomycetes. Mycosphere 2017, 8, 697–796. [Google Scholar] [CrossRef]
- Staats, M.; van Baarlen, P.; van Kan, J.A. Molecular phylogeny of the plant pathogenic genus Botrytis and the evolution of host specificity. Mol. Biol. Evol. 2005, 22, 333–346. [Google Scholar] [CrossRef] [PubMed]
- Park, H.G.; Managbanag, J.R.; Stamenova, E.K.; Jong, S.C. Comparative analysis of common indoor Cladosporium species based on molecular data and conidial characters. Mycotaxon 2004, 89, 441–451. [Google Scholar]
- Chen, Q.; Hou, L.W.; Duan, W.J.; Crous, P.W.; Cai, L. Didymellaceae revisited. Stud. Mycol. 2017, 87, 105–159. [Google Scholar] [CrossRef] [PubMed]
- Wuczkowski, M.; Passoth, V.; Turchetti, B.; Andersson, A.C.; Olstorpe, M.; Laitila, A.; Theelen, B.; van Broock, M.; Buzzini, P.; Prillinger, H.; et al. Description of Holtermanniella gen. nov., including Holtermanniella takashimae sp. nov. and four new combinations, and proposal of the order Holtermanniales to accommodate tremellomycetous yeasts of the Holtermannia clade. Int. J. Syst. Evol. Microbiol. 2011, 61, 680–689. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J.; Basehoar, E.; Ward, T.J. Four new species of Metschnikowia and the transfer of seven Candida species to Metschnikowia and Clavispora as new combinations. Ant. Van Leeuwenhoek 2018, 111, 2017–2035. [Google Scholar] [CrossRef]
- Avis, T.J.; Caron, S.; Boekhout, T.; Hamelin, R.C.; Bélanger, R.R. Molecular and physiological analysis of the powdery mildew antagonist Pseudozyma flocculosa and related fungi. Phytopathology 2001, 91, 249–254. [Google Scholar] [CrossRef]
- Li, C.H.; Cervantes, M.; Springer, D.J.; Boekhout, T.; Ruiz-Vazquez, R.M.; Torres-Martinez, S.R.; Heitman, J.; Lee, S.C. Sporangiospore size dimorphism is linked to virulence of Mucor circinelloides. PLoS Path. 2011, 7, e1002086. [Google Scholar] [CrossRef]
- Jankowiak, R.; Strzałka, B.; Bilański, P.; Kacprzyk, M.; Wieczorek, P.; Linnakoski, R. Ophiostomatoid fungi associated with hardwood-infesting bark and ambrosia beetles in Poland: Taxonomic diversity and vector specificity. Fungal Ecol. 2019, 39, 152–167. [Google Scholar] [CrossRef]
- Houbraken, J.; Visagie, C.M.; Meijer, M.; Frisvad, J.C.; Busby, P.E.; Pitt, J.I.; Seifert, K.A.; Louis-Seize, G.; Demirel, R.; Yilmaz, N.; et al. A taxonomic and phylogenetic revision of Penicillium section Aspergilloides. Stud. Mycol. 2014, 78, 373–451. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.-M.; Boekhout, T.; Bai, F.-Y. Cryptococcus foliicola sp. nov. and Cryptococcus taibaiensis sp. nov., novel basidiomycetous yeast species from plant leaves. J. Gen. Appl. Microbiol. 2011, 57, 285–291. [Google Scholar] [CrossRef] [PubMed][Green Version]
| Genus | Isolates | Characters | Informative | % Informative | Model | -lnL |
|---|---|---|---|---|---|---|
| Aureobasidium | 14 | 517 | 44 | 8.51 | SYM+I | 1317.537 |
| Cladosporium | 36 | 482 | 45 | 9.34 | SYM+R3 | 1530.217 |
| Filobasidium | 21 | 619 | 170 | 27.46 | TPM2u+F+R3 | 3027.598 |
| Naganishia | 18 | 588 | 67 | 11.39 | TIM2+F+R3 | 1906.687 |
| Papliotrema | 29 | 507 | 143 | 28.21 | GTR+F+I+G4 | 2833.792 |
| Rhodotorula | 21 | 572 | 139 | 24.30 | GTR+F+R3 | 2888.392 |
| Sampaiozyma | 11 | 553 | 63 | 11.39 | TIM3+F+I+G4 | 1541.096 |
| Sporobolomyces | 34 | 560 | 151 | 26.96 | TIM2+F+I+G4 | 3160.923 |
| Symmetrospora | 17 | 582 | 131 | 22.51 | GTR+F+G4 | 2778.027 |
| Vishniacozyma | 25 | 493 | 157 | 31.85 | TIM2+F+G4 | 2545.886 |
| Genus | Species | Total Isolates | Conventional | Organic | Hydroponic |
|---|---|---|---|---|---|
| Acanthophysium | 1 | 1 | 1 | ||
| Alternaria | 6 | 6 | 5 | 1 | |
| Aureobasidium | 2 | 10 | 6 | 4 | |
| Beauveria | 1 | 1 | 1 | ||
| Botryotinia | 1 | 1 | 1 | ||
| Bullera | 2 | 2 | 2 | ||
| Bulleromyces | 1 | 1 | 1 | ||
| Cladosporium | 8 | 59 | 19 | 34 | 6 |
| Cystofilobasidium | 2 | 12 | 7 | 5 | |
| Epicoccum | 1 | 1 | 1 | ||
| Filobasidium | 6 | 33 | 16 | 17 | |
| Gibellulopsis | 1 | 3 | 2 | 1 | |
| Holtermanniella | 1 | 1 | 1 | ||
| Leucosporidium | 1 | 1 | 1 | ||
| Metschnikowia | 2 | 3 | 1 | 2 | |
| Moesziomyces | 1 | 1 | 1 | ||
| Mucor | 1 | 1 | 1 | ||
| Naganishia | 1 | 1 | 1 | ||
| Ophiostoma | 1 | 1 | 1 | ||
| Papiliotrema | 2 | 3 | 1 | 2 | |
| Penicillium | 2 | 3 | 1 | 2 | |
| Rhodotorula | 3 | 3 | 1 | 1 | 1 |
| Sampaiozyma | 2 | 2 | 1 | 1 | |
| Sporobolomyces | 2 | 67 | 36 | 31 | |
| Stemphylium | 1 | 2 | 2 | ||
| Symmetrospora | 1 | 2 | 2 | ||
| Tilletiopsis | 1 | 1 | 1 | ||
| Vishniacozyma | 5 | 27 | 9 | 18 | |
| Total | 59 | 249 | 110 | 128 | 11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haelewaters, D.; Urbina, H.; Brown, S.; Newerth-Henson, S.; Aime, M.C. Isolation and Molecular Characterization of the Romaine Lettuce Phylloplane Mycobiome. J. Fungi 2021, 7, 277. https://doi.org/10.3390/jof7040277
Haelewaters D, Urbina H, Brown S, Newerth-Henson S, Aime MC. Isolation and Molecular Characterization of the Romaine Lettuce Phylloplane Mycobiome. Journal of Fungi. 2021; 7(4):277. https://doi.org/10.3390/jof7040277
Chicago/Turabian StyleHaelewaters, Danny, Hector Urbina, Samuel Brown, Shannon Newerth-Henson, and M. Catherine Aime. 2021. "Isolation and Molecular Characterization of the Romaine Lettuce Phylloplane Mycobiome" Journal of Fungi 7, no. 4: 277. https://doi.org/10.3390/jof7040277
APA StyleHaelewaters, D., Urbina, H., Brown, S., Newerth-Henson, S., & Aime, M. C. (2021). Isolation and Molecular Characterization of the Romaine Lettuce Phylloplane Mycobiome. Journal of Fungi, 7(4), 277. https://doi.org/10.3390/jof7040277






