Genomic Studies of White-Rot Fungus Cerrena unicolor SP02 Provide Insights into Food Safety Value-Added Utilization of Non-Food Lignocellulosic Biomass
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain and Cultivation Media
2.2. Isolation of Total DNA and ITS Sequence Analysis
2.3. Genome Sequencing, Assembly and Annotation
2.4. Growth Characters on Lignocellulosic Materials
2.4.1. Growth Rate and Preference on Substrates
2.4.2. Solid-State Fermentation (SSF)
2.5. Data Availability and Accession Numbers
3. Results and Discussion
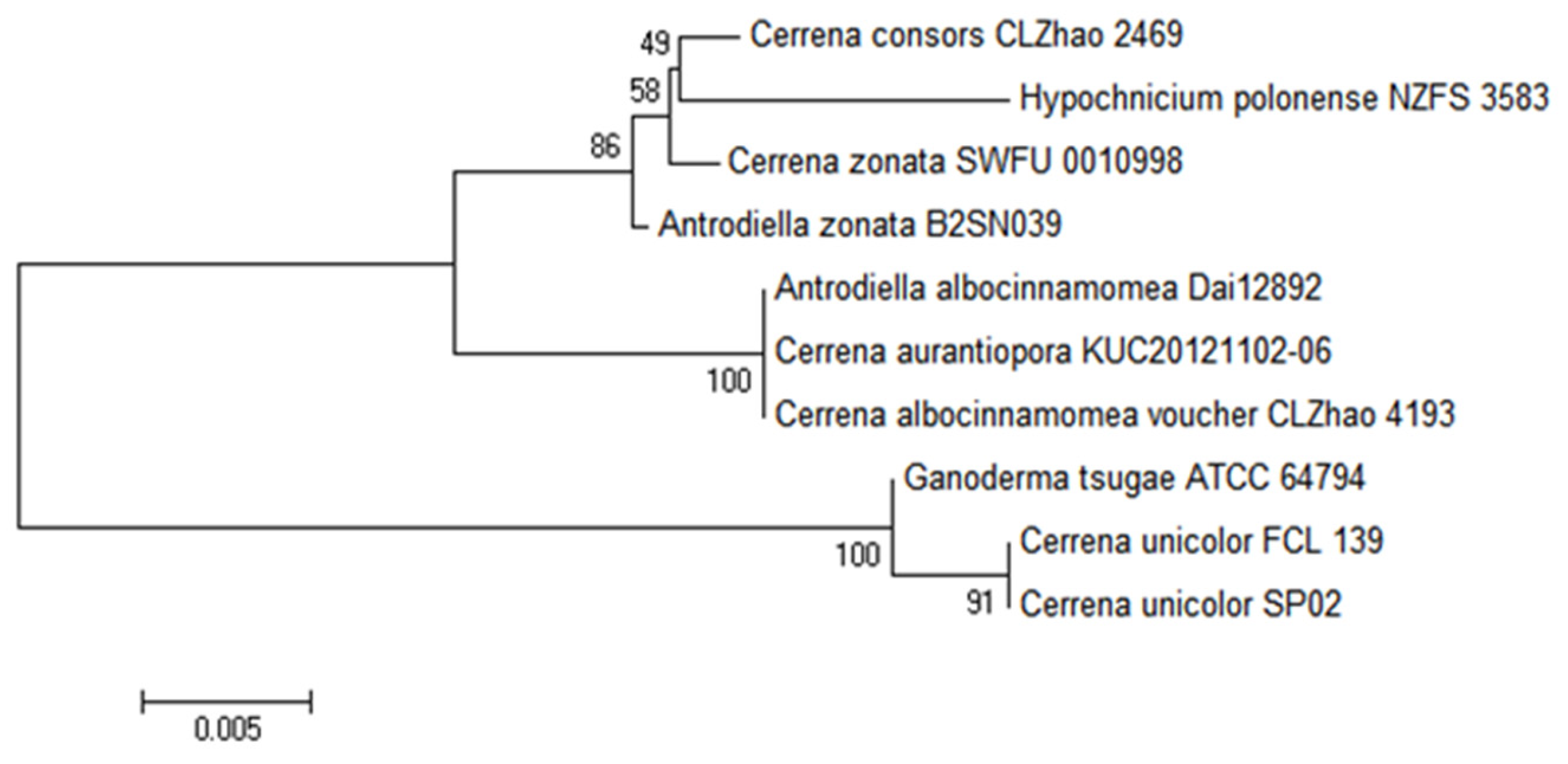
3.1. Phylogenetic Analysis of Strain SP02
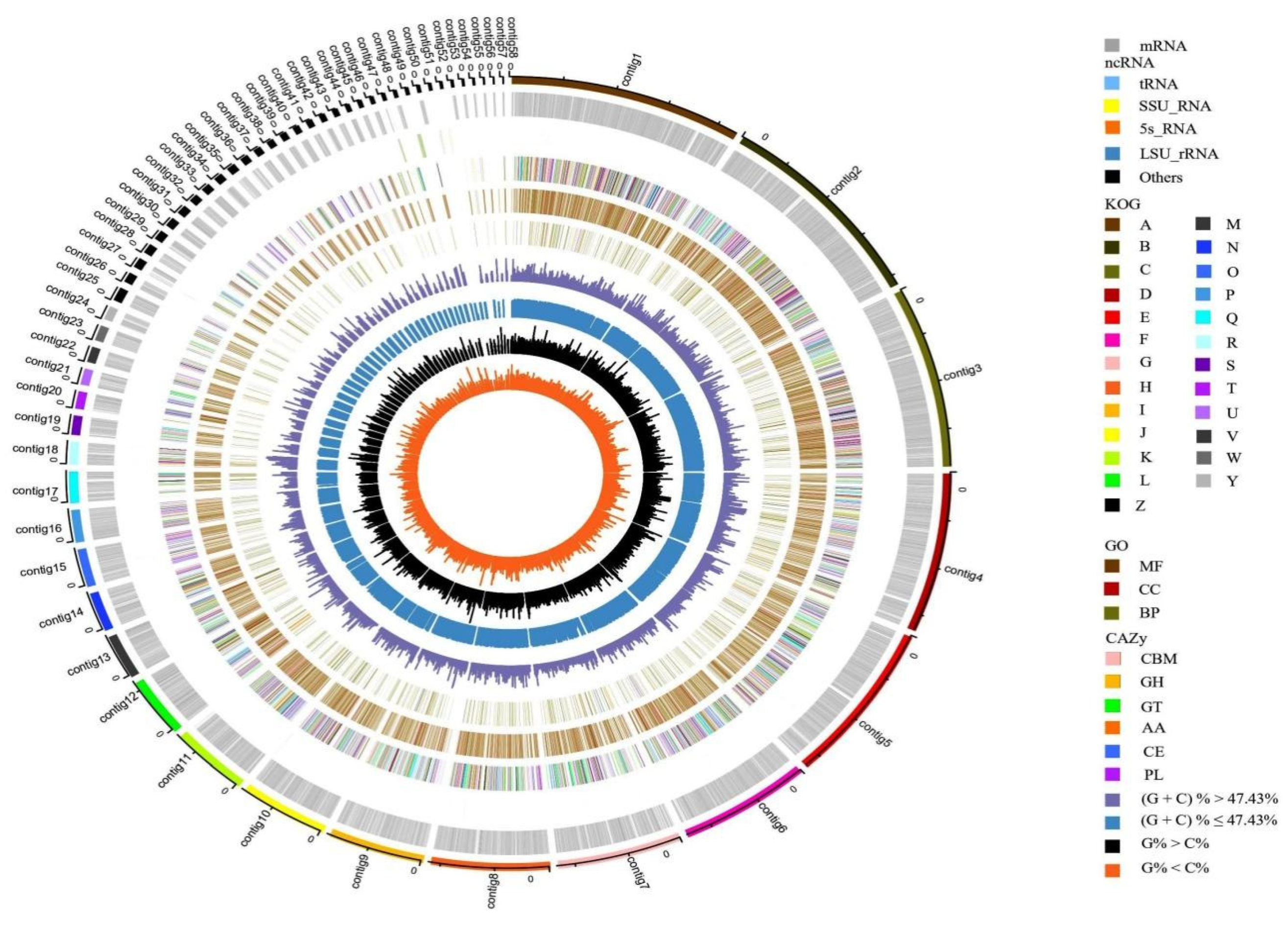
3.2. General Genome Characteristics of C. unicolor SP02
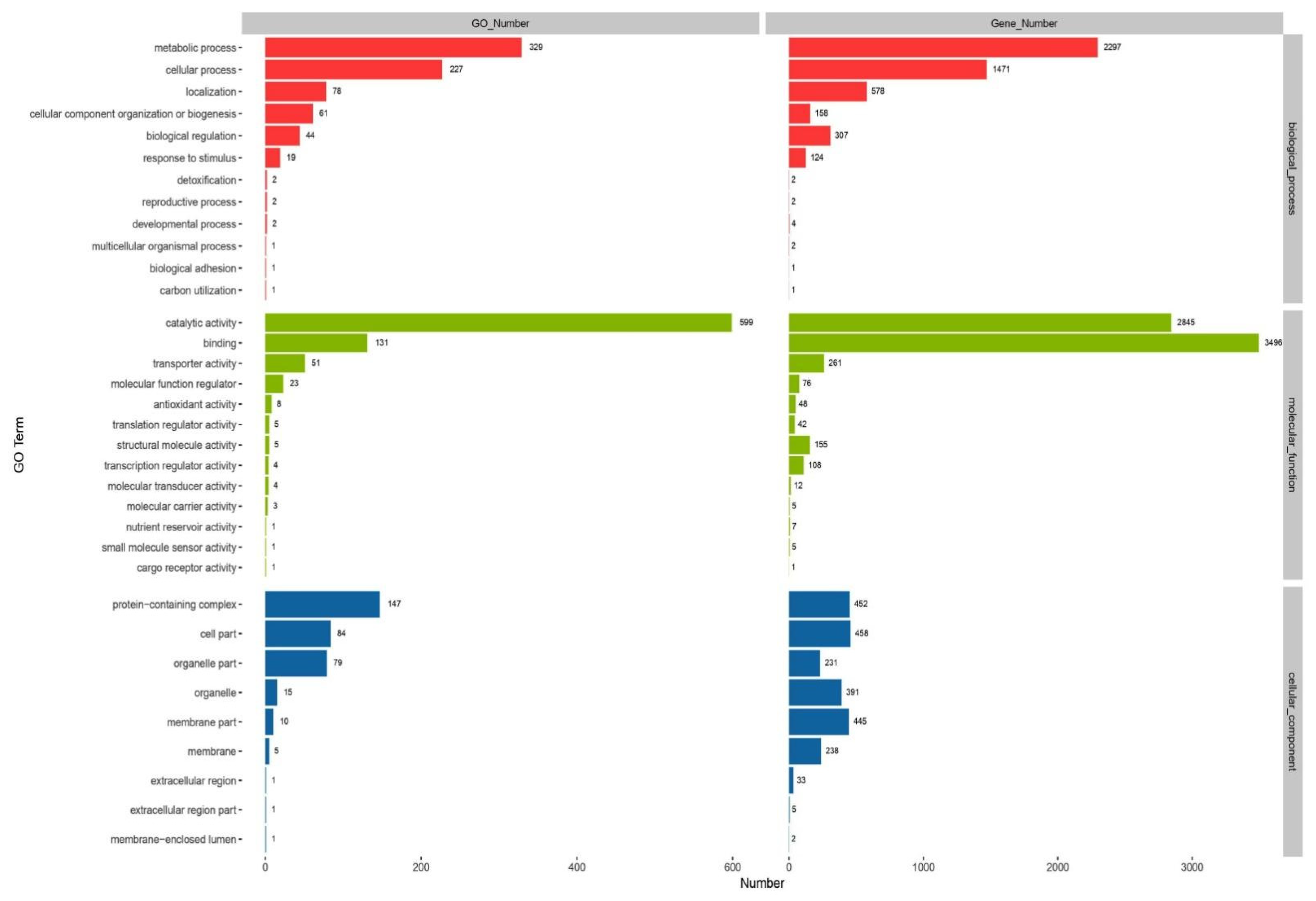
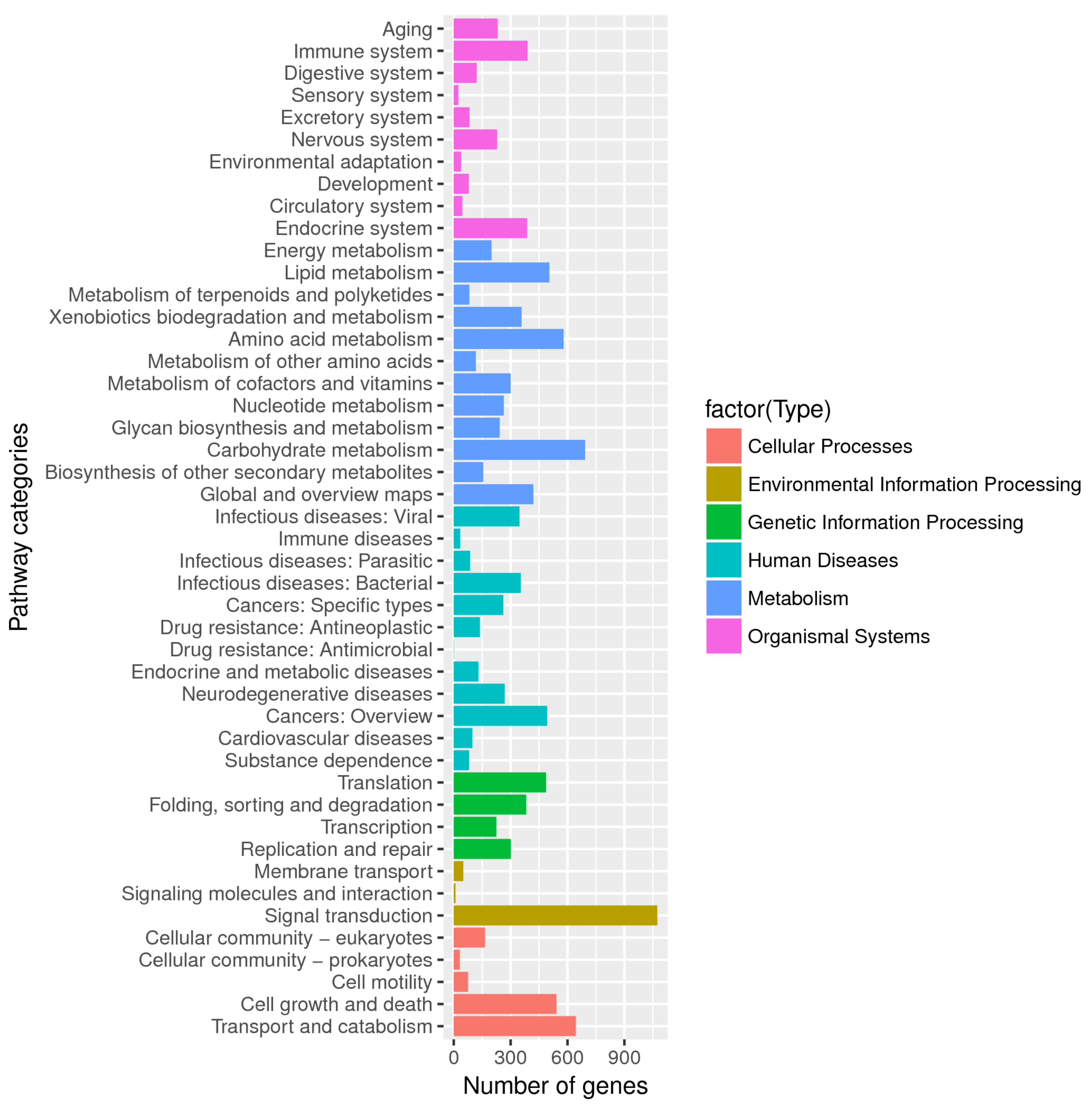
3.3. Gene Function of C. unicolor SP02
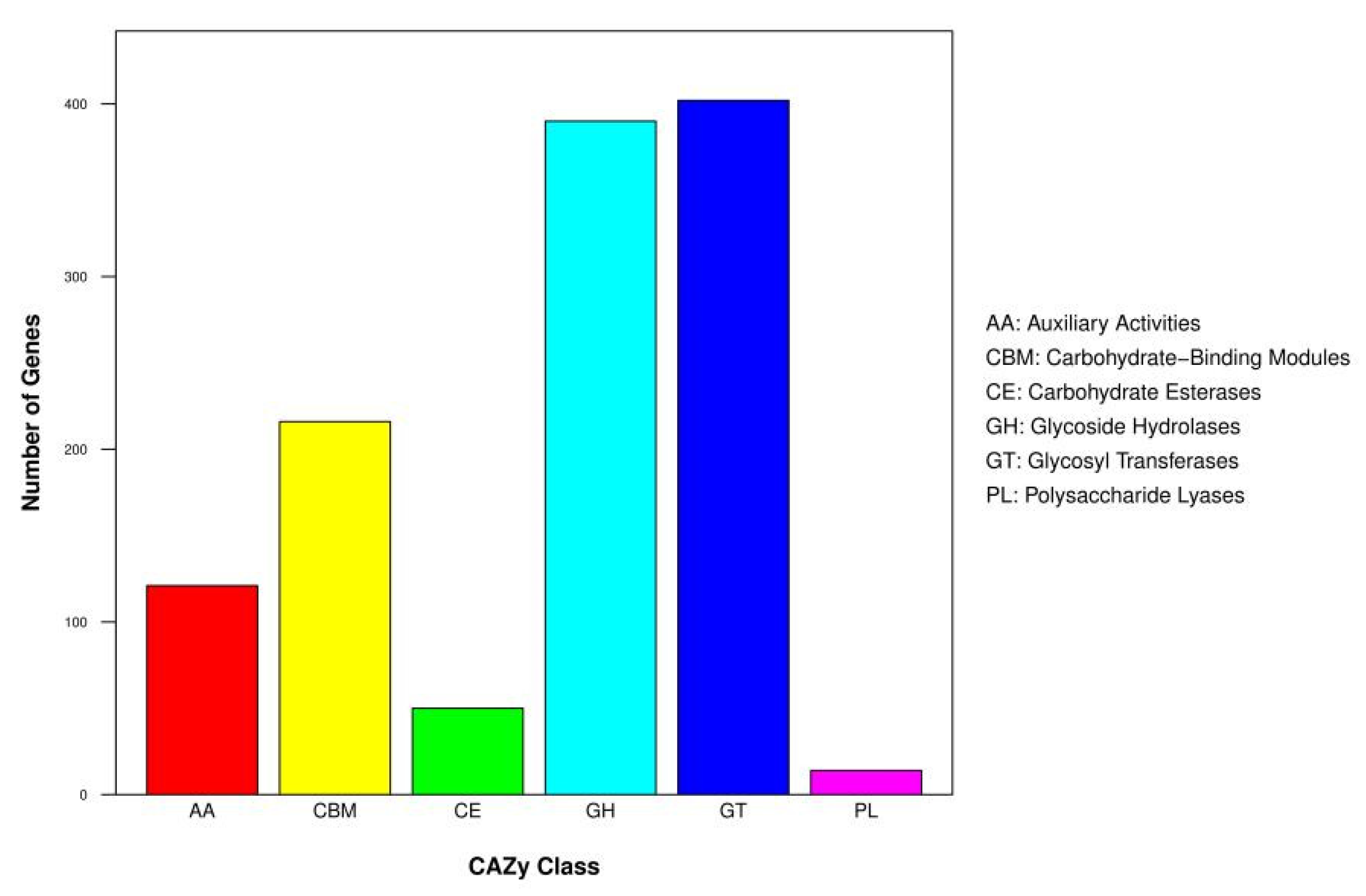
3.4. Genes Related to Biodegradation in C. unicolor SP02
3.4.1. Lignin-Degrading Enzymes
3.4.2. Cellulose Degrading Enzymes
3.4.3. Hemicellulose-Degrading Enzymes
3.4.4. Other Degrading Enzymes
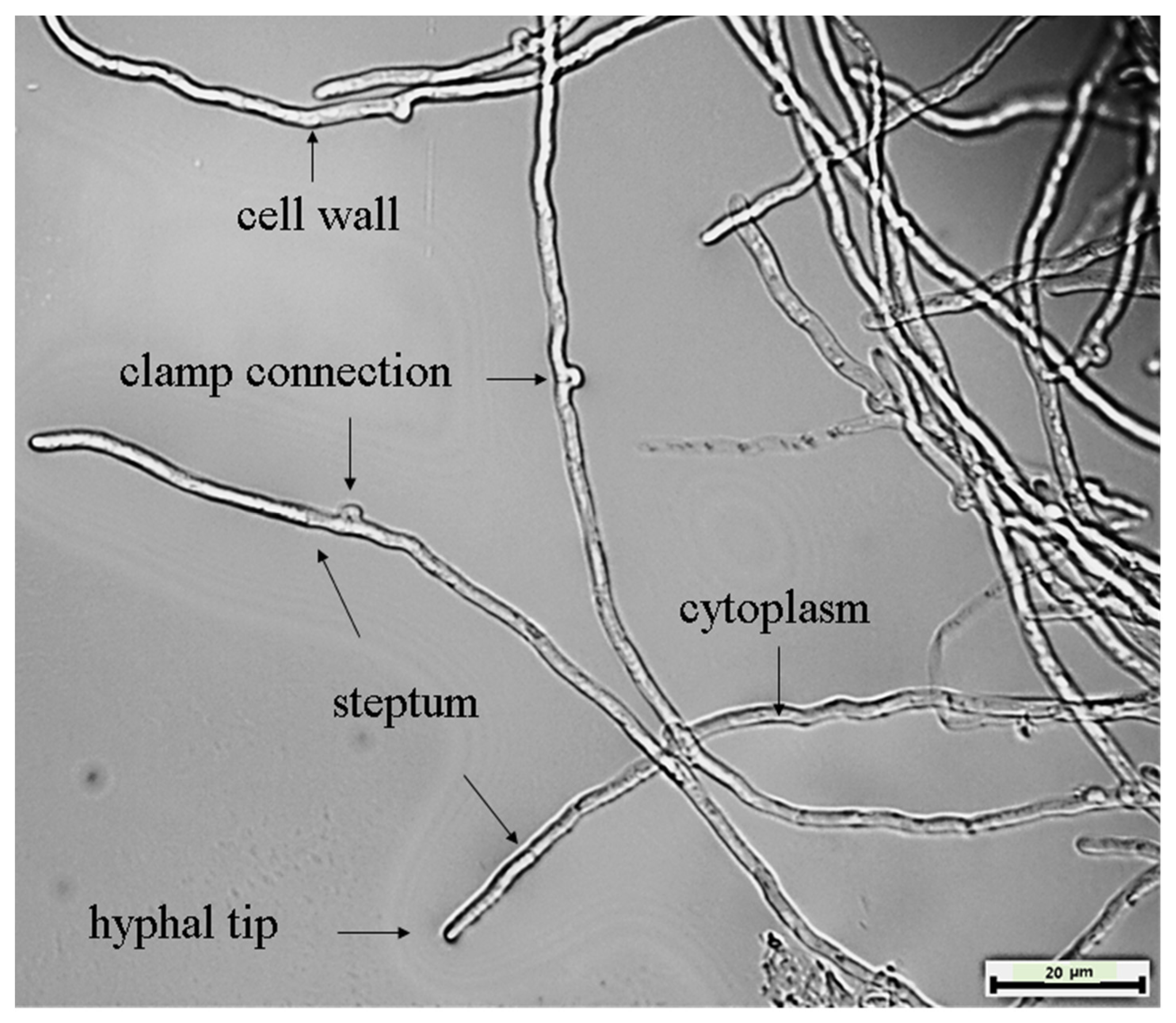
3.5. Cultural Characteristics
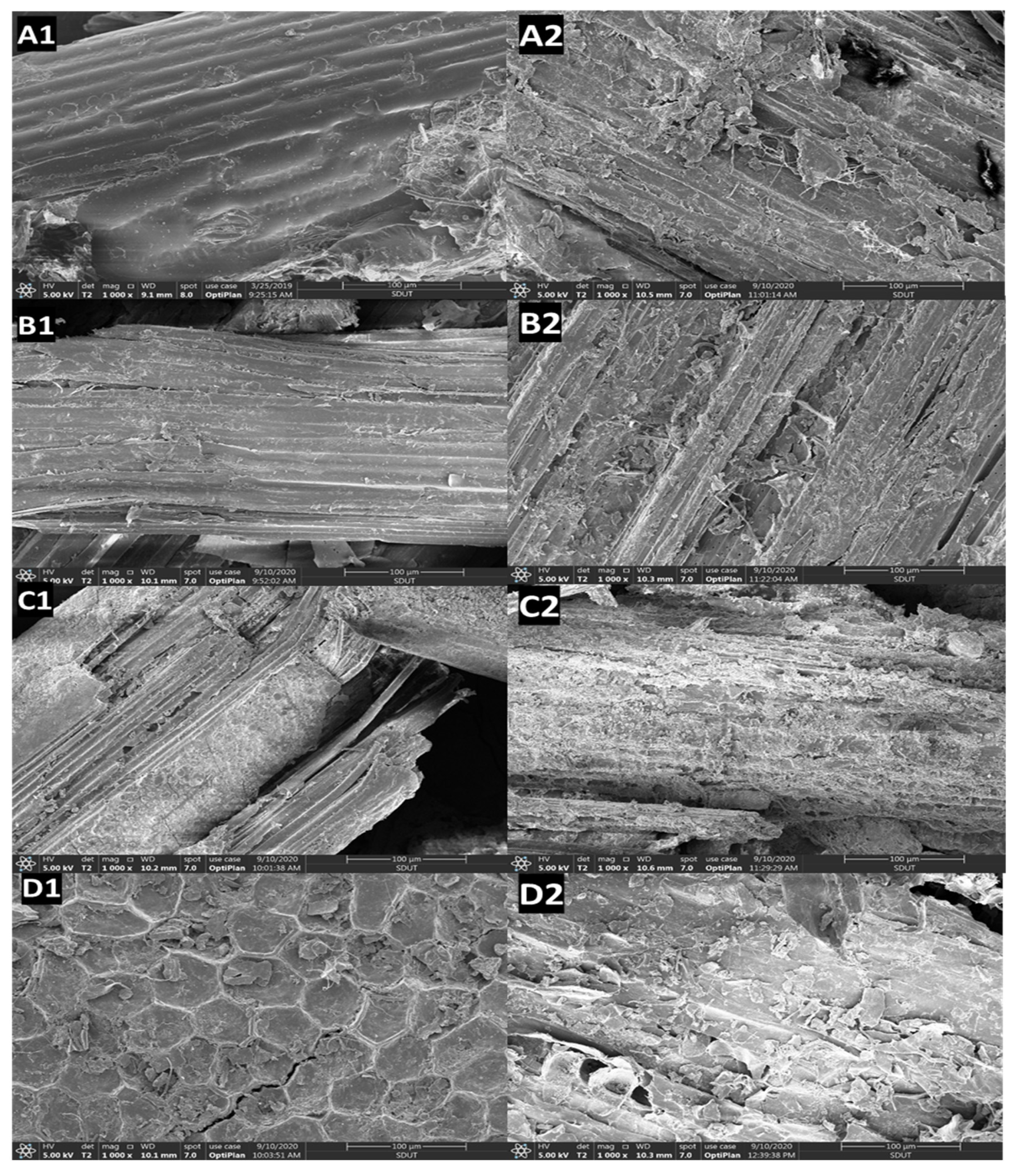
3.5.1. Growth of C. unicolor SP02 on Agro-Wastes
3.5.2. Lignocellulolytic SSF Characters
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pawlik, A.; Ruminowicz-Stefaniuk, M.; Frąc, M.; Mazur, A.; Wielbo, J.; Janusz, G. The wood decay fungus Cerrena unicolor adjusts its metabolism to grow on various types of wood and light conditions. PLoS ONE 2019, 14, e0211744. [Google Scholar] [CrossRef]
- Pinzari, F.; Ceci, A.; Abu-Samra, N.; Canfora, L.; Maggi, O.; Persiani, A. Phenotype MicroArray™ system in the study of fungal functional diversity and catabolic versatility. Res. Microbiol. 2016, 167, 710–722. [Google Scholar] [CrossRef]
- Singhvi, M.S.; Chaudhari, S.; Gokhale, D. Lignocellulose processing: A current challenge. RSC Adv. 2014, 4, 8271–8277. [Google Scholar] [CrossRef]
- Blatkiewicz, M.; Antecka, A.; Boruta, T.; Gorak, A.; Ledakowicz, S. Partitioning of laccases derived from Cerrena unicolor and Pleurotus sapidus in polyethylene glycol—Phosphate aqueous two–phase systems. Process. Biochem. 2018, 67, 165–174. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, L.; Zhang, H.; Wang, S.; Zhang, X.; Geng, A. A novel homodimer laccase from Cerrena unicolor BBP6: Purification, characterization, and potential in dye decolorization and denim bleaching. PLoS ONE 2018, 13, e0202440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Zhang, J.; Zhang, X.; Geng, A. Purification and characterization of a novel manganese peroxidase from white-rot fungus Cerrena unicolor BBP6 and its application in dye decolorization and denim bleaching. Process. Biochem. 2018, 66, 222–229. [Google Scholar] [CrossRef]
- Hibi, M.; Hatahira, S.; Nakatani, M.; Yokozeki, K.; Shimizu, S.; Ogawa, J. Extracellular oxidases of Cerrena scomplementarily functioning in artificial dye decolorization including laccase, manganese peroxidase, and novel versatile peroxidases. Biocatal. Agric. Biotechnol. 2012, 1, 220–225. [Google Scholar] [CrossRef] [Green Version]
- Sulej, J.; Janusz, G.; Osińska-Jaroszuk, M.; Rachubik, P.; Mazur, A.; Komaniecka, I.; Choma, A.; Rogalski, J. Characterization of Cellobiose Dehydrogenase from a Biotechnologically Important Cerrena unicolor Strain. Appl. Biochem. Biotechnol. 2015, 176, 1638–1658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belova, O.V.; Lisov, A.V.; Vinokurova, N.G.; Kostenevich, A.A.; Sapunova, L.I.; Lobanok, A.G.; Leontievsky, A.A. Xylanase and cellulase of fungus Cerrena unicolor VKM F-3196: Production, properties, and applications for the saccharification of plant material. Appl. Biochem. Microbiol. 2014, 50, 148–153. [Google Scholar] [CrossRef]
- Mazur, I.; Rola, B.; Stolarczyk, K.; Nazaruk, E.; Bilewicz, R.; Rogalski, J.; Ohga, S. The Large Scale Production of Cerrena unicolor Laccase on Waste Agricultural Based Media. J. Fac. Agric. Kyushu Univ. 2015, 60, 297–302. [Google Scholar] [CrossRef]
- Filazzola, M.T.; Sannino, F.; Rao, M.; Gianfreda, L. Effect of Various Pollutants and Soil-Like Constituents on Laccase from Cerrena unicolor. J. Environ. Qual. 1999, 28, 1929–1938. [Google Scholar] [CrossRef]
- Kameshwar, A.K.S.; Qin, W. Recent Developments in Using Advanced Sequencing Technologies for the Genomic Studies of Lignin and Cellulose Degrading Microorganisms. Int. J. Biol. Sci. 2016, 12, 156–171. [Google Scholar] [CrossRef] [PubMed]
- Jha, H. Fungal Diversity and Enzymes Involved in Lignin Degradation; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Sigoillot, J.C.; Berrin, J.G.; Bey, M.; Lesage-Meessen, L.; Levasseur, A.; Lomascolo, A.; Uzan-Boukhris, E. Fungal Strategies for Lignin Degradation. Adv. Bot. Res. 2012, 61, 263–308. [Google Scholar]
- Lizardi-Jiménez, M.A.; Hernández-Martínez, R. Solid state fermentation (SSF): Diversity of applications to valorize waste and biomass. 3 Biotech 2017, 7, 44. [Google Scholar] [CrossRef]
- Mizerska-Dudka, M.; Jaszek, M.; Błachowicz, A.; Rejczak, T.P.; Matuszewska, A.; Osińska-Jaroszuk, M.; Stefaniuk, D.; Janusz, G.; Sulej, J.; Kandefer-Szerszeń, M. Fungus Cerrena unicolor as an effective source of new antiviral, immunomodulatory, and anticancer compounds. Int. J. Biol. Macromol. 2015, 79 Pt 5, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Matuszewska, A.; Stefaniuk, D.; Jaszek, M.; Pięt, M.; Zając, A.; Matuszewski, Ł.; Cios, I.; Graz, M.; Paduch, R.; Bancerz, R. Antitumor potential of new low molecular weight antioxidative preparations from the white rot fungus Cerrena unicolor against human colon cancer cells. Sci. Rep. 2019, 9, 1975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pięt, M.; Zając, A.; Paduch, R.; Jaszek, M.; Frant, M.; Stefaniuk, D.; Matuszewska, A.; Grzywnowicz, K. Chemopreventive activity of bioactive fungal fractions isolated from milk-supplemented cultures of Cerrena unicolor and Pycnoporus sanguineus on colon cancer cells. 3 Biotech 2021, 11, 5. [Google Scholar] [CrossRef]
- Matuszewska, A.; Jaszek, M.; Stefaniuk, D.; Ciszewski, T.; Matuszewski, Ł. Anticancer, antioxidant, and antibacterial activities of low molecular weight bioactive subfractions isolated from cultures of wood degrading fungus Cerrena unicolor. PLoS ONE 2018, 13, e0197044. [Google Scholar] [CrossRef] [Green Version]
- Jaszek, M.; Osińska-Jaroszuk, M.; Janusz, G.; Matuszewska, A.; Stefaniuk, D.; Sulej, J.; Polak, J.; Ruminowicz, M.; Grzywnowicz, K.; Jarosz-Wilkolazka, A. New Bioactive Fungal Molecules with High Antioxidant and Antimicrobial Capacity Isolated from Cerrena unicolor Idiophasic Cultures. BioMed Res. Int. 2013, 2013, 497492. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Chen, Q.-J.; Zhu, M.-J.; Wang, H.-X.; Zhang, G.-Q. An extracellular laccase with antiproliferative activity from the sanghuang mushroom Inonotus baumii. J. Mol. Catal. B Enzym. 2014, 99, 20–25. [Google Scholar] [CrossRef]
- Janusz, G.; Mazur, A.; Wielbo, J.; Koper, P.; Żebracki, K.; Pawlik, A.; Ciołek, B.; Paszczyński, A.; Kubik-Komar, A. Comparative transcriptomic analysis of Cerrena unicolor revealed differential expression of genes engaged in degradation of various kinds of wood. Microbiol. Res. 2018, 207, 256–268. [Google Scholar] [CrossRef] [PubMed]
- Kerezoudi, E.N.; Mitsou, E.K.; Gioti, K.; Terzi, E.; Avgousti, I.; Panagiotou, A.; Koutrotsios, G.; Zervakis, G.I.; Mountzouris, K.C.; Tenta, R.; et al. Fermentation of Pleurotus ostreatus and Ganoderma lucidum mushrooms and their extracts by the gut microbiota of healthy and osteopenic women: Potential prebiotic effect and impact of mushroom fermentation products on human osteoblasts. Food Funct. 2021, 12, 1529–1546. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Ma, Q.; Ren, M.; Liang, D.; Yu, Q.; Luo, J. Antitumorpharmacological mechanism of the oral liquid of Poriacocos polysaccharide. J. Ethnopharmacol. 2017, 209, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Rahim, I.; Suherman; Hakzah; Nasruddin, A. The Ability of Rot Fungi from Cocoa Plant in Producing Lignocellulosic Enzymes. IOP Conf. Ser. Earth Environ. Sci. 2019, 270, 012037. [Google Scholar] [CrossRef]
- Zhuo, R.; Fan, F. A comprehensive insight into the application of white rot fungi and their lignocellulolytic enzymes in the removal of organic pollutants. Sci. Total Environ. 2021, 778, 146132. [Google Scholar] [CrossRef]
- Worrall, J.J.; Anagnost, S.E.; Zabel, R.A. Comparison of wood decay among diverse lignicolous fungi. Mycologia 1997, 89, 199–219. [Google Scholar] [CrossRef]
- Grimm, D.; Wösten, H.A.B. Mushroom cultivation in the circular economy. Appl. Microbiol. Biotechnol. 2018, 102, 7795–7803. [Google Scholar] [CrossRef] [Green Version]
- Kurata, A.; Fukuta, Y.; Mori, M.; Kishimoto, N.; Shirasaka, N. Draft Genome Sequence of the Basidiomycetous Fungus Flamulina velutipes TR01. Genome Announc. 2016, 4, e00505-16. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.H.; Zhou, G.; Ma, J.; Jiang, W.; Jin, L.G.; Zhang, Z.; Qiu, L.J. De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat. Biotechnol. 2014, 32, 1045–1052. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Gao, W.; Wu, X.; Zhao, M.; Qu, J.; Huang, C.; Zhang, J. Genome-Wide Characterization and Expression Analyses of Pleurotus ostreatus MYB Transcription Factors during Developmental Stages and under Heat Stress Based on de novo Sequenced Genome. Int. J. Mol. Sci. 2018, 19, 2052. [Google Scholar] [CrossRef] [Green Version]
- Sharma, K.K. Fungal genome sequencing: Basic biology to biotechnology. Crit. Rev. Biotechnol. 2016, 36, 743–759. [Google Scholar] [CrossRef]
- Nagy, L.G.; Riley, R.; Bergmann, P.J.; Krizsán, K.; Martin, F.M.; Grigoriev, I.; Cullen, D.; Hibbett, D.S. Genetic Bases of Fungal White Rot Wood Decay Predicted by Phylogenomic Analysis of Correlated Gene-Phenotype Evolution. Mol. Biol. Evol. 2017, 34, 35–44. [Google Scholar] [CrossRef]
- Yang, J.; Li, W.; Ng, T.B.; Deng, X.; Lin, J.; Ye, X. Laccases: Production, Expression Regulation, and Applications in Pharmaceutical Biodegradation. Front. Microbiol. 2017, 8, 832. [Google Scholar] [CrossRef] [PubMed]
- Jain, K.K.; Kumar, A.; Shankar, A.; Pandey, D.; Chaudhary, B.; Sharma, K.K. De novo transcriptome assembly and protein profiling of copper-induced lignocellulolytic fungus Ganoderma lucidum MDU-7 reveals genes involved in lignocellulose degradation and terpenoid biosynthetic pathways. Genomics 2020, 112, 184–198. [Google Scholar] [CrossRef]
- Mc Carthy, A. Third Generation DNA Sequencing: Pacific Biosciences’ Single Molecule Real Time Technology. Chem. Biol. 2010, 17, 675–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myers, E.W.; Sutton, G.G.; Delcher, A.L.; Dew, I.M.; Fasulo, D.P.; Flanigan, M.J.; Kravitz, S.A.; Mobarry, C.M.; Reinert, K.; Remington, K.A.; et al. A Whole-Genome Assembly of Drosophila. Science 2000, 287, 2196–2204. [Google Scholar] [CrossRef]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Kalush, F. The sequence of the human genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Istrail, S.; Sutton, G.G.; Florea, L.; Halpern, A.L.; Mobarry, C.M.; Lippert, R.; Walenz, B.; Shatkay, H.; Dew, I.; Miller, J.R.; et al. Whole-genome shotgun assembly and comparison of human genome assemblies. Proc. Natl. Acad. Sci. USA 2004, 101, 1916–1921. [Google Scholar] [CrossRef] [Green Version]
- Levy, S.; Sutton, G.; Ng, P.C.; Feuk, L.; Halpern, A.L.; Walenz, B.P.; Axelrod, N.; Huang, J.; Kirkness, E.F.; Denisov, G.; et al. The Diploid Genome Sequence of an Individual Human. PLoS Biol. 2007, 5, e254. [Google Scholar] [CrossRef] [Green Version]
- Goldberg, S.M.D.; Johnson, J.; Busam, D.; Feldblyum, T.; Ferriera, S.; Friedman, R.; Halpern, A.; Khouri, H.; Kravitz, S.A.; Lauro, F.M.; et al. A Sanger/pyrosequencing hybrid approach for the generation of high-quality draft assemblies of marine microbial genomes. Proc. Natl. Acad. Sci. USA 2006, 103, 11240–11245. [Google Scholar] [CrossRef] [Green Version]
- Berlin, K.; Koren, S.; Chin, C.S.; Drake, J.P.; Landolin, J.M.; Phillippy, A.M. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 2015, 33, 623–630. [Google Scholar] [CrossRef]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Schöffmann, O.; Morgenstern, B.; Waack, S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinform. 2006, 7, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Staerfeldt, H.-H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, D258–D261. [Google Scholar] [CrossRef] [Green Version]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [Green Version]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khvedelidze, R.; Tsiklauri, N.; Aleksidze, T.; Kvesitadze, E. Enzymatic Hydrolysis of Lignocellulosic Agricultural Wastes to Fermentable glucose. Agric. Res. Technol. Open Access J. 2018, 17, 5. [Google Scholar] [CrossRef]
- Xu, X.; Xu, Z.; Shi, S.; Lin, M. Lignocellulose degradation patterns, structural changes, and enzyme secretion by Inonotus obliquus on straw biomass under submerged fermentation. Bioresour. Technol. 2017, 241, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Bourbonnais, R.; Paice, M.G. Oxidation of non-phenolic substrates: An expanded role for laccase in lignin biodegradation. FEBS Lett. 1990, 267, 99–102. [Google Scholar] [CrossRef] [Green Version]
- Gold, M.H.; Glenn, J.K. Manganese peroxidase from Phanerochaete chrysosporium. Methods Enzymol. 1988, 161, 258–264. [Google Scholar] [CrossRef]
- Tien, M.; Kirk, T.K. Lignin-degrading enzyme from Phanerochaete chrysosporium: Purification, characterization, and catalytic properties of a unique H2O2-requiring oxygenase. Proc. Natl. Acad. Sci. USA 1984, 81, 2280–2284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugars. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Berghem, L.E.R.; Pettersson, L.G.; Axiö-Fredriksson, U. The Mechanism of Enzymatic Cellulose Degradation. Purification and Some Properties of Two Different 1,4-beta-Glucan Glucanohydrolases from Trichoderma viride. Eur. J. Biol. Chem. 1976, 61, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Sluiter, A.; Hames, B.; Ruiz, R.; Scarlata, C.; Sluiter, J.; Templeton, D.; Crocker, D.L.A.P. Determination of structural carbohydrates and lignin in biomass. Lab. Anal. Proced. 2004, 1617, 1–16. [Google Scholar]
- Roody, W.C. Mushrooms of West Virginia and the Central Appalachians; University Press of Kentucky: Lexington, KY, USA, 2003; ISBN 978-0-8131-9039. [Google Scholar]
- Pawlik, A.; Jaszek, M.; Stefaniuk, D.; Świderska-Burek, U.; Mazur, A.; Wielbo, J.; Koper, P.; Żebracki, K.; Janusz, G. Combined Effect of Light and Nutrients on the Micromorphology of the White rot Fungus Cerrena unicolor. Int. J. Mol. Sci. 2020, 21, 1678. [Google Scholar] [CrossRef] [Green Version]
- Madej, M.G.; Sun, L.; Yan, N.; Kaback, H.R. Functional architecture of MFS D-glucose transporters. Proc. Natl. Acad. Sci. USA 2014, 111, E719–E727. [Google Scholar] [CrossRef] [Green Version]
- Nehls, U.; Grunze, N.; Willmann, M.; Reich, M.; Küster, H. Sugar for my honey: Carbohydrate partitioning in ectomycorrhizal symbiosis. Phytochemistry 2007, 68, 82–91. [Google Scholar] [CrossRef]
- Strachan, C.R.; Singh, R.; VanInsberghe, D.; Ievdokymenko, K.; Budwill, K.; Mohn, W.W.; Eltis, L.; Hallam, S.J. Metagenomic scaffolds enable combinatorial lignin transformation. Proc. Natl. Acad. Sci. USA 2014, 111, 10143–10148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, C.; Powlowski, J.; Wu, M.; Butler, G.; Tsang, A. Curation of characterized glycoside hydrolases of Fungal origin. Database 2011, 2011, bar020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundell, T.K.; Mäkelä, M.; Hilden, K. Lignin-modifying enzymes in filamentous basidiomycetes—Ecological, functional and phylogenetic review. J. Basic Microbiol. 2010, 50, 5–20. [Google Scholar] [CrossRef]
- Sánchez, C. Lignocellulosic residues: Biodegradation and bioconversion by fungi. Biotechnol. Adv. 2009, 27, 185–194. [Google Scholar] [CrossRef]
- Chang, H.-X.; Yendrek, C.R.; Caetano-Anolles, G.; Hartman, G.L. Erratum to: Genomic characterization of plant cell wall degrading enzymes and in silico analysis of xylanases and polygalacturonases of Fusarium virguliforme. BMC Microbiol. 2017, 17, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hori, C.; Gaskell, J.; Igarashi, K.; Samejima, M.; Hibbett, D.; Henrissat, B.; Cullen, D. Genomewide analysis of polysaccharides degrading enzymes in 11 white- and brown-rot Polyporales provides insight into mechanisms of wood decay. Mycology 2013, 105, 1412–1427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kameshwar, A.K.S.; Qin, W. Understanding the structural and functional properties of carbohydrate esterases with a special focus on hemicellulose deacetylating acetyl xylan esterases. Mycology 2018, 9, 273–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rytioja, J.; Hildén, K.; Yuzon, J.; Hatakka, A.; de Vries, R.P.; Mäkelä, M.R. Plant-Polysaccharide-Degrading Enzymes from Basidiomycetes. Microbiol. Mol. Biol. Rev. 2014, 78, 614–649. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Fueyo, E.; Ruiz-Dueñas, F.J.; Ferreira, P.; Floudas, D.; Hibbett, D.S.; Canessa, P.; Larrondo, L.F.; James, T.Y.; Seelenfreund, D.; Lobos, S.; et al. Comparative genomics of Ceriporiopsis subvermispora and Phanerochaete chrysosporium provide insight into selective ligninolysis. Proc. Natl. Acad. Sci. USA 2012, 109, 5458–5463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Floudas, D.; Binder, M.; Riley, R.; Barry, K.; Blanchette, R.A.; Henrissat, B.; Martínez, A.T.; Otillar, R.; Spatafora, J.W.; Yadav, J.; et al. The Paleozoic Origin of Enzymatic Lignin Decomposition Reconstructed from 31 Fungal Genomes. Science 2012, 336, 1715–1719. [Google Scholar] [CrossRef] [Green Version]
- Zane, N.R.; Chen, Y.; Wang, M.Z.; Thakker, D.R. Cytochrome P450 and flavin-containing monooxygenase families: Age-dependent differences in expression and functional activity. Pediatr. Res. 2017, 83, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Reis, R.A.; Li, H.; Johnson, M.; Sobrado, P. New frontiers in flavin-dependent monooxygenases. Arch. Biochem. Biophys. 2021, 699, 108765. [Google Scholar] [CrossRef]
- Chen, W.; Lee, M.-K.; Jefcoate, C.; Kim, S.-C.; Chen, F.; Yu, J.-H. Fungal Cytochrome P450 Monooxygenases: Their Distribution, Structure, Functions, Family Expansion, and Evolutionary Origin. Genome Biol. Evol. 2014, 6, 1620–1634. [Google Scholar] [CrossRef] [Green Version]
- Durairaj, P.; Hur, J.-S.; Yun, H. Versatile biocatalysis of fungal cytochrome P450 monooxygenases. Microb. Cell Factories 2016, 15, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, S.; Xie, S.; Chen, H.; Cheng, Y.; Shi, Y.; Qin, X.; Dai, S.Y.; Zhang, X.; Yuan, J.S. Genomic and molecular mechanisms for efficient biodegradation of aromatic dye. J. Hazard. Mater. 2016, 302, 286–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jawallapersand, P.; Mashele, S.; Kovačič, L.; Stojan, J.; Komel, R.; Pakala, S.B.; Kraševec, N.; Syed, K. Cytochrome P450 Monooxygenase CYP53 Family in Fungi: Comparative Structural and Evolutionary Analysis and Its Role as a Common Alternative Anti-Fungal Drug Target. PLoS ONE 2014, 9, e107209. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Wu, Y.; Zhang, Y.; Yang, X.; Yang, E.; Xu, H.; Yang, Q.; Chagan, I.; Cui, X.; Chen, W.; et al. Lignin degradation potential and draft genome sequence of Trametes trogii S. Biotechnol. Biofuels 2019, 12, 256. [Google Scholar] [CrossRef] [PubMed]
- Doddapaneni, H.; Subramanian, V.; Fu, B.; Cullen, D. A comparative genomic analysis of the oxidative enzymes potentially involved in lignin degradation by Agaricus bisporus. Fungal Genet. Biol. 2013, 55, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Marinović, M.; Nousiainen, P.; Dilokpimol, A.; Kontro, J.; Moore, R.; Sipila, J.; Hildén, K. Selective Cleavage of Lignin beta-O-4 Aryl Ether Bond by beta-Etherase of the White-Rot Fungus Dichomitus squalens. ACS Sustain. Chem. Eng. 2018, 6, 2878–2882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kersten, P.; Cullen, D. Copper radical oxidases and related extracellular oxidoreductases of wood-decay Agaricomycetes. Fungal Genet. Biol. 2014, 72, 124–130. [Google Scholar] [CrossRef]
- Motoda, T.; Yamaguchi, M.; Tsuyama, T.; Kamei, I. Down-regulation of pyruvate decarboxylase gene of white-rot fungus Phlebia sMG-60 modify the metabolism of sugars and productivity of extracellular peroxidase activity. J. Biosci. Bioeng. 2019, 127, 66–72. [Google Scholar] [CrossRef]
- Mori, T.; Kako, H.; Sumiya, T.; Kawagishi, H.; Hirai, H. Direct lactic acid production from beech wood by transgenic white-rot fungus Phanerochaete sordida YK-1. J. Biotechnol. 2016, 239, 83–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kersten, P.; Cullen, D. Extracellular oxidative systems of the lignin-degrading Basidiomycete Phanerochaete chrysosporium. Fungal Genet. Biol. 2007, 44, 77–87. [Google Scholar] [CrossRef]
- Glass, N.L.; Schmoll, M.; Cate, J.H.; Coradetti, S. Plant Cell Wall Deconstruction by Ascomycete Fungi. Annu. Rev. Microbiol. 2013, 67, 477–498. [Google Scholar] [CrossRef] [PubMed]
- Courtade, G.; Wimmer, R.; Røhr, Å.K.; Preims, M.; Felice, A.K.; Dimarogona, M.; Aachmann, F.L. Interactions of a fungal lytic polysaccharide monooxygenase with beta-glucan substrates and cellobiose dehydrogenase. Proc. Natl. Acad. Sci. USA 2016, 113, 5922–5927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kameshwar, A.K.S.; Qin, W. Comparative study of genome-wide plant biomass-degrading CAZymes in white rot, brown rot and soft rot fungi. Mycology 2018, 9, 93–105. [Google Scholar] [CrossRef] [Green Version]
- De Vries, R.P.; Wiebenga, A.; Coutinho, P.M.; Henrissat, B. Plant polysaccharide degradation by fungi. In Proceedings of the 7th International Conference on Mushroom Biology and Mushroom Products, Arcachon, France, 4–7 October 2011. [Google Scholar]
- Knoch, E.; Dilokpimol, A.; Geshi, N. Arabinogalactan proteins: Focus on carbohydrate active enzymes. Front. Plant Sci. 2014, 5, 198. [Google Scholar] [CrossRef] [Green Version]
- Polizeli, M.L.T.M.; Rizzatti, A.C.S.; Monti, R.; Terenzi, H.F.; Jorge, J.A.; Amorim, D.S. Xylanases from fungi: Properties and industrial applications. Appl. Microbiol. Biotechnol. 2005, 67, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Van den Brink, J.; de Vries, R.P. Fungal enzyme sets for plant polysaccharide degradation. Appl. Microbiol. Biotechnol. 2011, 91, 1477–1492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, J.; Schäfer, D.; Eichen, N.V.D.; Haimerl, C.; Harth, S.; Oreb, M.; Benz, J.P.; Weuster-Botz, D. D-Galacturonic acid reduction by S. cerevisiae for L-galactonate production from extracted sugar beet press pulp hydrolysate. Appl. Microbiol. Biotechnol. 2021, 105, 5795–5807. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Ma, H.; Ren, S.; Gao, X.; He, X.; Zhu, S.; Deng, R.; Zhang, S. Insights into the mechanism of cyanobacteria removal by the algicidal fungi Bjerkandera adusta and Trametes versicolor. Microbiology 2020, 9, e1042. [Google Scholar] [CrossRef] [PubMed]
- Dhakar, K.; Jain, R.; Tamta, S.; Pandey, A. Prolonged Laccase Production by a Cold and pH Tolerant Strain of Penicillium pinophilum (MCC 1049) Isolated from a Low Temperature Environment. Enzym. Res. 2014, 2014, 120708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arora, D.S.; Rampal, P. Laccase production by some Phlebia species. J. Basic Microbiol. 2002, 42, 295–301. [Google Scholar] [CrossRef]
- Fricker, M.; Boddy, L.; Bebber, D. Network Organisation of Mycelial Fungi. Biol. Fungal Cell 2007, 8, 309–330. [Google Scholar] [CrossRef]
- Levasseur, A.; Lomascolo, A.; Chabrol, O.; Ruiz-Dueñas, F.J.; Boukhris-Uzan, E.; Piumi, F.; Kües, U.; Ram, A.F.J.; Murat, C.; Haon, M.; et al. The genome of the white-rot fungus Pycnoporus cinnabarinus: A basidiomycete model with a versatile arsenal for lignocellulosic biomass breakdown. BMC Genom. 2014, 15, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Janusz, G.; Czuryło, A.; Frąc, M.; Rola, B.; Sulej, J.; Pawlik, A.; Siwulski, M.; Rogalski, J. Laccase production and metabolic diversity among Flammulina velutipes strains. World J. Microbiol. Biotechnol. 2015, 31, 121–133. [Google Scholar] [CrossRef] [Green Version]
- Rinu, K.; Pandey, A.; Palni, L.M.S. Utilization of Psychrotolerant Phosphate Solubilizing Fungi under Low Temperature Conditions of the Mountain Ecosystem. In Microorganisms in Sustainable Agriculture and Biotechnology; Springer: Dordrecht, The Netherlands, 2012. [Google Scholar]
- Hölker, U.; Lenz, J. Solid-state fermentation—Are there any biotechnological advantages? Curr. Opin. Microbiol. 2005, 8, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Barrios-González, J. Solid-state fermentation: Physiology of solid medium, its molecular basis and applications. Process. Biochem. 2012, 47, 175–185. [Google Scholar] [CrossRef]
- Wang, F.; Ma, A.-Z.; Guo, C.; Zhuang, G.-Q.; Liu, C.-Z. Ultrasound-intensified laccase production from Trametes versicolor. Ultrason. Sonochem. 2013, 20, 118–124. [Google Scholar] [CrossRef]
- Dhakar, K.; Pandey, A. Laccase Production from a Temperature and pH Tolerant Fungal Strain of Trametes hirsuta (MTCC 11397). Enzym. Res. 2013, 2013, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Jaszek, M.; Miłek, J.; Żuchowski, J.; Stefaniuk, D.; Prendecka, M. Effective and complex stimulation of the biodegradation system of fungus Cerrena unicolor by rapeseed meal fermentation. Acta Biochim. Pol. 2016, 63, 549–554. [Google Scholar] [CrossRef] [Green Version]
- Osma, J.F.; Moilanen, U.; Toca-Herrera, J.L.; Rodríguez-Couto, S. Morphology and laccase production of white-rot fungi grown on wheat bran flakes under semi-solid-state fermentation conditions. FEMS Microbiol. Lett. 2011, 318, 27–34. [Google Scholar] [CrossRef]
- Li, X.; La, G.; Cheng, Q.; Wang, F.; Feng, F.; Zhang, B.; Zhang, Z. Profile of Natural Redox Mediators Production of Laccase-Producing Fungus Pleurotus ostreatus. Bull. Environ. Contam. Toxicol. 2014, 93, 478–482. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Singh, A.K.; Bilal, M.; Chandra, R. Sustainable Production of Thermostable Laccase from Agro-Residues Waste by Bacillus aquimaris AKRC02. Catal. Lett. 2021, 21, 1–17. [Google Scholar] [CrossRef]
- Meng, L.; Bai, X.; Wang, Q.; Li, X.; Zhang, S.; Wang, L.; Li, Z. Optimizing Laccase Production in Auricularia Cornea by Submerged Fermentation with Wheat Bran Extract: Applications in Decolorization of Malachite Green Dye; Research Square: Durham, NC, USA, 2021. [Google Scholar]
- Sharma, A.; Gupta, V.; Khan, M.; Balda, S.; Gupta, N.; Capalash, N.; Sharma, P. Flavonoid-rich agro-industrial residues for enhanced bacterial laccase production by submerged and solid-state fermentation. 3 Biotech 2017, 7, 200. [Google Scholar] [CrossRef] [PubMed]
- De Souza, C.G.M.; Zilly, A.; Peralta, R.M. Production of laccase as the sole phenoloxidase by a Brazilian strain of Pleurotus pulmonarius in solid state fermentation. J. Basic Microbiol. 2002, 42, 83–90. [Google Scholar] [CrossRef]
- Jasińska, A.; Góralczyk-Bińkowska, A.; Soboń, A.; Długoński, J. Lignocellulose resources for the Myrothecium roridum laccase production and their integrated application for dyes removal. Int. J. Environ. Sci. Technol. 2019, 16, 4811–4822. [Google Scholar] [CrossRef] [Green Version]
- Couto, S.R. Exploitation of a waste from the brewing industry for laccase production by two Trametes species. J. Food Eng. 2004, 64, 423–428. [Google Scholar] [CrossRef]
- Winquist, E.; Moilanen, U.; Mettälä, A.; Leisola, M.; Hatakka, A. Production of lignin modifying enzymes on industrial waste material by solid-state cultivation of fungi. Biochem. Eng. J. 2008, 42, 128–132. [Google Scholar] [CrossRef]
- Pu, Y.; Hu, F.; Huang, F.; Davison, B.H.; Ragauskas, A.J. Assessing the molecular structure basis for biomass recalcitrance during dilute acid and hydrothermal pretreatments. Biotechnol. Biofuels 2013, 6, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Behera, S.; Arora, R.; Nandhagopal, N.; Kumar, S. Importance of chemical pretreatment for bioconversion of lignocellulosic biomass. Renew. Sustain. Energy Rev. 2014, 36, 91–106. [Google Scholar] [CrossRef]
- Van Dyk, J.S.; Pletschke, B.I. A review of lignocellulose bioconversion using enzymatic hydrolysis and synergistic cooperation between enzymes–factors affecting enzymes, conversion and synergy. Biotechnol. Adv. 2012, 30, 1458–1480. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Yamaguchi, M.; Eendo, H.; Erejab, N.A.; Eohtani, M. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 2015, 6, 288. [Google Scholar] [CrossRef] [Green Version]
- Azizi, N.; Najafpour, G.; Younesi, H. Acid pretreatment and enzymatic saccharification of brown seaweed for polyhydroxybutyrate (PHB) production using Cupriavidus necator. Int. J. Biol. Macromol. 2017, 101, 1029–1040. [Google Scholar] [CrossRef]
- MacAskill, J.; Suckling, I.; Lloyd, J.; Manley-Harris, M. Unravelling the effect of pretreatment severity on the balance of cellulose accessibility and substrate composition on enzymatic digestibility of steam-pretreated softwood. Biomass-Bioenergy 2018, 109, 284–290. [Google Scholar] [CrossRef]
- Wang, K.; Yang, H.; Wang, W.; Sun, R.-C. Structural evaluation and bioethanol production by simultaneous saccharification and fermentation with biodegraded triploid poplar. Biotechnol. Biofuels 2013, 6, 42. [Google Scholar] [CrossRef] [Green Version]



| Feature Items | Statistics |
|---|---|
| Contig number | 58 |
| Length of genome assembly | 42,794,305 bp |
| C + G content | 47.43% |
| N50 | 2,483,375 bp |
| Longest contig | 4,365,256 bp |
| Shortest contig | 33,827 bp |
| Average (bp) | 737,832.84 bp |
| Number of protein-coding genes (CDS) | 12,277 |
| Total CDS length | 21,069,806 bp |
| Average CDS length (bp) | 1716.2 bp |
| rRNA number | 49 |
| tRNA number | 230 |
| other ncRNA number | 26 |
| Enzymes | Day | Corn Stalk | Wheat Straw | Rice Straw | Pine Bark |
|---|---|---|---|---|---|
| Laccase | 3 | ++ | ++ | ++ | ++ |
| 6 | +++ | ++ | ++ | ++ | |
| 9 | ++++ | +++ | ++ | ++ | |
| 12 | ++++ | +++ | ++++ | ++ | |
| 15 | ++++ | +++ | +++ | ++ | |
| 18 | ++++ | +++ | +++ | ++ | |
| MnP | 3 | ++ | + | +/− | − |
| 6 | ++ | ++ | +/− | − | |
| 9 | ++ | ++ | +/− | +/− | |
| 12 | ++ | ++ | ++ | − | |
| 15 | ++ | ++ | ++ | − | |
| 18 | ++ | ++ | ++ | +/− | |
| LiP | 3 | + | +/− | +/− | +/− |
| 6 | + | + | + | +/− | |
| 9 | + | + | + | +/− | |
| 12 | +/− | + | +/− | + | |
| 15 | + | + | +/− | + | |
| 18 | +/− | +/− | + | +/− | |
| CMCase | 3 | ++ | ++ | ++ | + |
| 6 | ++ | ++ | ++ | + | |
| 9 | ++ | ++ | + | + | |
| 12 | ++ | ++ | ++ | + | |
| 15 | ++ | ++ | ++ | + | |
| 18 | ++ | ++ | ++ | + | |
| FPase | 3 | + | ++ | + | + |
| 6 | ++ | + | + | + | |
| 9 | + | + | + | + | |
| 12 | + | + | + | + | |
| 15 | + | + | + | + | |
| 18 | + | + | + | + |
| Enzyme (U/g) | Corn Stalk | Wheat Straw | Rice Straw | Pine Bark |
|---|---|---|---|---|
| Lac | 177.26 ± 34.33 (9th day) | 15.88 ± 3.83 (15th day) | 113.36 ± 37.05 (12th day) | 8.91 ± 1.44 (6th day) |
| MnP | 7.04 ± 1.62 (6th day) | 2.69 ± 0.47 (9th day) | 6.53 ± 2.06 (15th day) | 0.07 ± 0.06 (9th day) |
| LiP | 0.73 ± 0.19 (9th day) | 0.33 ± 0.04 (12th day) | 0.68 ± 0.08 (6th day) | 0.43 ± 0.24 (12th day) |
| CMCase | 7.14 ± 1.85 (3rd day) | 7.88 ± 1.75 (3rd day) | 6.10 ± 2.70 (15th day) | 0.22 ± 0.01 (6th day) |
| FPA | 1.10 ± 0.29 (6th day) | 1.76 ± 0.35 (3rd day) | 0.38 ± 0.16 (3rd day) | 0.52 ± 0.11 (3rd day) |
| Final Consumption (%) | Corn Stalk | Wheat Straw | Rice Straw | Pine Bark |
|---|---|---|---|---|
| Total weight loss | 28.52 ± 2.58 | 24.76 ± 0.81 | 20.82 ± 0.98 | 9.23 ± 0.14 |
| Lignin loss | 53.82 ± 3.02 | 46.72 ± 0.93 | 27.34 ± 4.72 | 10.45 ± 1.23 |
| Cellulose loss | 43.45 ± 0.80 | 33.76 ± 3.64 | 36.89 ± 3.39 | 10.51 ± 1.57 |
| Hemicellulose loss | 53.90 ± 1.11 | 44.00 ± 3.83 | 31.88 ± 3.91 | 16.38 ± 2.90 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Shah, A.M.; Mohamed, H.; Zhang, Y.; Tsiklauri, N.; Song, Y. Genomic Studies of White-Rot Fungus Cerrena unicolor SP02 Provide Insights into Food Safety Value-Added Utilization of Non-Food Lignocellulosic Biomass. J. Fungi 2021, 7, 835. https://doi.org/10.3390/jof7100835
Zhang Z, Shah AM, Mohamed H, Zhang Y, Tsiklauri N, Song Y. Genomic Studies of White-Rot Fungus Cerrena unicolor SP02 Provide Insights into Food Safety Value-Added Utilization of Non-Food Lignocellulosic Biomass. Journal of Fungi. 2021; 7(10):835. https://doi.org/10.3390/jof7100835
Chicago/Turabian StyleZhang, Zichen, Aabid Manzoor Shah, Hassan Mohamed, Yao Zhang, Nino Tsiklauri, and Yuanda Song. 2021. "Genomic Studies of White-Rot Fungus Cerrena unicolor SP02 Provide Insights into Food Safety Value-Added Utilization of Non-Food Lignocellulosic Biomass" Journal of Fungi 7, no. 10: 835. https://doi.org/10.3390/jof7100835
APA StyleZhang, Z., Shah, A. M., Mohamed, H., Zhang, Y., Tsiklauri, N., & Song, Y. (2021). Genomic Studies of White-Rot Fungus Cerrena unicolor SP02 Provide Insights into Food Safety Value-Added Utilization of Non-Food Lignocellulosic Biomass. Journal of Fungi, 7(10), 835. https://doi.org/10.3390/jof7100835







