1. Introduction
The fungal genus
Coccidioides contains two species:
Coccidioides immitis and
Coccidioides posadasii, which are the etiological agents of coccidioidomycosis (Valley Fever; VF).
Coccidioides spp. are dimorphic ascomycetes of the family Onygenaceae existing in a mycelial form in the soil (saprobic phase) and a yeast-like spherule form (parasitic phase) in infected mammalian hosts [
1]. There is no single serological assay for VF that serves as an ideal diagnostic assay, and all available methods have moderate sensitivity for detection of antibodies in the first few weeks. Sensitive detection of specific
Coccidioides spp. antigens released from spherules in the acute phase of Valley Fever infection (VF) could be the ideal approach for diagnosis of VF. Detection of antigen in a body fluid would allow for early and definitive laboratory diagnosis of VF. However, the only antigen assay commercially available has moderate sensitivity, cross-reacts with other fungi, and must be sent to a reference laboratory [
2]. Thus, there is much work to be done to truly advance the goal of early and accurate diagnosis of coccidioidomycosis.
With over 3500 plasma proteins in the Human Proteome Organization (HUPO) human plasma proteome database [
3,
4] and an estimated >500 proteins circulating at any one time in human plasma [
5], identification of low abundance biomarker proteins among high abundance plasma proteins like albumin and immunoglobulins is a difficult challenge in mass spectrometry-based biomarker discovery [
5,
6]. To be able to identify low abundance proteins, enrichment using selective antibodies or deletion of highly abundant proteins from serum using other affinity reagents are frequently employed options. However, in order to identify the correct affinity enrichment reagent or to produce a selective antibody, one must identify and produce target biomarkers.
Identification of proteins from pure cultures of an infectious microorganism growing in vitro is not difficult. However, the most abundant proteins produced during in vitro culture may not be fully representative of the in vivo proteome of organisms, particularly in the case of a dimorphic fungus such as
Coccidioides. Protein production is known to be influenced by a number of host pressures not available in vitro (immune cells, adhesion to host tissues, hypoxia, nutrient gradients/deprivation, osmotic stress, etc.) These factors likely explain the significant differences in phenotype, virulence, and survival that have been noted between fungi grown in vivo versus in vitro [
7,
8]. Additionally, even different in vitro culture media formulations can cause significant phenotypic changes in
Coccidioides spp. [
9].
One biomarker discovery platform which is able to evaluate protein abundance in vivo is laser capture microdissection (LCM) followed by mass spectrometry. LCM facilitates the sampling of selected cellular structures and regions from ex vivo tissues. Here we describe and reveal results of an LCM-assisted label-free quantitative proteomic technique for the identification and relative quantification of VF protein biomarkers from extracted Coccidioides spherules from lung biopsies. In an effort to identify in vitro growth conditions that recapitulate growth conditions in human lungs, protein identifications and abundances from in vivo spherules were compared to the protein abundances produced from in vitro-grown spherules and mycelia in different media and conditions.
2. Materials and Methods
2.1. Coccidoides spp.-Infected Tissue Samples
Triplicate technical replicates of archived formalin-fixed paraffin embedded (FFPE) lung tissue blocks from each of 3 naturally infected human clinical cases were used in this study. Infections had been culture-confirmed cases of Coccidioides spp., and all three patients were immunosuppressed (2 HIV positive individuals and 1 treated with adalimumab). Tissue blocks were acquired from Mayo Clinic Arizona histology biobank in accordance with IRB# 12-000965 (human tissue). Ten micrometer thick sections were cut using a microtome and transferred to 1 mm polyethylene naphthalate (PEN) membrane slides (Carl Zeiss Microscopy; Jena, Germany), deparaffinized and stained with hematoxylin and eosin stain (H&E) using standard procedures.
2.2. LCM of FFPE Tissues
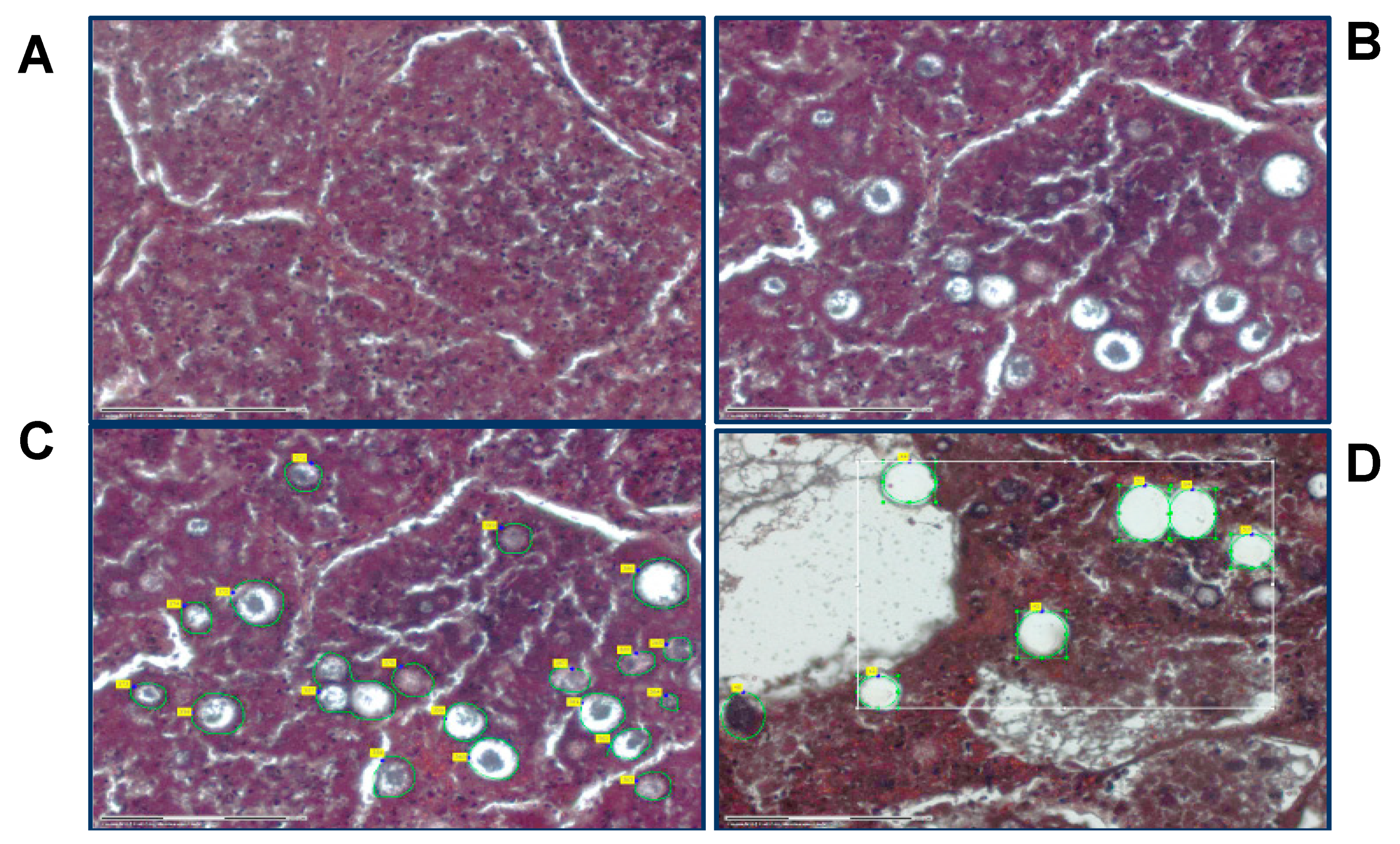
Laser capture microdissection (LCM) was performed using a Zeiss PALM MicroBeam scope with RoboPalm software (Carl Zeiss Microscopy). Approximately 500,000 μm
2 area of spherules in lung tissue was collected for each sample by laser capture. Tissue features (spherules) were selected, collected and catapulted into the cap of 0.5 mL Eppendorf tubes containing 35 µL of 0.1 M Tris-HCl (pH = 8.0), 0.002% Zwittergent Z3-16 (MilliporeSigma, Burlington, MA, USA) via laser pressure catapulting (
Figure 1). Immediately after capture, the tube was centrifuged at 14,000×
g for 2 min to collect the lysis solution and tissue into the bottom of the tube and was frozen at −80 °C until processing.
2.3. In-Solution Protein Digestion
Formalin–protein crosslinks were broken from the tissue fragments by heating the sample at 99 °C for 1 h. Proteins were reduced by the addition of 1.8 µL of 200 mM tris(2-carboxyethyl)phosphine (TCEP, MilliporeSigma) in 0.1 mM Tris, pH 8.2, (final conc. TCEP = 10 mM) and incubated at 56 °C for 30 min. Afterwards, proteins were alkylated by the addition of 1.9 µL of 200 mM iodoacetamide (IAA, MilliporeSigma) in 0.1 M Tris-HCl (final conc. = 10 mM) and incubated in the dark at RT for 30 min. Finally, proteins were digested by adding 2 μL of 0.01 µg/µL trypsin (Thermo, Waltham, MA, USA) and incubated at 37 °C for 16 h. The digestion was terminated by adding 4 μL of 2% formic acid (FA) prior to loading on the mass spectrometer. Loading was normalized to the area of tissue, optimally 500,000 square microns per sample provides ~500 ng protein.
2.4. Fungal Culture Preparation and Protein Lysis
The mycelial form of
Coccidioides posadasii strain Silveira (strain kindly provided by Dr. Bridget Barker of the University of Arizona) was grown in a shake flask rotating at 180 RPM in 30 °C for 7 days in one of the following six liquid medias: (1) Levine modification of Converse media [
10] with 0.5% N-Tamol (Dow Chemical Company, Midland MI), (2) RPMI-1640 medium (Corning) with 10% fetal bovine serum (FBS) or (3) RPMI-1640 medium with 0.1% Survanta (Beractant bovine lung surfactants; Abbot Laboratories, Chicago, IL, USA).
Spherule form of C. posadasii strain Silveira cultures were grown in vented shake flasks rotating at 180 RPM in 20% CO2 at 40 °C for 7 days in the same three media as above. Cultures were centrifuged at 4500 RPM for 30 min. in a Thermo Forma Multi RF tabletop centrifuge (Thermo). Supernatants were then sterilized by filtration through 0.45µm vacuum filtration units (MilliporeSigma). Culture pellets were resuspended 5:1 in fungal lysis buffer (50 mM Tris, pH 7.6, 100 mM NaCl, 50 mM EDTA, 5% SDS) prior to transfer into 1.5 mL silicone washer-sealed internally threaded cryovials containing ~350 µL of 0.5 mm acid-washed sterile glass beads. Spherule culture pellets in cryovials were subjected to two rounds of bead beating at maximum speed for 15 min followed by 3 cycles of flash freezing on dry ice. Sterility/viability was checked by plating 10% of the total volumes onto 2× glucose yeast extract agar (2 × GYE) for 3 weeks at RT.
2.5. Fungal Culture Protein Extraction and In-Gel Protein Digestion
Fungal culture proteins from filtered supernatants and pellet lysates were extracted using a modification of a previously published protocol [
11]. In brief, supernatants and pellets were centrifuged for 30 min. at 8000 RPM at 4 °C. The supernatants were put into fresh tubes and proteins were precipitated by the addition of 4 volumes of ice cold 10%
w/v trichloroacetic acid (TCA, MilliporeSigma) in acetone with 0.007%
w/v dithiothreitol (DTT, G Biosciences, St. Louis, MO, USA). Samples were centrifuged at 3000×
g for 10 min., and the resultant protein pellets were washed three times in ice cold acetone with 0.007% DTT. The final pellet was then resuspended in rehydration buffer [7 M urea, 2 M Thiourea, 4%
w/v 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate hydrate (CHAPS, all MilliporeSigma) and 20 mM DTT].
Samples in rehydration buffer were then filter exchanged with phosphate buffered saline (PBS), using Amicon® Ultra 0.5 mL 3 kDa filtration units (MilliporeSigma) prior to protein estimation using a micro bicinchoninic acid (BCA) protein assay kit (Thermo), and ~10 µg of protein from the supernatant samples and ~30 µg of corresponding pellet proteins were mixed with reducing Laemmli sample buffer (BioRad, Hercules, CA, USA), heated at 95 °C for 5 min. prior to loading on mini-Protean™ TGX precast gels (BioRad). Therefore, a total of 40 µg of combined supernatant and pellet proteins per sample were run in each sample lane. Following electrophoresis, gels were stained with Bio-Safe Coomassie G-250 Stain (BioRad) as per manufacturer’s instructions. Each sample lane of the SDS-PAGE gel was cut into six equal size slices. Band slices were then further reduced into cubes of 1–2 mm3 and put into low protein binding tubes (Eppendorf, Hamburg, Germany) prior to destaining. Proteins were reduced in 10 mM DTT for 30 min. at 60 °C, and alkylated with 55 mM ioadacetamide (IAA, MilliporeSigma) for 30 min. at room temperature in the dark, prior to 37 °C overnight digestion with Pierce™ MS Grade trypsin protease (Thermo) diluted to 20 ng/mL in 100 mM ammonium bicarbonate (MilliporeSigma). The peptides were then extracted from the gel pieces using 5% FA, ammonium bicarbonate and acetonitrile washes prior to being dried in a speed vacuum and stored at −80 °C until LC-MS analysis.
2.6. Proteomic Analysis of Fungal Pellets and Supernatants
Protein digests were reconstituted in 0.1% FA and analyzed using LC-MS/MS by loading onto a Dionex UltiMate® 3000 RSLC liquid chromatography (LC) system (Thermo, San Jose, CA, USA) using a PepMap RSLC C18 2 um, 75 um × 50 cm EASY-Spray™ column (Thermo). Peptides were separated using a 0.3 uL/min LC gradient comprised of 2–90% mobile phase B in 0–120 min. Mobile phase A and B were 0.1% FA in water and acetonitrile, respectively. Eluting peptides were directly injected into an Orbitrap Elite Velos mass spectrometer (Thermo) and ionized using collision-induced dissociation (CID) in positive ion mode. A “top 15″ data-dependent MS/MS analysis was performed (acquisition of a full scan spectrum followed by collision-induced dissociation mass spectra of the 15 most abundant ions in the survey scan).
2.7. Protein Identification and Label-Free Protein Quantification
Database searching was performed using Sequest (Thermo) in Proteome Discoverer v1.4.1.14 (Thermo) against a combined FASTA database of all the most recent Uniprot Coccidioides spp. proteomes (Coccidioides immitis RS, proteome ID: UP000001261, 12 June 2018 release date; Coccidioides immitis RMSCC 3703, proteome ID:UP000054559, 26 February 2018 release date; Coccidioides immitis H538.4, proteome ID: UP000054563, 26 February 2018 release date; Coccidioides immitis RMSCC 2394, UP000054565, 26 February 2018 release date; Coccidioides posadasii strain RMSCC 757/Silveira, proteome ID: UP000002497, 26 February 2018 release date, Coccidioides posadasii C735, proteome ID: UP000009084, 9 November 2018 release date; and Coccidioides posadasii RMSCC 3488, UP000054567, 26 October 2018 release date). Searches were performed using a fragment tolerance of 0.60 Da (Monoisotopic), parent tolerance of 10 ppm (Monoisotopic), with carbamidomethyl of cysteine as fixed and oxidation of methionine as variable modifications with maximum missed cleavages allowed of 2. Protein identifications were accepted if they achieved a minimum of 2 peptides per protein and a false discovery rate (FDR) of <1%. Label-free protein quantification was performed using normalization of spectral abundance factors (NSAF) in Scaffold (v4.8.7, Proteome Software Inc., Portland, OR, USA).
2.8. Biomarker Identification and Bioinformatics
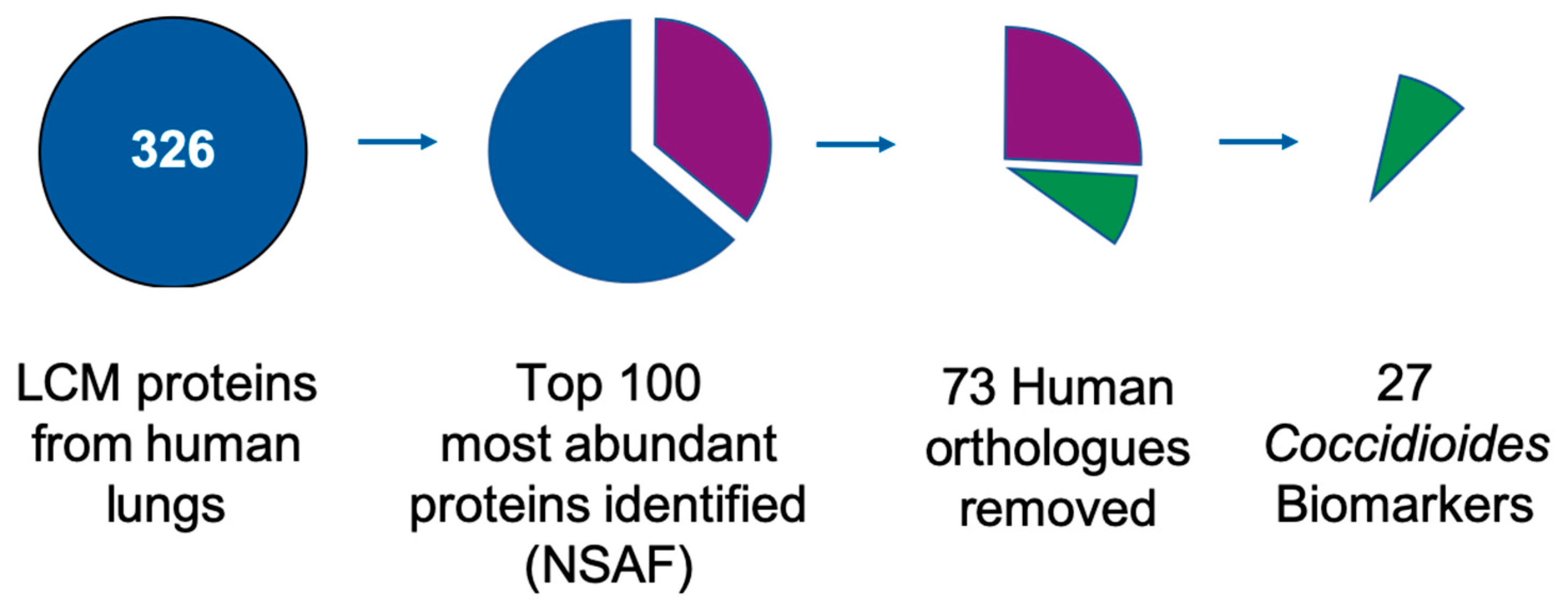
The top 100 most abundant
Coccidioides spp. proteins found in LCM lung tissues from the mean of triplicate technical replicates were identified. These “top 100” most abundant Coccidioidal proteins were subjected to a pBLAST search (
www.blast.ncbi.nlm.nih.gov) against the
Homo sapiens proteome (taxid:9606), to exclude human orthologues. An independent analysis was performed to detect
Coccidioides spp. peptides found in the LCM lung tissues and assess their homology against any peptides found in human and non-
Coccidioides spp. fungi proteins present in NCBI non-redundant (NR) database. Detected peptides were filtered to retain
Coccidioides spp. peptides that do not share 100% homology with either human or non-
Coccidioides spp. fungi proteins (
Supplementary Table S2). These peptides could be utilized to develop targeted mass spectrometry-based assays for detecting the presence of
Coccidioides spp. in human biological samples.
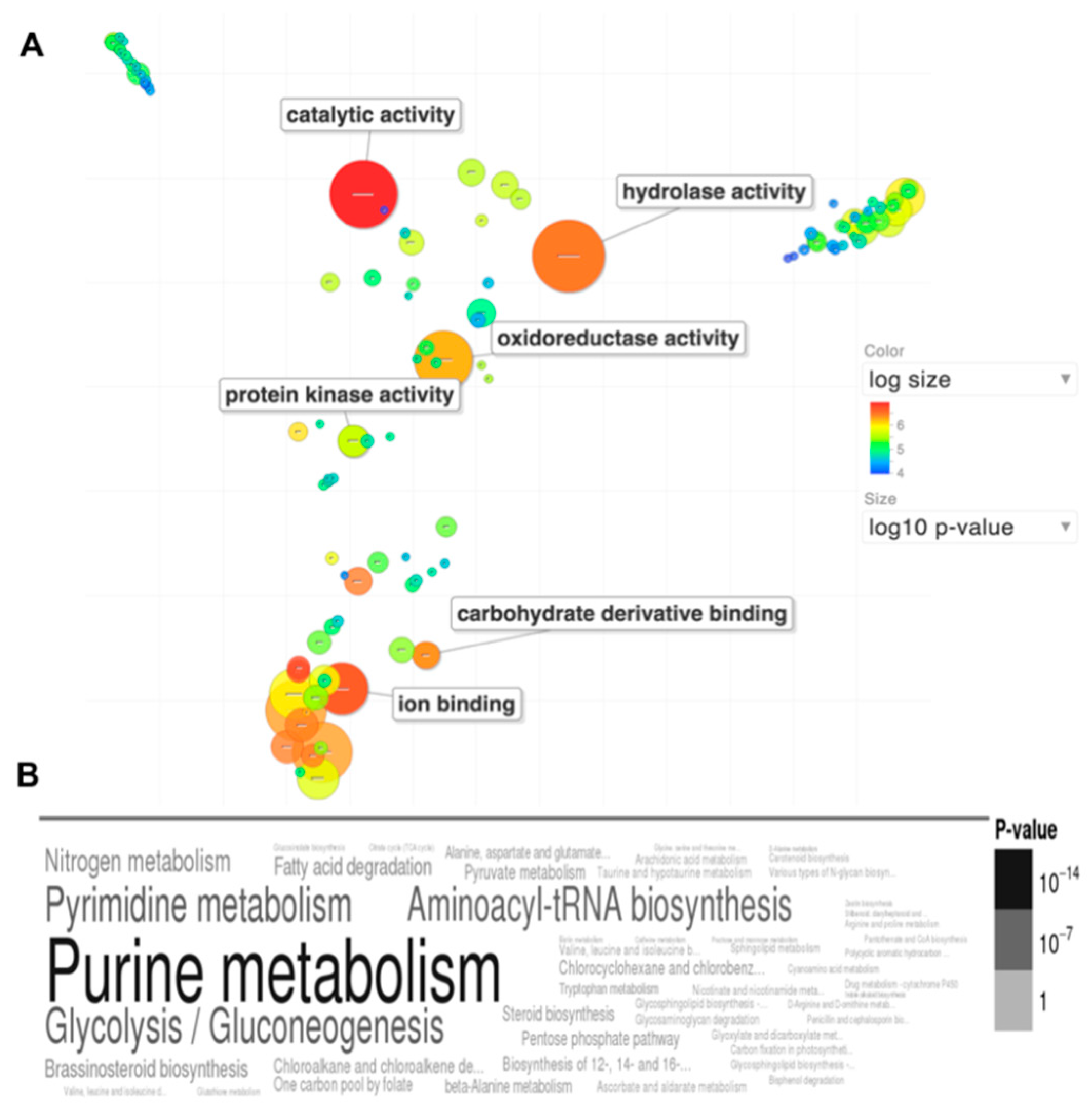
Gene ontology identifiers of each remaining protein were pulled from Uniprot (
www.uniprot.org). A gene ontology enrichment scatterplot was produced using the Revigo [
12] plugin in FungiDB (
www.fungidb.org) [
13]. The wordcloud of enriched KEGG metabololic pathways was produced using the GOSummaries [
14] plugin, also from FungiDB. Protein O-glycosylation predictions were made using NetOGlyc server 4.0 [
15]. N-glycosylation and signal peptide predictions were determined using NetNGlyc 1.0 server [
16].
2.9. Statistical Analyses
GraphPad Prism v.8.0 was used for production and statistical analyses, including those used in creating volcano plots and box and whisker plots. Volcano plots were created using multiple t tests of Normalized Spectral Abundance Factor (NSAF), corrected for multiple comparisons using Holm–Sidak method. NSAF is a unitless, arbitrary value used to rank abundance of proteins across samples and experiments [
17]. Analysis of in vivo versus in vitro grown spherule abundances was calculated using a one-way ANOVA followed by Tukey test and adjusted for multiple comparisons using Sidak’s method.
4. Discussion
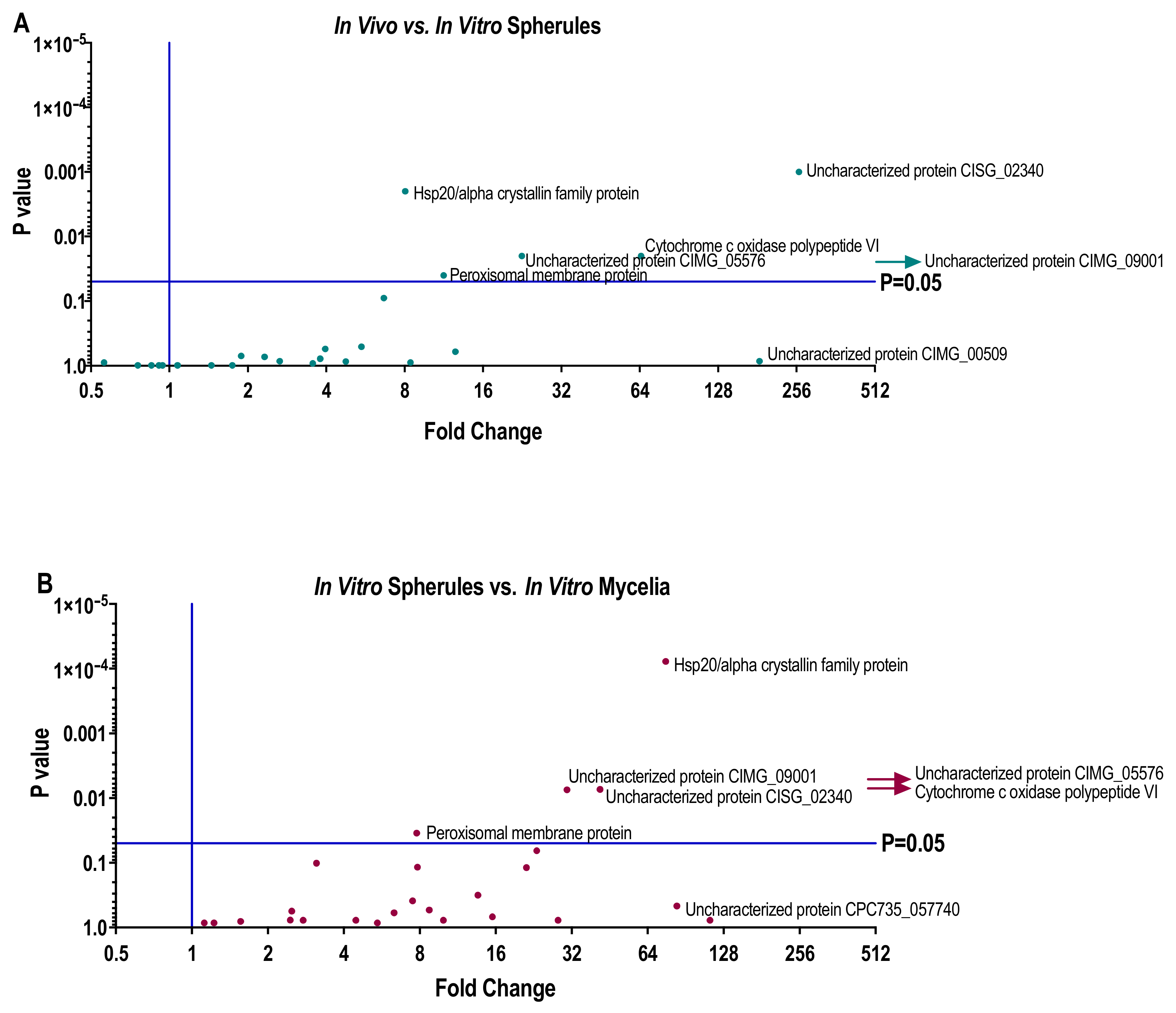
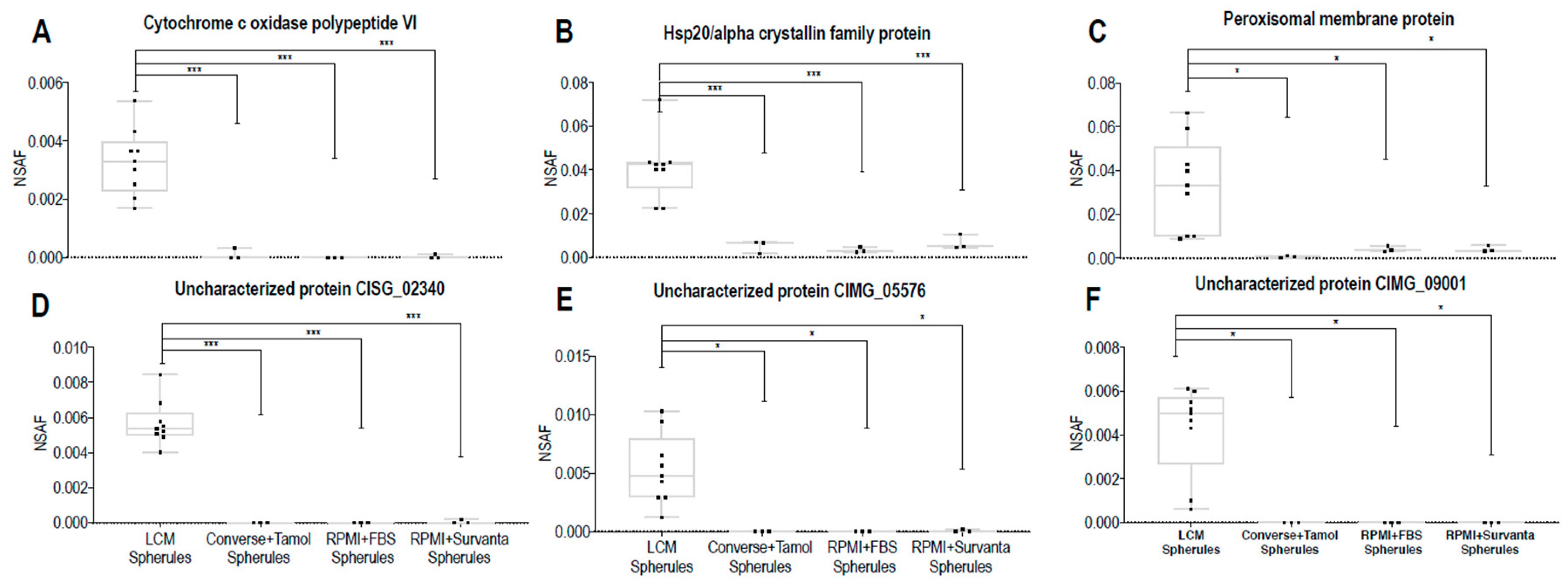
In this study, biomarker candidate proteins were identified from Coccidioides spp.-infected lung tissue using LCM coupled with mass spectrometry. The 100 most abundant proteins in LCM-captured spherules were identified, and their expression in vitro was measured in three different culture media conditions. Of the one-hundred, 73 human orthologues sharing ≥40% identity to human proteins were removed, leaving 27 possible Coccidioides spp. biomarker proteins. All 27 (100%) of the biomarker candidate proteins were expressed in vitro in at least one growth phase-medium combination; however, protein abundance depended on both growth phase and medium type. Additionally, three of the 27 biomarker candidates were found to be Coccidioides-specific biomarkers (Uncharacterized proteins CIMG_00509, CIMG_05576, CIMG_09001), as they do not share significant sequence identity to other fungal lung pathogens. Subsequent studies may help characterize the functions of these proteins and further determine if they are unique among all fungal pathogens.
Limitations of this study include the low number (n = 3) different human lungs tissue from which we performed LCM-mass spectrometry. Although this number is low, it likely represents more host-pathogen interaction diversity than examining infected lungs from 12 inbred mice. To date, there has only been one other proteomic evaluation of
Coccidioides spp. proteins from in vivo produced samples. In this study by Lewis et al. (2015), who evaluated
Coccidoides spp. proteins collected from bronchiolar lavage fluid (BALF) of laboratory-infected mice [
18], only 8
Coccidioides spp. proteins were identified. In fact, all 8 of the
Coccidioides spp. proteins identified in BALF were also identified in our LCM lung tissue dataset. Remarkably, 2 of the 8 (25%) BALF proteins they identified (Peroxisomal membrane protein and Uncharacterized protein CIMG_09001) were in our 27 candidate biomarkers dataset. However, the other 6 proteins they identified in BALF either shared too much identity with human proteins or were not in the top 100 most abundant proteins from LCM lung tissues. It is interesting that Uncharacterized protein CIMG_09001, which we identified as a
Coccidioides-specific biomarker candidate, was produced in BALF. This data gives credence to the LCM-assisted biomarker discovery methodology used in this study.
Barring the Lewis et al. study, all other published proteomic studies of
Coccidioides spp. have evaluated proteins produced from in vitro cultures, which are not truly representative of in vivo expression [
4,
19,
20,
21]. Additionally, all previous studies used only one culture medium: Converse media supplemented with Tamol. Interestingly, of the 27 biomarker candidates identified in this study as abundantly produced in vivo, 4 (14.9%) were not produced in the spherule form, and 9 (33.3%) were not produced in the mycelial form, when grown in Converse with Tamol in vitro. Previous studies which only used in vitro growth in Converse are likely to have missed these proteins. Other factors which may result in different findings include species and provenance of the
Coccidioides isolate, as well as protein extraction methods employed.
One of the aims of this study was to find a single growth medium which causes spherule protein expression to most closely resemble in vivo growth. However, none of the media tested closely mimics in vivo growth as spherules. Different in vitro culture media and conditions allowed for different subsets of
Coccidioides proteins to be produced. Regardless, of the 3 media evaluated in this study, the maximum number of the 27 biomarker candidates was produced using RPMI+ Survanta (26/27; 96.3%) in spherule cultures. RPMI media supplemented with Survanta was recently published by our group for in vitro culture of
Coccidioides spp. [
22]. We chose Survanta as an alternative surfactant to Tamol, which is a synthetic anionic surfactant made from sulfonic acid salts, more commonly used as a paint dispersant. Tamol has been shown to induce faster spherulation in in vitro culture and cause reversion of mycelia into spherules [
23,
24]. Although the exact mechanism of Tamol-induced spherulation is not entirely known, it is possibly due to its ability to emulsify media components and reduce surface tension around the spherule cell walls. Survanta is a natural bovine lung extract used in the prevention and treatment of respiratory distress syndrome in premature infants. In addition to containing surfactant proteins B and C (SP-B and SP-C), it contains phospholipids, neutral lipids, fatty acids, and surfactant-associated proteins which mimic the surface-tension lowering properties of natural lung surfactant. Additionally, as a working solution of 0.1% Survanta in RPMI, it contains only 0.1 mg/mL protein, and is much more compatible with mass spectrometry than RPMI supplemented with 10% FBS. While RPMI with Survanta is not the ideal in vitro proxy to represent spherule growth in the lung, our results in protein expression and glycan expression [
22] suggest that further investigation of media types could yield further insights into the biology of
Coccidioides spp.
In vivo grown spherules also produced protein abundances that differed from in vitro grown spherules. Peroxisomal matrix protein, Hsp20/alpha crystallin family protein, Cytochrome c oxidase polypeptide VI, Uncharacterized protein CISG_02340, Uncharacterized protein CIMG_09001, and Uncharacterized protein CIMG_05576 were all significantly more abundant in vivo. While the function of the Uncharacterized proteins is unknown, small heat shock proteins like the Hsp20/alpha crystallin family protein have been implicated in the pathogenesis of the plant fungal pathogen
Ustilago maydis [
25], as well as the pathogenicity and persistence of the bacterial lung pathogen
Mycobacterium tuberculosis [
26]. Peroxisomal matrix protein, in addition to being found in mouse BALF, has also been investigated as a recombinant vaccine candidate for VF [
4,
27]. Although it showed protection against intraperitoneal challenge, only modest protection was afforded following high dose intranasal challenge. In addition to being biomarker candidates, any of the other 27 proteins have the potential to also be vaccine candidates or targets for therapeutic drugs.
Taken together, the findings reported in this study suggest that Coccidioidal protein expression in vivo is distinct from Coccidioidal protein expression in vitro amongst its two growth phases in each of 3 different culture media. Furthermore, these findings form a foundation upon which to select relevant biomarkers for antigen-based detection of Coccidioides in potentially infected patients.