The High Osmolarity Glycerol (HOG) Pathway Functions in Osmosensing, Trap Morphogenesis and Conidiation of the Nematode-Trapping Fungus Arthrobotrys oligospora
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Deletion of the KU70, HOG1 and MSB2 Homologs in A. oligospora
2.2. Generation of Target Gene Deletion Mutants
2.3. Phenotypical Characterization
2.4. Statistics
3. Results
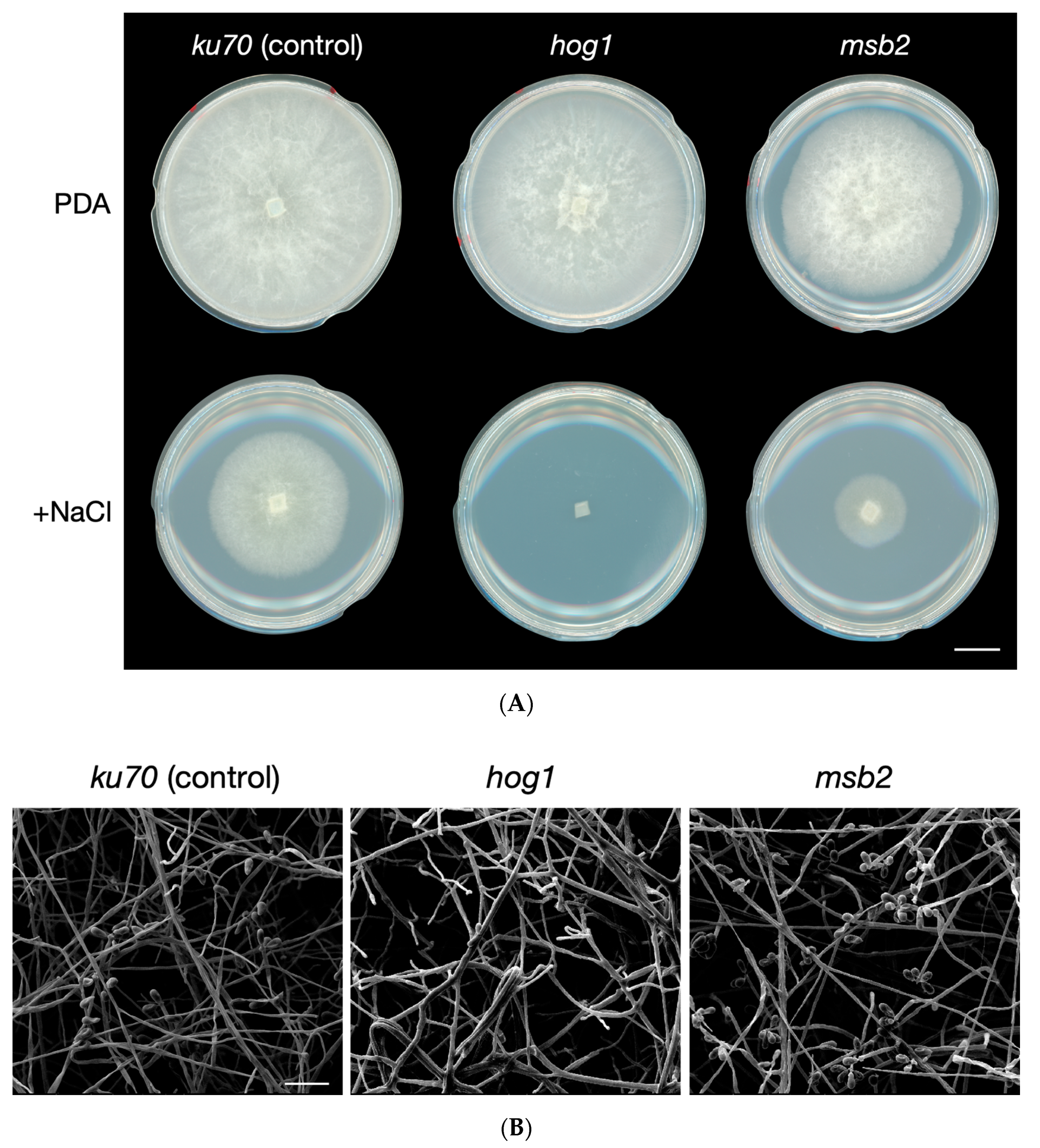
3.1. Identification of the KU70 Gene in A. oligospora
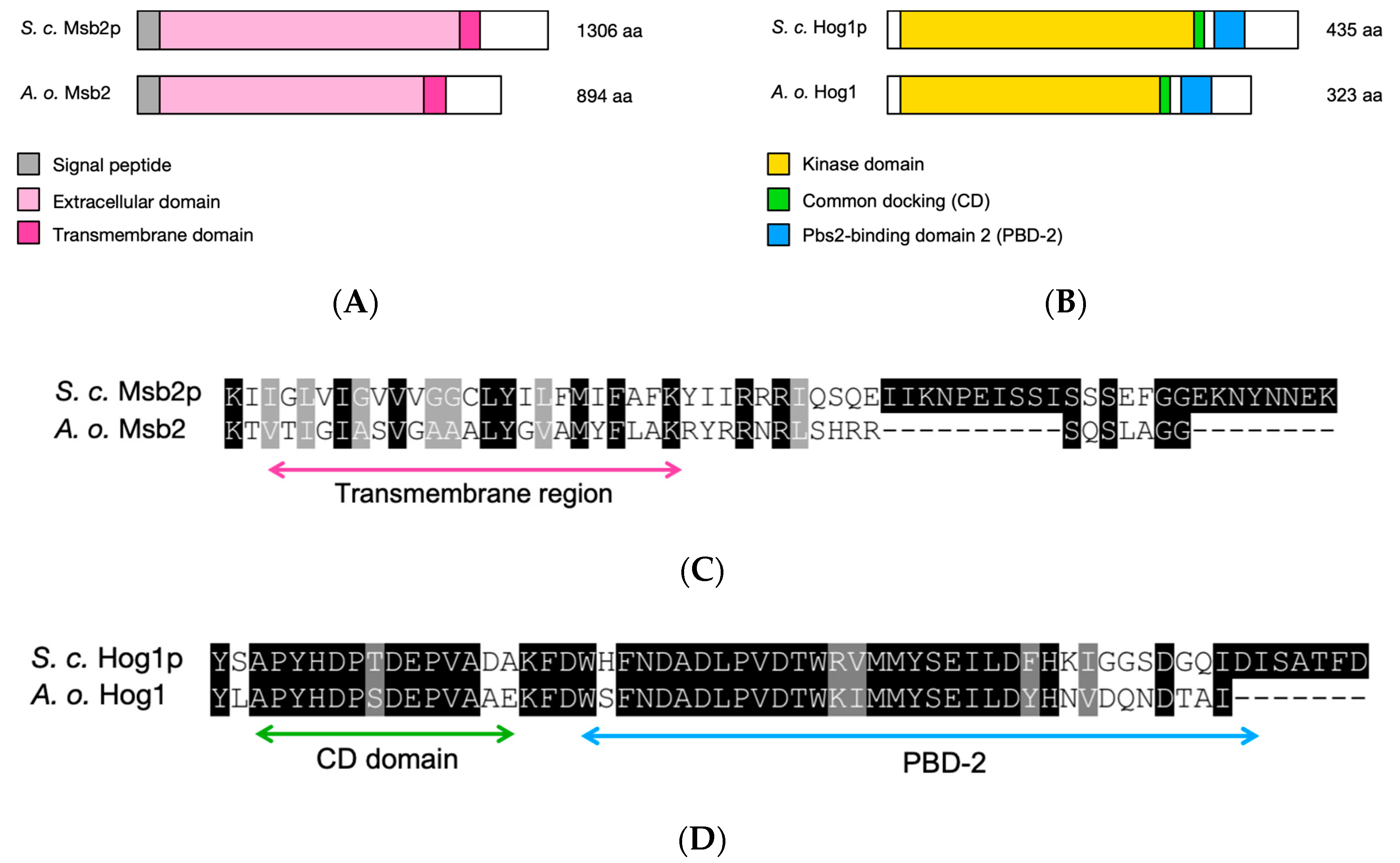
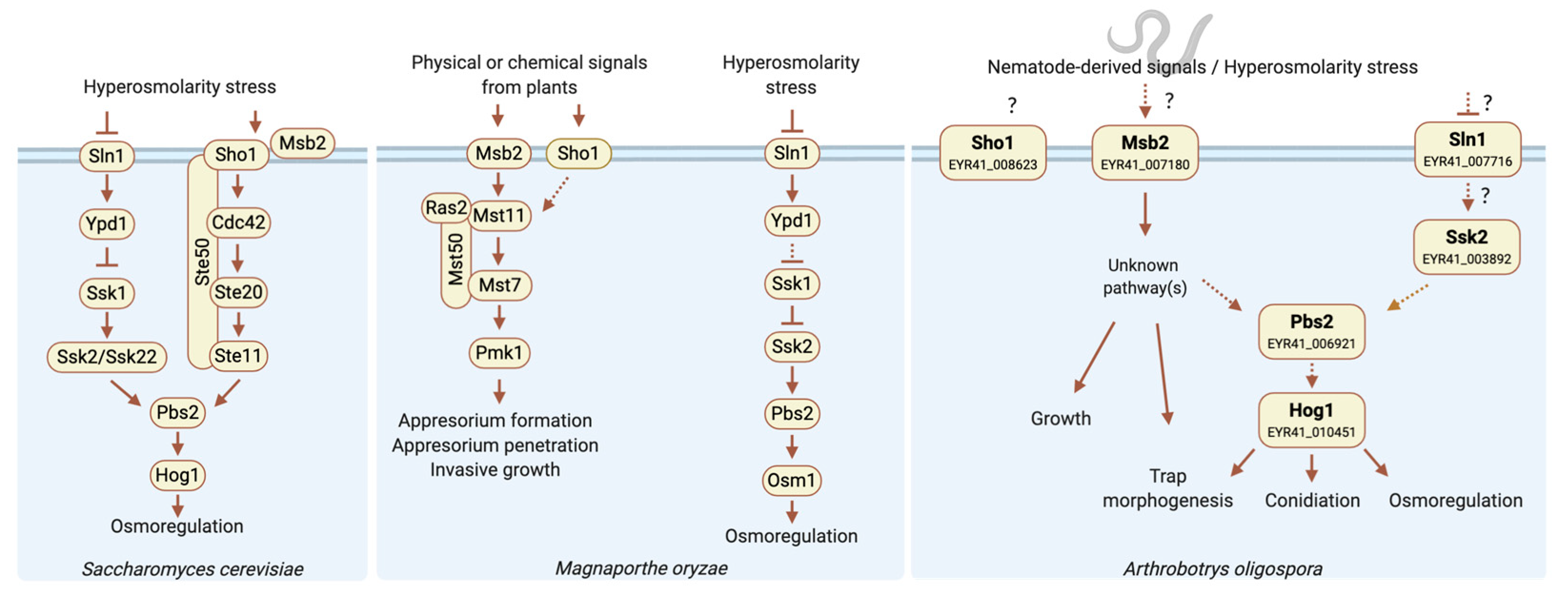
3.2. Identification of the HOG1 and MSB2 Genes in A. oligospora Suggests a Conserved Role in Osmosensing
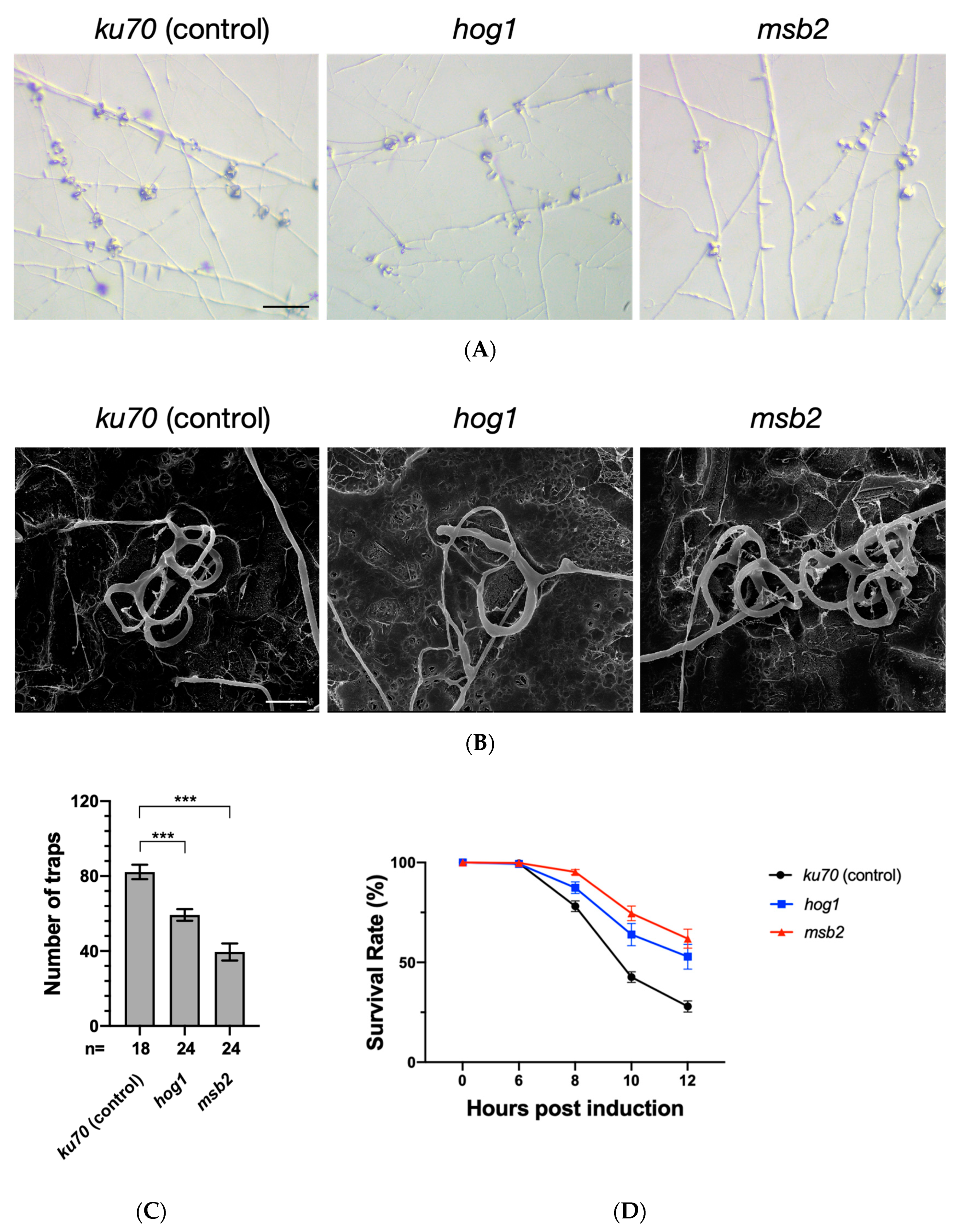
3.3. The Hog1 Pathway Is Required for Proper Formation of Traps during Nematode Induction
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Reed, R.H.; Chudek, J.; Foster, R.; Gadd, G. Osmotic significance of glycerol accumulation in exponentially growing yeasts. Appl. Environ. Microbiol. 1987, 53, 2119–2123. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Zhang, X.; Liu, H.; Xu, J.-R. Mitogen-activated protein kinase signaling in plant pathogenic fungi. PLoS Pathog. 2018, 14, e1006875. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Zhang, S.; Zhou, X.; Wang, C.; Xiang, P.; Zheng, Q.; Xu, J.-R. The FgHOG1 pathway regulates hyphal growth, stress responses, and plant infection in Fusarium graminearum. PLoS ONE 2012, 7, e49495. [Google Scholar] [CrossRef] [PubMed]
- Heller, J.; Ruhnke, N.; Espino, J.J.; Massaroli, M.; Collado, I.G.; Tudzynski, P. The mitogen-activated protein kinase BcSak1 of Botrytis cinerea is required for pathogenic development and has broad regulatory functions beyond stress response. Mol. Plant Microbe Interact. 2012, 25, 802–816. [Google Scholar] [CrossRef]
- Kreuzwieser, J.; Scheerer, U.; Kruse, J.; Burzlaff, T.; Honsel, A.; Alfarraj, S.; Georgiev, P.; Schnitzler, J.-P.; Ghirardo, A.; Kreuzer, I. The Venus flytrap attracts insects by the release of volatile organic compounds. J. Exp. Bot. 2014, 65, 755–766. [Google Scholar] [CrossRef]
- Cooke, R.C. Ecological characteristics of nematode-trapping Hyphomycetes: I. Preliminary studies. Ann. Appl. Biol. 1963, 52, 431–437. [Google Scholar] [CrossRef]
- Pramer, D. Nematode-trapping fungi. Science 1964, 144, 382–388. [Google Scholar] [CrossRef]
- Tunlid, A.; Jansson, H.-B.; Nordbring-Hertz, B. Fungal attachment to nematodes. Mycol. Res. 1992, 96, 401–412. [Google Scholar] [CrossRef]
- Pearson, G.; Robinson, F.; Beers Gibson, T.; Xu, B.-E.; Karandikar, M.; Berman, K.; Cobb, M.H. Mitogen-activated protein (MAP) kinase pathways: Regulation and physiological functions. Endocr. Rev. 2001, 22, 153–183. [Google Scholar]
- Zhao, X.; Mehrabi, R.; Xu, J.-R. Mitogen-activated protein kinase pathways and fungal pathogenesis. Eukaryot. Cell 2007, 6, 1701–1714. [Google Scholar] [CrossRef]
- Posas, F.; Wurgler-Murphy, S.M.; Maeda, T.; Witten, E.A.; Thai, T.C.; Saito, H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1–YPD1–SSK1 “two-component” osmosensor. Cell 1996, 86, 865–875. [Google Scholar] [CrossRef]
- Maeda, T.; Takekawa, M.; Saito, H. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 1995, 269, 554–558. [Google Scholar] [CrossRef]
- Tanaka, K.; Tatebayashi, K.; Nishimura, A.; Yamamoto, K.; Yang, H.-Y.; Saito, H. Yeast osmosensors Hkr1 and Msb2 activate the Hog1 MAPK cascade by different mechanisms. Sci. Signal. 2014, 7, ra21. [Google Scholar] [CrossRef]
- Tatebayashi, K.; Tanaka, K.; Yang, H.Y.; Yamamoto, K.; Matsushita, Y.; Tomida, T.; Imai, M.; Saito, H. Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. EMBO J. 2007, 26, 3521–3533. [Google Scholar] [CrossRef]
- O’Rourke, S.M.; Herskowitz, I. A third osmosensing branch in Saccharomyces cerevisiae requires the Msb2 protein and functions in parallel with the Sho1 branch. Mol. Cell. Biol. 2002, 22, 4739–4749. [Google Scholar] [CrossRef]
- Tatebayashi, K.; Yamamoto, K.; Nagoya, M.; Takayama, T.; Nishimura, A.; Sakurai, M.; Momma, T.; Saito, H. Osmosensing and scaffolding functions of the oligomeric four-transmembrane domain osmosensor Sho1. Nat. Commun. 2015, 6, 1–15. [Google Scholar] [CrossRef]
- Huberman, L.B.; Coradetti, S.T.; Glass, N.L. Network of nutrient-sensing pathways and a conserved kinase cascade integrate osmolarity and carbon sensing in Neurospora crassa. Proc. Natl. Acad. Sci. USA 2017, 114, E8665–E8674. [Google Scholar] [CrossRef]
- Bahn, Y.-S.; Kojima, K.; Cox, G.M.; Heitman, J. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol. Biol. Cell 2006, 17, 3122–3135. [Google Scholar] [CrossRef]
- Alonso-Monge, R.; Navarro-Garcia, F.; Molero, G.; Diez-Orejas, R.; Gustin, M.; Pla, J.; Sanchez, M.; Nombela, C. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J. Bacteriol. 1999, 181, 3058–3068. [Google Scholar] [CrossRef]
- Dixon, K.P.; Xu, J.-R.; Smirnoff, N.; Talbot, N.J. Independent signaling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea. Plant Cell 1999, 11, 2045–2058. [Google Scholar] [CrossRef]
- Moriwaki, A.; Kihara, J.; Mori, C.; Arase, S. A MAP kinase gene, BMK1, is required for conidiation and pathogenicity in the rice leaf spot pathogen Bipolaris oryzae. Microbiol. Res. 2007, 162, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Nordbring-Hertz, B.; Jansson, H.B.; Tunlid, A. Nematophagous fungi. eLS 2001. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genom. 2008. [Google Scholar] [CrossRef]
- Jin, X.; Ming-He, M.; Xiao-Wei, H.; Ke-Qin, Z. Improvement on genetic transformation in the nematode-trapping fungus Arthrobotrys oligospora and its quantification on dung samples. Mycopathologia 2005, 159, 533–538. [Google Scholar] [CrossRef]
- Tunlid, A.; Åhman, J.; Oliver, R. Transformation of the nematode-trapping fungus Arthrobotrys oligospora. Fems Microbiol. Lett. 1999, 173, 111–116. [Google Scholar] [CrossRef]
- Yang, C.-T.; de Ulzurrun, G.V.-D.; Gonçalves, A.P.; Lin, H.-C.; Chang, C.-W.; Huang, T.-Y.; Chen, S.-A.; Lai, C.-K.; Tsai, I.J.; Schroeder, F.C. Natural diversity in the predatory behavior facilitates the establishment of a robust model strain for nematode-trapping fungi. Proc. Natl. Acad. Sci. USA 2020, 117, 6762–6770. [Google Scholar] [CrossRef]
- Koh, C.M.J.; Liu, Y.; Du, M.; Ji, L. Molecular characterization of KU70 and KU80 homologues and exploitation of a KU70-deficient mutant for improving gene deletion frequency in Rhodosporidium toruloides. BMC Microbiol. 2014, 14, 50. [Google Scholar] [CrossRef]
- Choquer, M.; Robin, G.; Le Pêcheur, P.; Giraud, C.; Levis, C.; Viaud, M. Ku70 or Ku80 deficiencies in the fungus Botrytis cinerea facilitate targeting of genes that are hard to knock out in a wild-type context. FEMS Microbiol. Lett. 2008, 289, 225–232. [Google Scholar] [CrossRef]
- Pöggeler, S.; Kück, U. Highly efficient generation of signal transduction knockout mutants using a fungal strain deficient in the mammalian ku70 ortholog. Gene 2006, 378, 1–10. [Google Scholar] [CrossRef]
- Punt, P.J.; Oliver, R.P.; Dingemanse, M.A.; Pouwels, P.H.; van den Hondel, C.A. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 1987, 56, 117–124. [Google Scholar] [CrossRef]
- Chee, M.K.; Haase, S.B. New and redesigned pRS plasmid shuttle vectors for genetic manipulation of Saccharomyces cerevisiae. G3 Genes Genomes Genet. 2012, 2, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Murakami, Y.; Tatebayashi, K.; Saito, H. Two adjacent docking sites in the yeast Hog1 mitogen-activated protein (MAP) kinase differentially interact with the Pbs2 MAP kinase kinase and the Ptp2 protein tyrosine phosphatase. Mol. Cell. Biol. 2008, 28, 2481–2494. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Z.; Xing, X.; Xie, M.; Yang, L.; Yang, X.; Zheng, Y.; Chen, Y.; Ma, N.; Li, Q.; Zhang, K.-Q. MAP kinase Slt2 orthologs play similar roles in conidiation, trap formation, and pathogenicity in two nematode-trapping fungi. Fungal Genet. Biol. 2018, 116, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Bai, N.; Yang, J.; Jiang, K.; Zhou, D.; Zhao, Y.; Li, D.; Niu, X.; Zhang, K.-Q.; Yang, J. Protein Kinase Ime2 Is Required for Mycelial Growth, Conidiation, Osmoregulation, and Pathogenicity in Nematode-Trapping Fungus Arthrobotrys oligospora. Front. Microbiol. 2020, 10, 3065. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, K.; Zhang, X.; Song, W.; Zhao, Q.; Dong, Y.; Guo, M.; Zheng, X.; Zhang, Z. A two-component histidine kinase, MoSLN1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Curr. Genet. 2010, 56, 517–528. [Google Scholar] [CrossRef]
- Liu, W.; Zhou, X.; Li, G.; Li, L.; Kong, L.; Wang, C.; Zhang, H.; Xu, J.-R. Multiple plant surface signals are sensed by different mechanisms in the rice blast fungus for appressorium formation. PLoS Pathog. 2011, 7, e1001261. [Google Scholar] [CrossRef]
- Lanver, D.; Mendoza-Mendoza, A.; Brachmann, A.; Kahmann, R. Sho1 and Msb2-related proteins regulate appressorium development in the smut fungus Ustilago maydis. Plant Cell 2010, 22, 2085–2101. [Google Scholar] [CrossRef]
- Pérez-Nadales, E.; Di Pietro, A. The membrane mucin Msb2 regulates invasive growth and plant infection in Fusarium oxysporum. Plant Cell 2011, 23, 1171–1185. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, J.; Fang, W.; Zhang, J.; Luo, Z.; Zhang, M.; Fan, Y.; Pei, Y. Mitogen-activated protein kinase hog1 in the entomopathogenic fungus Beauveria bassiana regulates environmental stress responses and virulence to insects. Appl. Environ. Microbiol. 2009, 75, 3787–3795. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuo, C.-Y.; Chen, S.-A.; Hsueh, Y.-P. The High Osmolarity Glycerol (HOG) Pathway Functions in Osmosensing, Trap Morphogenesis and Conidiation of the Nematode-Trapping Fungus Arthrobotrys oligospora. J. Fungi 2020, 6, 191. https://doi.org/10.3390/jof6040191
Kuo C-Y, Chen S-A, Hsueh Y-P. The High Osmolarity Glycerol (HOG) Pathway Functions in Osmosensing, Trap Morphogenesis and Conidiation of the Nematode-Trapping Fungus Arthrobotrys oligospora. Journal of Fungi. 2020; 6(4):191. https://doi.org/10.3390/jof6040191
Chicago/Turabian StyleKuo, Chih-Yen, Sheng-An Chen, and Yen-Ping Hsueh. 2020. "The High Osmolarity Glycerol (HOG) Pathway Functions in Osmosensing, Trap Morphogenesis and Conidiation of the Nematode-Trapping Fungus Arthrobotrys oligospora" Journal of Fungi 6, no. 4: 191. https://doi.org/10.3390/jof6040191
APA StyleKuo, C.-Y., Chen, S.-A., & Hsueh, Y.-P. (2020). The High Osmolarity Glycerol (HOG) Pathway Functions in Osmosensing, Trap Morphogenesis and Conidiation of the Nematode-Trapping Fungus Arthrobotrys oligospora. Journal of Fungi, 6(4), 191. https://doi.org/10.3390/jof6040191




