Molecular Epidemiology of Azole-Resistant Aspergillus fumigatus in Sawmills of Eastern France by Microsatellite Genotyping
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of A. fumigatus Strains
2.2. Short Tandem Repeat for A. fumigatus (STRAf) Typing and Analysis
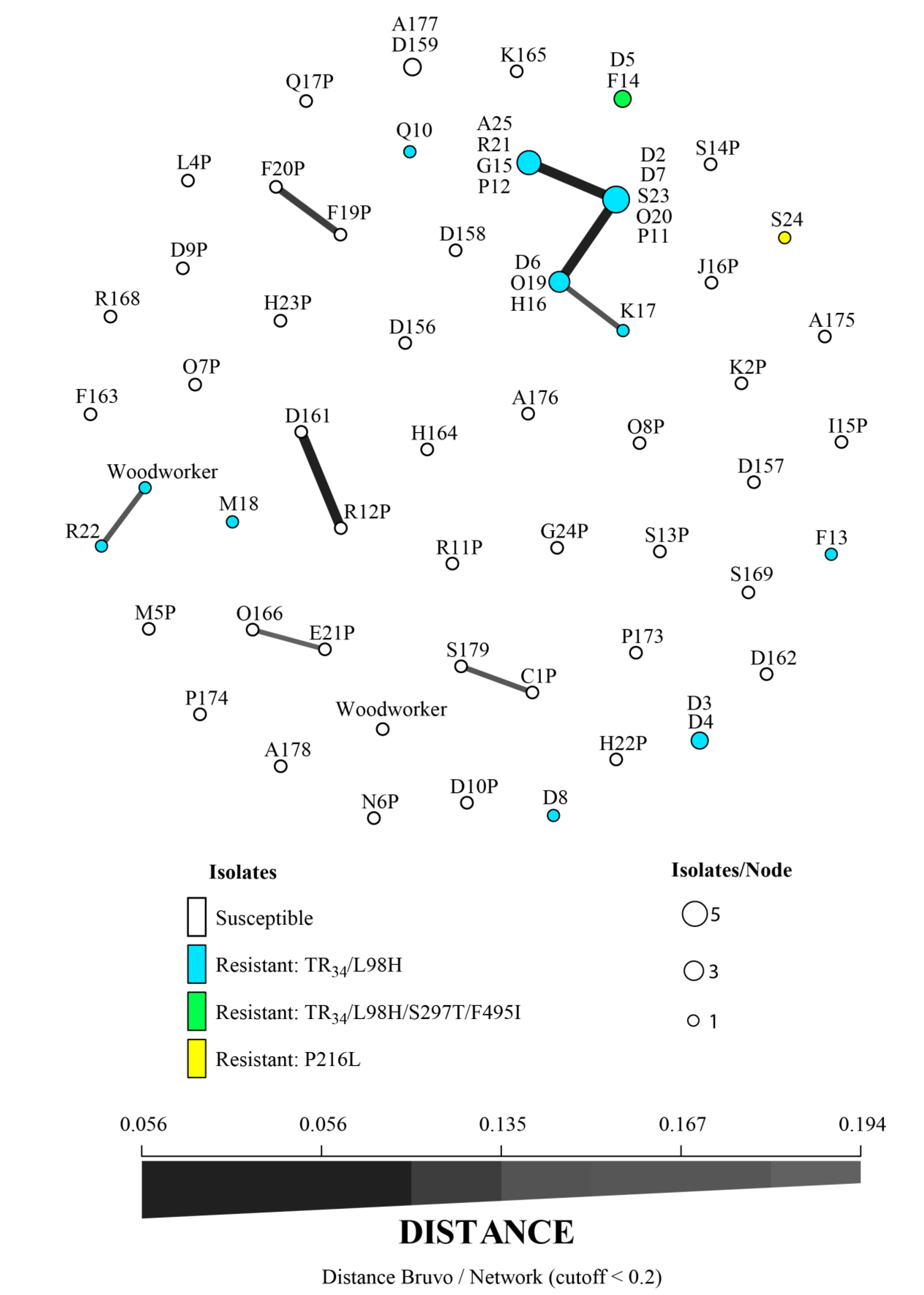
3. Results
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Snelders, E.; van der Lee, H.A.; Kuijpers, J.; Rijs, A.J.; Varga, J.; Samson, R.A.; Mellado, E.; Donders, A.R.T.; Melchers, W.J.G.; Verweij, P.E. Emergence of Azole Resistance in Aspergillus fumigatus and Spread of a Single Resistance Mechanism. PLoS Med. 2008, 5, e219. [Google Scholar] [CrossRef]
- Verweij, P.E.; Snelders, E.; Kema, G.H.; Mellado, E.; Melchers, W.J. Azole resistance in Aspergillus fumigatus: A side-effect of environmental fungicide use? Lancet Infect. Dis. 2009, 9, 789–795. [Google Scholar] [CrossRef]
- Verweij, P.E.; Kema, G.H.; Zwaan, B.; Melchers, W.J. Triazole fungicides and the selection of resistance to medical triazoles in the opportunistic mould Aspergillus fumigatus: The environmental route of azole resistance selection in Aspergillus fumigatus. Pest Manag. Sci. 2013, 69, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Jeanvoine, A.; Rocchi, S.; Bellanger, A.P.; Reboux, G.; Millon, L. Azole-resistant Aspergillus fumigatus: A global phenomenon originating in the environment? Med. Mal. Infect. 2019. [Google Scholar] [CrossRef]
- Verweij, P.E.; Chowdhary, A.; Melchers, W.J.; Meis, J.F. Azole Resistance in Aspergillus fumigatus: Can We Retain the Clinical Use of Mold-Active Antifungal Azoles? Clin. Infect. Dis. 2016, 62, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Chowdhary, A.; Meis, J.F. Emergence of azole resistant Aspergillus fumigatus and One Health: Time to implement environmental stewardship. Environ. Microbiol. 2018, 20, 1299–1301. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Snelders, E.; Zwaan, B.J.; Schoustra, S.E.; Meis, J.F.; van Dijk, K.; Hagen, F.; van der Beek, M.T.; Kampinga, G.A.; Zoll, J.; et al. A Novel Environmental Azole Resistance Mutation in Aspergillus fumigatus and a Possible Role of Sexual Reproduction in Its Emergence. mBio 2017, 8, e00791-17. [Google Scholar] [CrossRef]
- Schoustra, S.E.; Debets, A.J.; Rijs, A.J.; Zhang, J.; Snelders, E.; Leendertse, P.C.; Melchers, W.J.; Rietveld, A.G.; Zwaan, B.J.; Verweij, P.E. Environmental Hotspots for Azole Resistance Selection of Aspergillus fumigatus, the Netherlands. Emerg. Infect. Dis. 2019, 25, 1347–1353. [Google Scholar] [CrossRef]
- Sharma, C.; Hagen, F.; Moroti, R.; Meis, J.F.; Chowdhary, A. Triazole-resistant Aspergillus fumigatus harbouring G54 mutation: Is it de novo or environmentally acquired? J. Glob. Antimicrob. Resist. 2015, 3, 69–74. [Google Scholar] [CrossRef]
- Jeanvoine, A.; Rocchi, S.; Reboux, G.; Crini, N.; Crini, G.; Millon, L. Azole-resistant Aspergillus fumigatus in sawmills of Eastern France. J. Appl. Microbiol. 2017, 123, 172–184. [Google Scholar] [CrossRef]
- Ashu, E.E.; Hagen, F.; Chowdhary, A.; Meis, J.F.; Xu, J. Global Population Genetic Analysis of Aspergillus fumigatus. mSphere 2017, 2, e00019-17. [Google Scholar] [CrossRef] [PubMed]
- Abdolrasouli, A.; Rhodes, J.; Beale, M.A.; Hagen, F.; Rogers, T.R.; Chowdhary, A.; Meis, J.F.; Armstrong-James, D.; Fisher, M.C. Genomic Context of Azole Resistance Mutations in Aspergillus fumigatus Determined Using Whole-Genome Sequencing. mBio 2015, 6, e00536-15. [Google Scholar] [CrossRef] [PubMed]
- de Valk, H.A.; Meis, J.F.; Curfs, I.M.; Muehlethaler, K.; Mouton, J.W.; Klaassen, C.H. Use of a Novel Panel of Nine Short Tandem Repeats for Exact and High-Resolution Fingerprinting ofAspergillus fumigatus Isolates. J. Clin. Microbiol. 2005, 43, 4112–4120. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rubio, R.; Escribano, P.; Gomez, A.; Guinea, J.; Mellado, E. Comparison of Two Highly Discriminatory Typing Methods to Analyze Aspergillus fumigatus Azole Resistance. Front. Microbiol. 2018, 9, 1626. [Google Scholar] [CrossRef]
- Jeanvoine, A.; Rocchi, S.; Reboux, G.; Grenouillet, F.; Benassarou, M.; Chirouze, C.; Millon, L. Sinus aspergillosis due to an azole-resistant Aspergillus fumigatus strain carrying the TR34/L98H mutation in immunocompetent host. Infect. Dis. 2016, 48, 765–766. [Google Scholar] [CrossRef] [PubMed]
- European Committee for Antimicrobial Susceptibility Testing (EUCAST). Available online: https://www.eucast.org/astoffungi/ (accessed on 24 June 2020).
- Rocchi, S.; Ponçot, M.; Morin-Crini, N.; Laboissière, A.; Valot, B.; Godeau, C.; Léchenault-Bergerot, C.; Reboux, G.; Crini, G.; Millon, L. Determination of azole fungal residues in soils and detection of Aspergillus fumigatus-resistant strains in market gardens of Eastern France. Environ. Sci. Pollut. Res. 2018, 25, 32015–32023. [Google Scholar] [CrossRef]
- Chowdhary, A.; Kathuria, S.; Xu, J.; Sharma, C.; Sundar, G.; Singh, P.K.; Gaur, S.N.; Hagen, F.; Klaassen, C.H.; Meis, J.F. Clonal Expansion and Emergence of Environmental Multiple-Triazole-Resistant Aspergillus fumigatus Strains Carrying the TR34/L98H Mutations in the cyp51A Gene in India. PLoS ONE 2012, 7, e52871. [Google Scholar] [CrossRef]
- Chang, H.; Ashu, E.; Sharma, C.; Kathuria, S.; Chowdhary, A.; Xu, J. Diversity and origins of Indian multi-triazole resistant strains of Aspergillus fumigatus. Mycoses 2016, 59, 450–466. [Google Scholar] [CrossRef]
- de Groot, T.; Meis, J.F. Microsatellite Stability in STR Analysis Aspergillus fumigatus Depends on Number of Repeat Units. Front. Cell. Infect. Microbiol. 2019, 9, 82. [Google Scholar] [CrossRef]
- Badali, H.; Vaezi, A.; Haghani, I.; Yazdanparast, S.A.; Hedayati, M.T.; Mousavi, B.; Ansari, S.; Hagen, F.; Meis, J.F.; Chowdhary, A. Environmental study of azole-resistant Aspergillus fumigatus with TR34/L98H mutations in the cyp51A gene in Iran. Mycoses 2013, 56, 659–663. [Google Scholar] [CrossRef]
- Alvarez-Moreno, C.; Lavergne, R.A.; Hagen, F.; Morio, F.; Meis, J.F.; Le Pape, P. Azole-resistant Aspergillus fumigatus harboring TR34/L98H, TR46/Y121F/T289A and TR53 mutations related to flower fields in Colombia. Sci. Rep. 2017, 7, 45631. [Google Scholar] [CrossRef] [PubMed]
- Korfanty, G.A.; Teng, L.; Pum, N.; Xu, J. Contemporary Gene Flow is a Major Force Shaping the Aspergillus fumigatus Population in Auckland, New Zealand. Mycopathologia 2019, 184, 479–492. [Google Scholar] [CrossRef]
- Sewell, T.R.; Zhu, J.; Rhodes, J.; Hagen, F.; Meis, J.F.; Fisher, M.C.; Jombart, T. Nonrandom Distribution of Azole Resistance across the Global Population of Aspergillus fumigatus. mBio 2019, 10, e00392-19. [Google Scholar] [CrossRef] [PubMed]
- Meis, J.F.; Chowdhary, A.; Rhodes, J.L.; Fisher, M.C.; Verweij, P.E. Clinical implications of globally emerging azole resistance in Aspergillus fumigatus. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150460. [Google Scholar] [CrossRef] [PubMed]
| Isolate Identification | Date of Isolation | Origin | Susceptible or Resistant and Cyp51A Mutation |
|---|---|---|---|
| A25 | February 2015 | Substrate, sawmill A | Resistant, TR34/L98H |
| D2 | January 2016 | Substrate, sawmill D | Resistant, TR34/L98H |
| D3 | January 2016 | Substrate, sawmill D | Resistant, TR34/L98H |
| D4 | January 2016 | Substrate, sawmill D | Resistant, TR34/L98H |
| D5 | January 2016 | Substrate, sawmill D | Resistant, TR34/L98H/S297T/F495I |
| D6 | January 2016 | Substrate, sawmill D | Resistant, TR34/L98H |
| D7 | January 2016 | Substrate, sawmill D | Resistant, TR34/L98H |
| D8 | January 2016 | Substrate, sawmill D | Resistant, TR34/L98H |
| F13 | February 2016 | Substrate, sawmill F | Resistant, TR34/L98H |
| F14 | February 2016 | Substrate, sawmill F | Resistant, TR34/L98H/S297T/F495I |
| G15 | February 2016 | Substrate, sawmill G | Resistant, TR34/L98H |
| H16 | February 2016 | Substrate, sawmill H | Resistant, TR34/L98H |
| K17 | March 2016 | Substrate, sawmill K | Resistant, TR34/L98H |
| M18 | March 2016 | Substrate, sawmill M | Resistant, TR34/L98H |
| O19 | March 2016 | Substrate, sawmill O | Resistant, TR34/L98H |
| O20 | March 2016 | Substrate, sawmill O | Resistant, TR34/L98H |
| P11 | September 2014 | Substrate, sawmill P | Resistant, TR34/L98H |
| P12 | September 2014 | Substrate, sawmill P | Resistant, TR34/L98H |
| Q10 | January 2016 | Substrate, sawmill R | Resistant, TR34/L98H |
| R21 | April 2016 | Substrate, sawmill S | Resistant, TR34/L98H |
| R22 | April 2016 | Substrate, sawmill S | Resistant, TR34/L98H |
| S23 | April 2016 | Substrate, sawmill T | Resistant, TR34/L98H |
| S24 | April 2016 | Substrate, sawmill T | Resistant, TR34/L98H |
| Wood-worker | October 2013 | Clinical | Resistant, TR34/L98H |
| A175 | February 2015 | Substrate, sawmill A | Susceptible |
| A176 | February 2015 | Substrate, sawmill A | Susceptible |
| A177 | February 2015 | Substrate, sawmill A | Susceptible |
| A178 | February 2015 | Substrate, sawmill A | Susceptible |
| C1P | January 2016 | Substrate, sawmill C | Susceptible |
| D156 | January 2016 | Substrate, sawmill D | Susceptible |
| D157 | January 2016 | Substrate, sawmill D | Susceptible |
| D158 | January 2016 | Substrate, sawmill D | Susceptible |
| D159 | January 2016 | Substrate, sawmill D | Susceptible |
| D161 | January 2016 | Substrate, sawmill D | Susceptible |
| D162 | January 2016 | Substrate, sawmill D | Susceptible |
| D9P | January 2016 | Substrate, sawmill D | Susceptible |
| D10P | January 2016 | Substrate, sawmill D | Susceptible |
| E21P | January 2016 | Substrate, sawmill E | Susceptible |
| F19P | February 2016 | Substrate, sawmill F | Susceptible |
| F20P | February 2016 | Substrate, sawmill F | Susceptible |
| F163 | February 2016 | Substrate, sawmill F | Susceptible |
| G24P | February 2016 | Substrate, sawmill G | Susceptible |
| H22P | February 2016 | Substrate, sawmill H | Susceptible |
| H23P | February 2016 | Substrate, sawmill H | Susceptible |
| H164 | February 2016 | Substrate, sawmill H | Susceptible |
| I15P | February 2016 | Substrate, sawmill I | Susceptible |
| J16P | March 2016 | Substrate, sawmill J | Susceptible |
| K2P | March 2016 | Substrate, sawmill K | Susceptible |
| K165 | March 2016 | Substrate, sawmill K | Susceptible |
| L4P | March 2016 | Substrate, sawmill L | Susceptible |
| M5P | March 2016 | Substrate, sawmill M | Susceptible |
| N6P | March 2016 | Substrate, sawmill N | Susceptible |
| O7P | March 2016 | Substrate, sawmill O | Susceptible |
| O8P | March 2016 | Substrate, sawmill O | Susceptible |
| O166 | March 2016 | Substrate, sawmill O | Susceptible |
| P173 | September 2014 | Substrate, sawmill P | Susceptible |
| P174 | September 2014 | Substrate, sawmill P | Susceptible |
| Q17P | January 2016 | Substrate, sawmill R | Susceptible |
| R11P | April 2016 | Substrate, sawmill S | Susceptible |
| R12P | April 2016 | Substrate, sawmill S | Susceptible |
| R168 | April 2016 | Substrate, sawmill S | Susceptible |
| S13P | April 2016 | Substrate, sawmill T | Susceptible |
| S14P | April 2016 | Substrate, sawmill T | Susceptible |
| S169 | April 2016 | Substrate, sawmill T | Susceptible |
| S179 | April 2016 | Substrate, sawmill T | Susceptible |
| Wood-worker | July 2013 | Clinical | Susceptible |
| Azole-Resistant A. fumigatus (ARAf, n = 24) | Azole-Susceptible A. fumigatus (ASAf, n = 42) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Microsatellite Marker | N Alleles | Repeat Range | Median Size | Diversity (D) | Microsatellite Marker | N Alleles | Repeat Range | Median Size | Diversity (D) |
| 2A | 5 | 13–26 | 14 | 0.507 | 2A | 11 | 10–27 | 18 | 0.768 |
| 2B | 4 | 10–24 | 21 | 0.462 | 2B | 9 | 12–25 | 19 | 0.766 |
| 2C | 4 | 8–16 | 8 | 0.552 | 2C | 11 | 8–20 | 12 | 0.868 |
| 3A | 11 | 10–119 | 32 | 0.872 | 3A | 19 | 10–49 | 26 | 0.909 |
| 3B | 3 | 8–11 | 8 | 0.403 | 3B | 10 | 8–22 | 9 | 0.771 |
| 3C | 7 | 6–32 | 6 | 0.649 | 3C | 19 | 6–45 | 18 | 0.924 |
| 4A | 5 | 5–18 | 8 | 0.361 | 4A | 13 | 7–26 | 9 | 0.774 |
| 4B | 3 | 7–10 | 10 | 0.392 | 4B | 8 | 5–26 | 9 | 0.746 |
| 4C | 5 | 5–30 | 20 | 0.465 | 4C | 9 | 5–36 | 7 | 0.770 |
| Total alleles | 47 | NR | NR | NR | Total alleles | 109 | NR | NR | NR |
| Average D | NR | NR | NR | 0.518 | Average D | NR | NR | NR | 0.811 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeanvoine, A.; Godeau, C.; Laboissière, A.; Reboux, G.; Millon, L.; Rocchi, S. Molecular Epidemiology of Azole-Resistant Aspergillus fumigatus in Sawmills of Eastern France by Microsatellite Genotyping. J. Fungi 2020, 6, 120. https://doi.org/10.3390/jof6030120
Jeanvoine A, Godeau C, Laboissière A, Reboux G, Millon L, Rocchi S. Molecular Epidemiology of Azole-Resistant Aspergillus fumigatus in Sawmills of Eastern France by Microsatellite Genotyping. Journal of Fungi. 2020; 6(3):120. https://doi.org/10.3390/jof6030120
Chicago/Turabian StyleJeanvoine, Audrey, Chloé Godeau, Audrey Laboissière, Gabriel Reboux, Laurence Millon, and Steffi Rocchi. 2020. "Molecular Epidemiology of Azole-Resistant Aspergillus fumigatus in Sawmills of Eastern France by Microsatellite Genotyping" Journal of Fungi 6, no. 3: 120. https://doi.org/10.3390/jof6030120
APA StyleJeanvoine, A., Godeau, C., Laboissière, A., Reboux, G., Millon, L., & Rocchi, S. (2020). Molecular Epidemiology of Azole-Resistant Aspergillus fumigatus in Sawmills of Eastern France by Microsatellite Genotyping. Journal of Fungi, 6(3), 120. https://doi.org/10.3390/jof6030120






