Diversity of Fungi Associated with Diseases of Cultivated Brassicaceae in Southern Italy
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Fungal Isolation
2.2. DNA Extraction
2.3. MSP-PCR Analysis and Molecular Characterization
| Locus | Primer | Primer DNA Sequence 5′→3′ | PCR Conditions | Reference |
|---|---|---|---|---|
| ITS | ITS1 | TCCGTAGGTGAACCTGCGG | 94 °C–3 min; [94 °C–30 s, 55 °C–30 s, 72 °C–30 s] × 35; 72 °C–10 min. | [67] |
| ITS4 | GCTGCGTTCTT ATCGATGC | |||
| tef-1α | EF1-728F | CATCGAGAAGTTCGAGAAGG | 94 °C–3 min; [94 °C–30 s, 58 °C–30 s, 72 °C–30 s] × 35; 72 °C–10 min. | [68] |
| EF1-986R | TACTTGAAGGAACCCTTACC | |||
| EF-983F | GCYCCYGGHCAYCGTGAYTTYA | 94 °C–5 min; [94 °C–45 s, 54 °C–45 s, 72 °C–60 s] × 36; 72 °C–10 min. | [74] | |
| EF-2218R | ATGACACCRACRGCRACRGTYT | |||
| EF1 | ATGGGTAAGGARGACAAGAC | 94 °C–3 min; [94 °C–60 s, 53 °C–60 s, 72 °C–120 s] × 35; 72 °C–10 min. | [73] | |
| EF2 | GGARGTACCAGTSATCATGTT | |||
| Alt-a1 | Dir5cAlta1 | GAGAACAGCTTCATGGACTTCTCTTT | 94 °C–1 min; [94 °C–30 s, 55–30 s, 72 °C–45 s] × 35; 72 °C–5 min. | [69] |
| Inv4Alta1 | CGCGGCAGTAGTTGGGAA | |||
| rpb2 | fRPB2-5f2 | GAYGAYMGWGATCAYTTYGG | 94 °C–2.3 min; [94 °C–60 s, 50–53 °C–30 s, 72 °C–120 s] × 35; 72 °C–10 min. | [70] |
| fRPB2-7cR | CCCATRGCTTGYTTRCCCAT | |||
| gapdh | gpd1 | CAACGGCTTCGGTCGCATTG | 96 °C for 2.00 min, followed by 30 cycles of denaturation at 96 °C for 60 s, annealing at 48 °C for 1 min, and elongation at 72 °C for 45 s. With each cycle, the time at 72 °C was extended by 4 s. Usually, the products from the first amplification were purified and then reamplified with the same primers using another 25 PCR cycles like the initial 30 but with 54 °C of annealing temperature. | [71] |
| gpd2 | GCCAAGCAGTTGGTTGTGC |
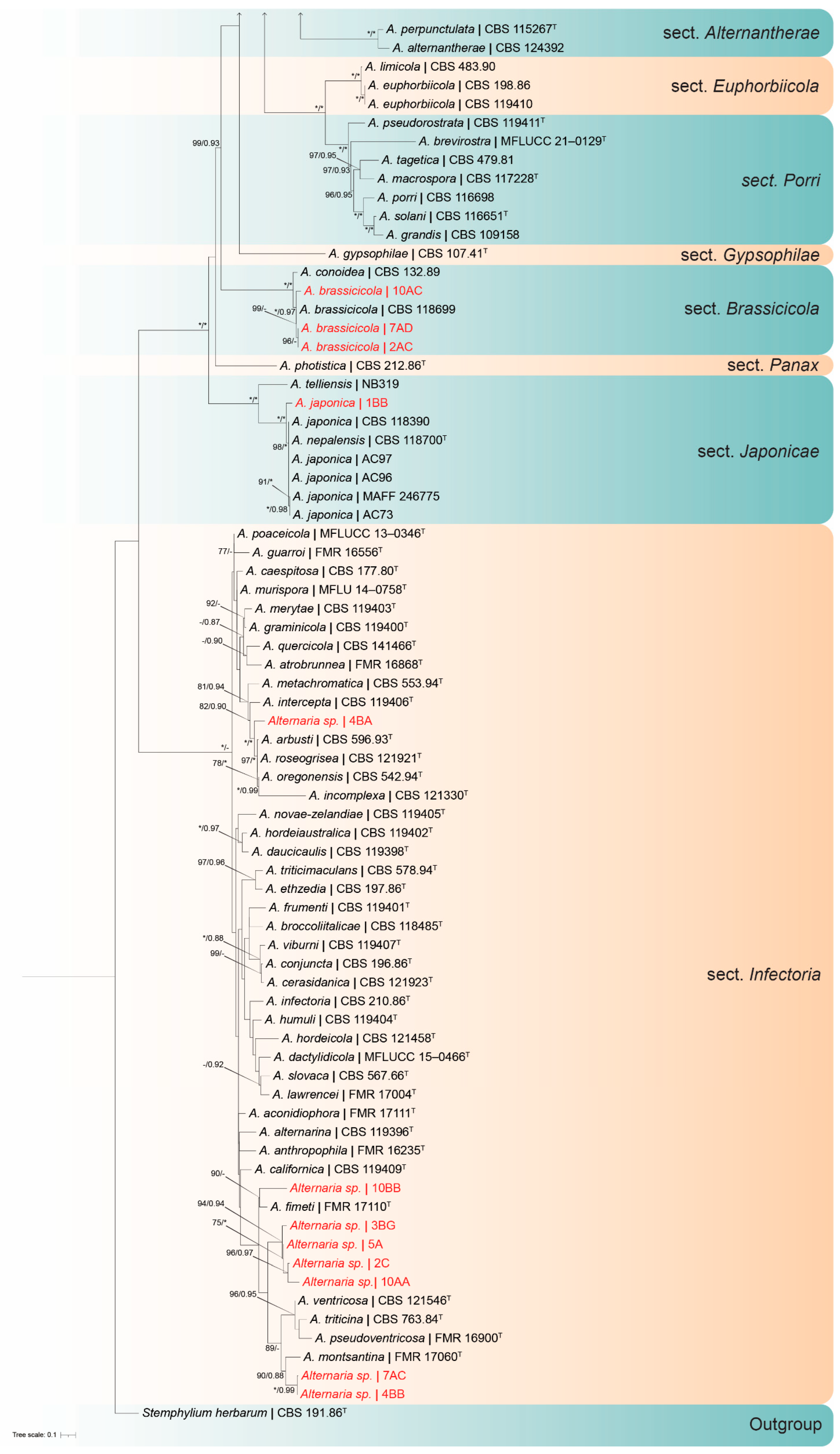
2.4. Phylogenetic Analyses
3. Results
3.1. Fungal Isolation
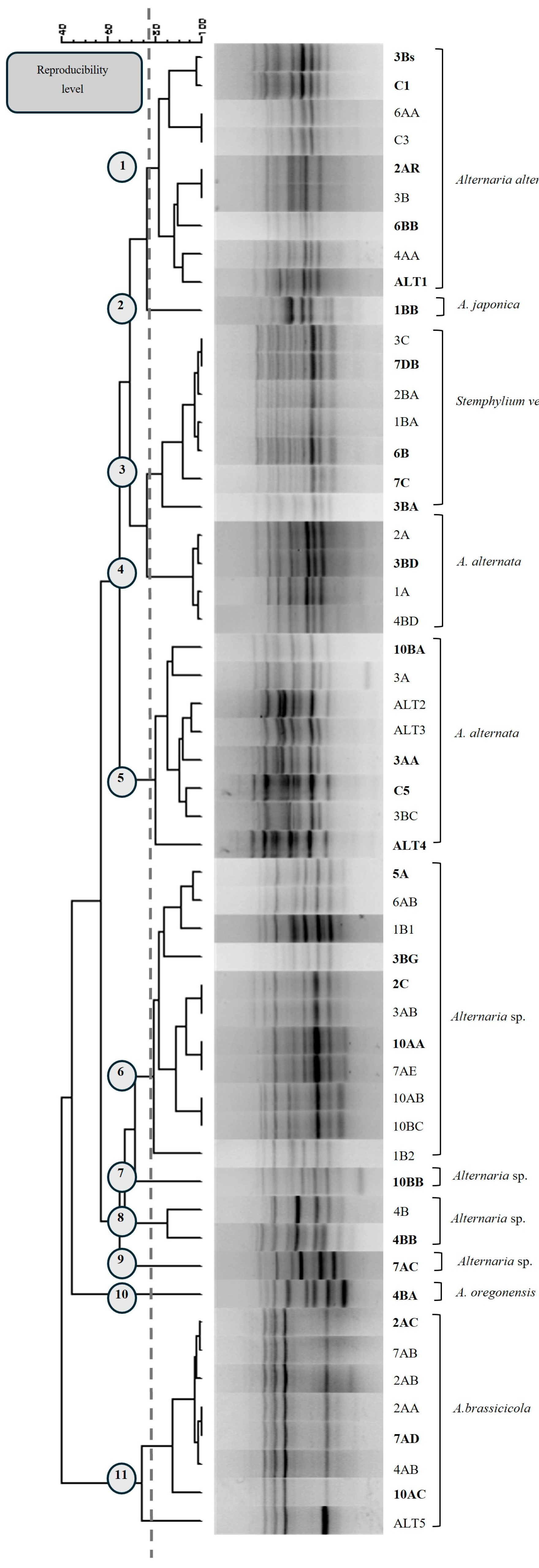
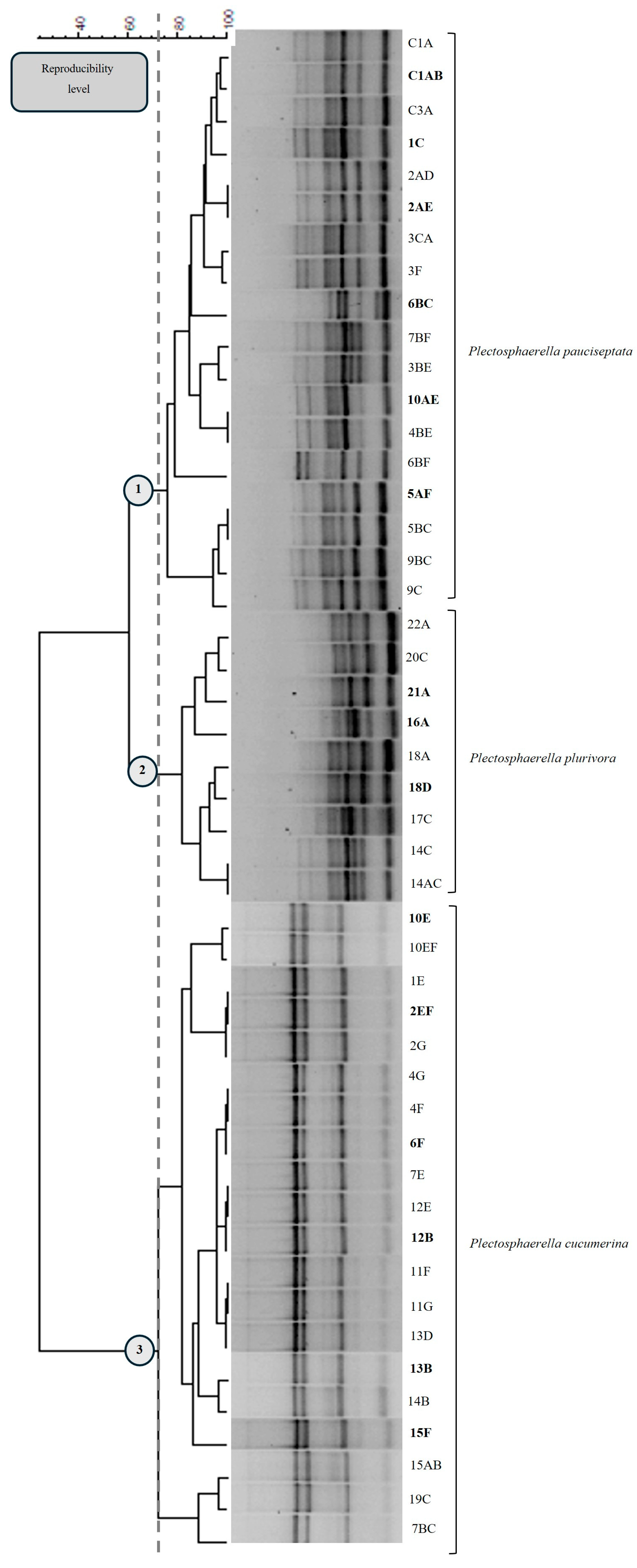
3.2. Microsatellite PCR Profiles and Molecular Characterization
3.3. Molecular Identification of Representative Isolated Fungi
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gómez-Campo, C. Taxonomy. In Biology of Brassica Coenospecies; Gómez-Campo, C., Ed.; Elsevier: Amsterdam, The Netherlands, 1999; pp. 3–32. [Google Scholar]
- Kapusta-Duch, J.; Kopeć, A.; Piątkowska, E.; Borczak, B.; Leszczyńska, T. The beneficial effects of Brassica vegetables on human health. Rocz. Państw. Zakł. Hig. 2012, 63, 389–395. [Google Scholar] [PubMed]
- Brennan, E.B.; Acosta-Martínez, V. Cover cropping frequency is the main driver of soil microbial changes during six years of organic vegetable production. Soil Biol. Biochem. 2017, 109, 188–204. [Google Scholar] [CrossRef]
- Rakow, G. Species origin and economic importance of Brassica. In Brassica; Pua, E.C., Douglas, C.J., Eds.; Springer: Berlin/Heidelberg, Germany, 2004; Volume 54, pp. 3–11. [Google Scholar] [CrossRef]
- Subramanian, P.; Kim, S.H.; Hahn, B.S. Brassica biodiversity conservation: Prevailing constraints and future avenues for sustainable distribution of plant genetic resources. Front. Plant Sci. 2023, 14, 1220134. [Google Scholar] [CrossRef]
- Istituto Nazionale di Statistica (ISTAT). Available online: https://esploradati.istat.it/databrowser/#/it/dw/categories/IT1,Z1000AGR,1.0/AGR_CRP/DCSP_COLTIVAZIONI/ (accessed on 17 March 2025).
- Mourou, M.; Raimondo, M.L.; Lops, F.; Carlucci, A. Fungi and Chromista diseases: Molecular detection and host–plant interaction. Plants 2023, 12, 1033. [Google Scholar] [CrossRef]
- Akram, W.; Li, G.; Ahmad, A.; Anjum, T.; Ali, B.; Luo, W.; Guo, J.; Xie, D.; Wang, Q. Leaf spot disease caused by Alternaria arborescens, A. tenuissima, and A. infectoria on Brassica rapa subsp. parachinensis in China. Plant Dis. 2019, 103, 2480. [Google Scholar] [CrossRef]
- Hinchliffe, E.R.; Keinath, A.P.; Meadows, I.M. First report of Alternaria leaf spot caused by Alternaria brassicae on broccoli in North Carolina, U.S.A. Plant Dis. 2024, 108, 3659. [Google Scholar] [CrossRef]
- Matić, S.; Gilardi, G.; Varveri, M.; Garibaldi, A.; Gullino, M.L. Molecular diversity of Alternaria spp. from leafy vegetable crops, and their sensitivity to azoxystrobin and boscalid. Phytopathol. Mediterr. 2019, 58, 519–533. [Google Scholar] [CrossRef]
- Nishikawa, J.; Nakashima, C. Japanese species of Alternaria and their species boundaries based on host range. Fungal Syst. Evol. 2020, 5, 197–282. [Google Scholar] [CrossRef]
- Ramirez-Villacis, D.X.; Barriga-Medina, N.; Llerena-Llerena, S.; Pazmino-Guevara, C.; Leon-Reyes, A. First report of Alternaria alternata causing leaf spot on broccoli in Ecuador. Plant Dis. 2023, 107, 2866. [Google Scholar] [CrossRef]
- Salybekova, N.; Basim, E.; Basim, H.; Zhusupovna, G. Characterization of Alternaria brassicae causing black leaf spot disease of cabbage (Brassica oleracea var. capitata) in the southern part of Kazakhstan. Acta Sci. Pol. Hortorum Cultus 2019, 18, 3–13. [Google Scholar] [CrossRef]
- Park, K.; Kim, C.-H. Occurrence of anthracnose on cabbage caused by Colletotrichum dematium. Mycobiology 2001, 29, 61–62. [Google Scholar] [CrossRef]
- Afroz, T.; Jee, S.; Aktaruzzaman, M.; Choi, M.H.-W.; Kim, J.H.; Assefa, A.D.; Hahn, B.S.; Lee, H.-S. First report of Fusarium wilt caused by Fusarium equiseti on cabbage (Brassica oleracea var. capitata L.) in Korea. Plant Dis. 2021, 105, 1198. [Google Scholar] [CrossRef]
- Gaetán, S.A. Occurrence of Fusarium wilt on canola caused by Fusarium oxysporum f. sp. conglutinans in Argentina. Plant Dis. 2005, 89, 432. [Google Scholar] [CrossRef] [PubMed]
- Garibaldi, A.; Gilardi, G.; Gullino, M.L. Evidence for an expanded host range of Fusarium oxysporum f. sp. raphani. Phytoparasitica 2006, 34, 115–121. [Google Scholar] [CrossRef]
- Li, P.L.; Shi, Y.X.; Guo, M.Y.; Xie, X.W.; Chai, A.L.; Li, B.J. Fusarium wilt of cauliflower caused by Fusarium equiseti in China. Can. J. Plant Pathol. 2017, 39, 77–82. [Google Scholar] [CrossRef]
- Robak, J.; Czubatka, A.; Czajka, A.; Smolinska, U. First report of cabbage head rot caused by Fusarium avenaceum in Poland. Plant Dis. 2014, 98, 1741. [Google Scholar] [CrossRef]
- Claassen, B.J.; Berry, P.A.; Thomas, W.J.; Mallory-Smith, C.; Ocamb, C.M. Black leg and chlorotic leaf spot occurrence on Brassicaceae crop and weed hosts. Plant Dis. 2021, 105, 3418–3425. [Google Scholar] [CrossRef]
- Gautam, A.K.; Avasthi, S.; Prasher, I.B.; Sushma; Verma, R.K. Diversity and distribution of cercosporoid fungi in Himachal Pradesh: An annotated checklist. Stud. Fung. 2020, 5, 17–49. [Google Scholar] [CrossRef]
- Gunasinghe, N.; Barbetti, M.J.; You, M.P.; Burrell, D.; Neate, S. White leaf spot caused by Neopseudocercosporella capsellae: A re-emerging disease of Brassicaceae. Front. Cell. Infect. Microbiol. 2020, 10, 588090. [Google Scholar] [CrossRef]
- Videira, S.I.R.; Groenewald, J.Z.; Nakashima, C.; Braun, U.; Barreto, R.W.; de Wit, P.J.G.M.; Crous, P.W. Mycosphaerellaceae—Chaos or clarity? Stud. Mycol. 2017, 87, 257–421. [Google Scholar] [CrossRef] [PubMed]
- Garibaldi, A.; Gilardi, G.; Ortu, G.; Gullino, M.L. First report of Plectosphaerella cucumerina on greenhouse-cultured wild rocket (Diplotaxis tenuifolia) in Italy. Plant Dis. 2012, 96, 1825. [Google Scholar] [CrossRef] [PubMed]
- Li, P.L.; Chai, A.-L.; Shi, Y.-X.; Xie, X.-W.; Li, B.-J. First report of root rot caused by Plectosphaerella cucumerina on cabbage in China. Mycobiology 2017, 45, 110–113. [Google Scholar] [CrossRef] [PubMed]
- Misawa, T.; Aoki, M. First report of Rhizoctonia solani AG-1 IC causing head rot of cabbage in Japan. New Dis. Rep. 2017, 36, 12. [Google Scholar] [CrossRef]
- Turkkan, M.; Kilicoglu, M.C.; Erper, I. Characterization and pathogenicity of Rhizoctonia isolates collected from Brassica oleracea var. acephala in Ordu, Turkey. Phytoparasitica 2020, 48, 273–286. [Google Scholar] [CrossRef]
- Yang, G.H.; Chen, J.Y.; Pu, W.Q. First report of head rot of cabbage and web blight of snap bean caused by Rhizoctonia solani AG-4 HGI. Plant Pathol. 2007, 56, 351. [Google Scholar] [CrossRef]
- Mahalingam, T.; Guruge, B.M.A.; Somachandra, K.P.; Rajapakse, C.S.; Attanayake, R.N. First report of white mold caused by Sclerotinia sclerotiorum on cabbage in Sri Lanka. Plant Dis. 2017, 101, 249. [Google Scholar] [CrossRef]
- Shrestha, U.; Swilling, K.J.; Butler, D.M.; Ownley, B.H. First report of basal drop and white mold on lettuce, broccoli, and mustard caused by Sclerotinia sclerotiorum in Tennessee, USA. Plant Dis. 2018, 102, 249. [Google Scholar] [CrossRef]
- Sanogo, S.; Lujan, P.A.; Baucom, D. First report of Sclerotinia sclerotiorum on cabbage in New Mexico. Plant Dis. 2015, 99, 891. [Google Scholar] [CrossRef]
- Hattab, Z.; Ben Lasfar, N.; Abid, M.; Bellazreg, F.; Fathallah, A.; Letaief, A. Alternaria alternata infection causing rhinosinusitis and orbital involvement in an immunocompetent patient. New Microbes New Infect. 2019, 32, 100561. [Google Scholar] [CrossRef]
- Pastor, F.J.; Guarro, J. Alternaria infections: Laboratory diagnosis and relevant clinical features. Clin. Microbiol. Infect. 2008, 14, 734–746. [Google Scholar] [CrossRef]
- Hongsanan, S.; Hyde, K.; Phookamsak, R.; Wanasinghe, D.; McKenzie, E.; Sarma, V.V.; Boonmee, S.; Lücking, R.; Bhat, J.D.; Liu, N.; et al. Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere 2020, 11, 2107. [Google Scholar] [CrossRef]
- Wijayawardene, N.; Hyde, K.; Dai, D.; Sánchez-García, M.; Goto, B.; Saxena, R.; Erdoğdu, M.; Selçuk, F.; Rajeshkumar, K.; Aptroot, A.; et al. Outline of fungi and fungus-like taxa—2021. Mycosphere 2022, 13, 53–453. [Google Scholar] [CrossRef]
- De Britto, S.; Kapera, D.; Jogaiah, S. First report of leaf spot disease caused by Alternaria brassicicola on broccoli in Papua New Guinea. Plant Dis. 2020, 104, 3073. [Google Scholar] [CrossRef]
- Gannibal, P.B.; Gasich, E.L. Causal agents of the alternariosis of cruciferous plants in Russia: Species composition, geography, and ecology. Mikol. Fitopatol. 2009, 43, 447–456. [Google Scholar]
- Nowicki, M.; Nowakowska, M.; Niezgoda, A.; Kozik, E. Alternaria black spot of crucifers: Symptoms, importance of disease, and perspectives of resistance breeding. J. Fruit Ornam. Plant Res. 2012, 76, 5–19. [Google Scholar] [CrossRef]
- Humpherson-Jones, F.M.; Maude, R.B. Studies on the epidemiology of Alternaria brassicicola in Brassica oleracea seed production crops. Ann. Appl. Biol. 1982, 100, 61–71. [Google Scholar] [CrossRef]
- Rimmer, S.R.; Shattuck, V.I.; Buchwaldt, L. Compendium of Brassica Diseases; American Phytopathological Society: St. Paul, MN, USA, 2007; p. 117. ISBN 978-0-89054-344-3. [Google Scholar]
- Arie, T. Fusarium diseases of cultivated plants: Control, diagnosis, and molecular and genetic studies. J. Pestic. Sci. 2019, 44, 275–281. [Google Scholar] [CrossRef]
- Chai, A.; Li, X.; Kang, H.J.; Zhao, Q.; Shi, Y.; Li, B. First report of Fusarium cucurbiticola causing vascular wilt on cauliflower in China. Plant Dis. 2022, 106, 2993. [Google Scholar] [CrossRef]
- Akram, W.; Ahmad, A.; Guo, J.; Yasin, N.A.; Akbar, M.; Luo, W.; Wu, T.; Wang, Q.; Lu, M.; Xie, D.; et al. Occurrence of head rot disease caused by Fusarium verticillioides on Chinese flowering cabbage (Brassica rapa L. subsp. parachinensis) in China. Crop Prot. 2020, 134, 105180. [Google Scholar] [CrossRef]
- Howlett, B.J.; Idnurm, A.; Pedras, M.S.C. Leptosphaeria maculans, the causal agent of blackleg disease of Brassicas. Fungal Genet. Biol. 2001, 33, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Humpherson-Jones, F.M. The incidence of Alternaria spp. and Leptosphaeria maculans in commercial Brassica seed in the United Kingdom. Plant Pathol. 1985, 34, 385–390. [Google Scholar] [CrossRef]
- Šašek, V.; Nováková, M.; Dobrev, P.I.; Valentová, O.; Burketová, L. β-Aminobutyric acid protects Brassica napus plants from infection by Leptosphaeria maculans: Resistance induction or a direct antifungal effect? Eur. J. Plant Pathol. 2011, 133, 279–289. [Google Scholar] [CrossRef]
- Fitt, B.D.L.; Brun, H.; Barbetti, M.J.; Rimmer, S.R. Worldwide importance of phoma stem canker (Leptosphaeria maculans and L. biglobosa) on oilseed rape (Brassica napus). Eur. J. Plant Pathol. 2006, 114, 3–15. [Google Scholar] [CrossRef]
- Kutcher, H.R.; Melfort, S.; Rimmer, S.R. Determination of Pathogenic Variability of Leptosphaeria maculans in Western Canada and Resistance in Canadian Brassica napus Cultivars; Agriculture and Agri-Food Canada: Saskatoon, SK, Canada, 2009. [Google Scholar]
- Gunasinghe, N.; You, M.P.; Barbetti, M.J. Phenotypic and phylogenetic studies associated with the crucifer white leaf spot pathogen, Pseudocercosporella capsellae, in Western Australia. Plant Pathol. 2016, 65, 205–217. [Google Scholar] [CrossRef]
- Inman, A.J. The Biology and Epidemiology of White Leaf Spot (Pseudocercosporella capsellae) on Oilseed Rape. Ph.D. Thesis, University of London, London, UK, 1992. [Google Scholar]
- Ocamb, C. Disease Alert—White Leaf Spot and Gray Stem in Crucifer Seed Crops in Western Oregon; Oregon State University: Corvallis, OR, USA, 2014. [Google Scholar]
- Ocamb, C. A Clinic Close-Up: Blackleg, Light Leaf Spot, and White Leaf Spot in Western Oregon; Oregon State University Extension Service: Corvallis, OR, USA, 2016. [Google Scholar]
- Murtza, T.; You, M.; Barbetti, M. Geographic location and year determine virulence, and yearly genetic change, in populations of Neopseudocercosporella capsellae. Plant Pathol. 2019, 68, 1706–1718. [Google Scholar] [CrossRef]
- Van de Wouw, A.P.; Idnurm, A.; Davidson, J.A.; Sprague, S.J.; Khangura, R.K.; Ware, A.; Lindbeck, K.D.; Marcroft, S.J. Fungal diseases of canola in Australia: Identification of trends, threats and potential therapies. Australas. Plant Pathol. 2016, 45, 415–423. [Google Scholar] [CrossRef]
- Dixelius, C.; Bohman, S.; Wretblad, S. Disease resistance. In Biotechnology in Agriculture and Forestry; Douglas, C., Pua, E., Eds.; Springer: Berlin, Germany, 2004; pp. 253–271. [Google Scholar]
- Siciliano, I.; Gilardi, G.; Ortu, G.; Gisi, U.; Gullino, M.L.; Garibaldi, A. Identification and characterization of Alternaria species causing leaf spot on cabbage, cauliflower, wild and cultivated rocket by using molecular and morphological features and mycotoxin production. Eur. J. Plant Pathol. 2017, 149, 401–413. [Google Scholar] [CrossRef]
- Garibaldi, A.; Gilardi, G.; Bertoldo, C.; Gullino, M.L. First report of leaf spot of wild (Diplotaxis tenuifolia) and cultivated (Eruca vesicaria) rocket caused by Alternaria japonica in Italy. Plant Dis. 2011, 95, 1316. [Google Scholar] [CrossRef] [PubMed]
- Gilardi, G.; Gullino, M.L.; Garibaldi, A. New diseases of wild and cultivated rocket in Italy. Acta Hortic. 2013, 1005, 569–572. [Google Scholar] [CrossRef]
- Fisher, P.J.; Petrini, O.; Petrini, L.E.; Descals, E. A preliminary study of fungi inhabiting xylem and whole stems of Olea europaea. Sydowia 1992, 44, 117–121. [Google Scholar]
- Carlucci, A.; Raimondo, M.L.; Santos, J.; Phillips, A.J.L. Plectosphaerella species associated with root and collar rots of horticultural crops in southern Italy. Persoonia 2012, 28, 34–48. [Google Scholar] [CrossRef] [PubMed]
- De Hoog, G.S.; Guarro, J.; Gené, J.; Figueras, M.J. Atlas of Clinical Fungi, 3rd ed.; Centraalbureau voor Schimmelcultures: Utrecht, The Netherlands, 2000. [Google Scholar]
- Sousa Melo, B.; Voltan, A.R.; Arruda, W.; Lopes, F.A.C.; Georg, R.C.; Ulhoa, C.J. Morphological and molecular aspects of sclerotial development in the phytopathogenic fungus Sclerotinia sclerotiorum. Microbiol. Res. 2019, 229, 126326. [Google Scholar] [CrossRef] [PubMed]
- Woudenberg, J.H.C.; Hanse, B.; van Leeuwen, G.C.M.; Groenewald, J.Z.; Crous, P.W. Stemphylium revisited. Stud. Mycol. 2017, 87, 77–103. [Google Scholar] [CrossRef]
- Carlucci, A.; Raimondo, M.L.; Cibelli, F.; Phillips, A.J.L.; Lops, F. Pleurostomophora richardsiae, Neofusicoccum parvum and Phaeoacremonium aleophilum associated with a decline of olives in southern Italy. Phytopathol. Mediterr. 2013, 52, 517–527. [Google Scholar] [CrossRef]
- Meyer, W.; Mitchell, T.G.; Freedman, E.Z.; Vilgalys, R. Hybridization probes for conventional DNA fingerprinting used as single primers in the polymerase chain reaction to distinguish strains of Cryptococcus neoformans. J. Clin. Microbiol. 1993, 31, 2274–2280. [Google Scholar] [CrossRef]
- Santos, J.M.; Phillips, A.J.L. Resolving the complex of Diaporthe (Phomopsis) species occurring on Foeniculum vulgare in Portugal. Fungal Divers. 2009, 34, 111–125. [Google Scholar]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous Ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Pavón, M.Á.; González, I.; Pegels, N.; Martín, R.; García, T. PCR detection and identification of Alternaria species-groups in processed foods based on the genetic marker Alt a 1. Food Control 2010, 21, 1745–1756. [Google Scholar] [CrossRef]
- Sung, G.H.; Sung, J.M.; Hywel-Jones, N.L.; Spatafora, J.W. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenet. Evol. 2007, 44, 1204–1223. [Google Scholar] [CrossRef]
- Berbee, M.L.; Pirseyedi, M.; Hubbard, S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 1999, 91, 964–977. [Google Scholar] [CrossRef]
- Giraldo, A.; Hernández-Restrepo, M.; Crous, P.W. New plectosphaerellaceous species from Dutch garden soil. Mycol. Prog. 2019, 18, 1135–1154. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef] [PubMed]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L.; Sullivan, J. Phylogeny inference based on parsimony and other methods using PAUP*. In The Phylogenetic Handbook: A Practical Approach to DNA and Protein Phylogeny; Cambridge University Press: Cambridge, UK, 2003; pp. 160–206. [Google Scholar]
- Page, R.D.M. TreeView: An application to display phylogenetic trees on personal computers. Bioinformatics 1996, 12, 357–358. [Google Scholar] [CrossRef]
- O’Donnell, K.; Whitaker, B.K.; Laraba, I.; Proctor, R.H.; Brown, D.W.; Broders, K.; Kim, H.S.; McCormick, S.P.; Busman, M.; Aoki, T.; et al. DNA sequence-based identification of Fusarium: A work in progress. Plant Dis. 2022, 106, 1597–1609. [Google Scholar] [CrossRef]
- El-Gremi, S.; Attia, S.M. Production and characterization of microbial dye produced by the saprophytic fungus Epicoccum sp. J. Agric. Chem. Biotechnol. 2007, 32, 7685–7692. [Google Scholar] [CrossRef]
- Martins, L.; Hartmann, I.; Alves, D.O.; Martins, P.C.; Garcia, C.H.; Leclercq, C.C.; Ferreira, R.; He, J.; Renaut, J.; Becker, J.D.; et al. Elucidating how the saprophytic fungus Aspergillus nidulans uses the plant polyester suberin as a carbon source. BMC Genom. 2014, 15, 613. [Google Scholar] [CrossRef] [PubMed]
- Ziaee, A.; Zia, M.; Goli, M. Identification of saprophytic and allergenic fungi in indoor and outdoor environments. Environ. Monit. Assess. 2018, 190, 574. [Google Scholar] [CrossRef]
- Saharan, G.S.; Mehta, N.; Meena, P.D. Alternaria Diseases of Crucifers: Biology, Ecology and Disease Management; Springer: Singapore; Berlin/Heidelberg, Germany; New York, NY, USA, 2016; p. 299. [Google Scholar] [CrossRef]
- Blagojević, J.D.; Vukojević, J.B.; Ivanović, Ž.S. Occurrence and characterization of Alternaria species associated with leaf spot disease in rapeseed in Serbia. Plant Pathol. 2020, 69, 883–900. [Google Scholar] [CrossRef]
- Al-Lami, H.F.D.; You, M.P.; Barbetti, M.J. Incidence, pathogenicity and diversity of Alternaria spp. associated with Alternaria leaf spot of canola (Brassica napus) in Australia. Plant Pathol. 2019, 68, 492–503. [Google Scholar] [CrossRef]
- Ren, X.X.; Zhang, G.Z.; Dai, W.A. First report of damping-off caused by Alternaria japonica on Chinese cabbage seedlings in China. Plant Dis. 2012, 96, 1378. [Google Scholar] [CrossRef] [PubMed]
- Bassimba, D.D.M.; Mira, J.L.; Vicent, A. First report of Alternaria japonica causing black spot of turnip in Spain. Plant Dis. 2013, 97, 1505. [Google Scholar] [CrossRef] [PubMed]
- Keinath, A.P.; Toporek, S.M.; DuBose, V.B.; Zardus, S.H.; Ballew, J.B. First report of Alternaria japonica, a causal agent of black spot, on kale in South Carolina, U.S.A. Plant Dis. 2021, 105, 2016. [Google Scholar] [CrossRef]
- Gilardi, G.; Demarchi, S.; Ortu, G.; Gullino, M.L.; Garibaldi, A. Occurrence of Alternaria japonica on seeds of wild and cultivated rocket. J. Phytopathol. 2014, 163, 419–422. [Google Scholar] [CrossRef]
- Tidwell, T.E.; Blomquist, C.L.; Rooney-Latham, S.; Scheck, H.J. Leaf spot of arugula, caused by Alternaria japonica, in California. Plant Dis. 2014, 98, 1272. [Google Scholar] [CrossRef]
- Hong, S.G.; Cramer, R.A.; Lawrence, C.B.; Pryor, B.M. Alt a 1 allergen homologs from Alternaria and related taxa: Analysis of phylogenetic content and secondary structure. Fungal Genet. Biol. 2005, 42, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Chalbi, A.; Sghaier-Hammami, B.; Meca, G.; Quiles, J.M.; Abdelly, C.; Masiello, M. Characterization of mycotoxigenic Alternaria species isolated from the Tunisian halophyte Cakile maritima. Phytopathol. Mediterr. 2020, 59, 107–108. [Google Scholar] [CrossRef]
- Lawrence, D.P.; Gannibal, P.B.; Peever, T.L.; Pryor, B.M. The sections of Alternaria: Formalizing species-group concepts. Mycologia 2013, 105, 530–546. [Google Scholar] [CrossRef]
- Woudenberg, J.H.C.; Seidl, M.F.; Groenewald, J.Z.; de Vries, M.; Stielow, J.B.; Thomma, B.P.H.J.; Crous, P.W. Alternaria section Alternaria: Species, formae speciales or pathotypes? Stud. Mycol. 2015, 82, 1–21. [Google Scholar] [CrossRef]
- Köhl, J.; Groenenboom-de Haas, B.; Goossen-van de Geijn, H.; Speksnijder, A.; Kastelein, P.; de Hoog, S.; Ende, B.G.v.D. Pathogenicity of Stemphylium vesicarium from different hosts causing brown spot in pear. Eur. J. Plant Pathol. 2008, 124, 151–162. [Google Scholar] [CrossRef]
- Prados-Ligero, A.M.; González-Andújar, J.L.; Melero-Vara, J.M.; Basallote-Ureba, M.J. Development of Pleospora allii on garlic debris infected by Stemphylium vesicarium. Eur. J. Plant Pathol. 1998, 104, 861–870. [Google Scholar] [CrossRef]
- Aveling, T.A.S.; Snyman, H.G. Infection studies of Stemphylium vesicarium on onion leaves. Mycol. Res. 1993, 97, 984–988. [Google Scholar] [CrossRef]
- Pei, Y.-F.; Wang, Y.; Geng, Y.; Zhang, X.-G. Three new species of Stemphylium from Sinkiang, China. Mycotaxon 2010, 111, 167–173. [Google Scholar] [CrossRef]
- Ponti, I.; Cavanni, P.; Brunelli, A. Maculatura bruna delle pere: Eziologia e difesa. Inf. Fitopatol. 1982, 32, 35–40. [Google Scholar]
- Singh, P.; Bugiani, R.; Cavanni, P.; Nakajima, H.; Kodama, M.; Otani, H.; Kohmoto, K. Purification and biological characterization of host-specific SV-toxins from Stemphylium vesicarium causing brown spot of European pear. Phytopathology 1999, 89, 947–953. [Google Scholar] [CrossRef]
- Basallote-Ureba, M.J.; Prados-Ligero, A.M.; Melero-Vara, J.M. Aetiology of leaf spot of garlic and onion caused by Stemphylium vesicarium in Spain. Plant Pathol. 1999, 48, 139–145. [Google Scholar] [CrossRef]
- Andersen, B.; Solfrizzo, M.; Visconti, A. Metabolite profiles of common Stemphylium species. Mycol. Res. 1995, 99, 672–676. [Google Scholar] [CrossRef]
- Tsuge, T.; Harimoto, Y.; Akimitsu, K.; Ohtani, K.; Kodama, M.; Akagi, Y.; Egusa, M.; Yamamoto, M.; Otani, H. Host-selective toxins produced by the plant pathogenic fungus Alternaria alternata. FEMS Microbiol. Rev. 2013, 37, 44–66. [Google Scholar] [CrossRef]
- U.S. Environmental Protection Agency. Chemistry Dashboard. Available online: https://comptox.epa.gov/dashboard/DTXSID10198982 (accessed on 8 June 2020).
- Abad, P.; Pérez, A.; Marqués, M.C.; Vicente, M.J.; Bruton, B.D.; García-Jiménez, J. Assessment of vegetative compatibility of Acremonium cucurbitacearum and Plectosphaerella cucumerina isolates from diseased melon plants. EPPO Bull. 2000, 30, 199–204. [Google Scholar] [CrossRef]
- Raimondo, M.L.; Carlucci, A. Characterization and pathogenicity assessment of Plectosphaerella species associated with stunting disease on tomato and pepper crops in Italy. Plant Pathol. 2018, 67, 626–641. [Google Scholar] [CrossRef]
- Xu, J.; Xu, X.-D.; Cao, Y.-Y.; Zhang, W.-M. First report of greenhouse tomato wilt caused by Plectosphaerella cucumerina in China. Plant Dis. 2014, 98, 158. [Google Scholar] [CrossRef]
- Carrieri, R.; Pizzolongo, G.; Carotenuto, G.; Tarantino, P.; Lahoz, E. First report of necrotic leaf spot caused by Plectosphaerella cucumerina on lamb’s lettuce in southern Italy. Plant Dis. 2014, 98, 998. [Google Scholar] [CrossRef]
- Raimondo, M.L.; Carlucci, A. Characterization and pathogenicity of Plectosphaerella spp. collected from basil and parsley in Italy. Phytopathol. Mediterr. 2018, 57, 284–295. [Google Scholar] [CrossRef]
- Usami, T.; Morii, S.; Matsubara, C.; Amemiya, Y. Plectosphaerella rot of lettuce, coriander, and chervil caused by Plectosphaerella pauciseptata. J. Gen. Plant Pathol. 2012, 78, 368–371. [Google Scholar] [CrossRef]
- Chen, Z.S.; Chen, Y.Y.; Zhou, Y.Y.; Chen, P.Z.; Zhang, W.; Liu, M.; Yan, J.Y.; Wang, C.F. First report of Plectosphaerella pauciseptata associated with tomato root rot disease in China. Plant Dis. 2025, 109, 1377. [Google Scholar] [CrossRef]
- Han, L.; Zhou, X.; Zhao, Y.; Wu, L.; Ping, X.; He, Y.; Peng, S.; He, X.; Du, Y. First report of Plectosphaerella plurivora causing root rot disease in Panax notoginseng in China. J. Phytopathol. 2020, 168, 375–379. [Google Scholar] [CrossRef]
- Gilardi, G.; Garibaldi, A.; Gullino, M.L. Seed transmission of Plectosphaerella cucumerina, causal agent of leaf spot of Diplotaxis tenuifolia in Italy. Phytoparasitica 2013, 41, 411–416. [Google Scholar] [CrossRef]




| Species | Isolate Number | Location | Host | Variety | Year | Part of Plant | GenBank Accession Number | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ITS | tef1-α | Alt-a1 | rpb2 | gapdh | |||||||
| Alternaria alternata | ALT1 | Lucera | Brassica oleracea var. botrytis | Trinacria | 2022 | Leaves | PV872872 | PV952645 | PV929800 | PV934163 | - |
| ALT2 | Lucera | Brassica oleracea var. botrytis | Trinacria | 2022 | Leaves | - | - | - | - | - | |
| ALT3 | Lucera | Brassica oleracea var. botrytis | Trinacria | 2022 | Leaves | - | - | - | - | - | |
| ALT4 | Lucera | Brassica oleracea var. botrytis | Akinen | 2023 | Stem | PV872878 | PV952651 | PV929806 | PV934169 | - | |
| C1 | Lucera | Brassica oleracea var. botrytis | Akinen | 2023 | Stem | PV872873 | PV952646 | PV929801 | PV934164 | - | |
| C5 | Lucera | Brassica oleracea var. botrytis | Aprilia | 2023 | Stem | PV872877 | PV952650 | PV929805 | PV934168 | - | |
| 3AA | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Leaves | PV872876 | PV952649 | PV929804 | PV934167 | - | |
| C3 | Lucera | Brassica oleracea var. italica | Parthenon | 2023 | Stem | - | - | - | - | - | |
| 1A | Cerignola | Brassica oleracea var. italica | Mugnoli | 2022 | Leaves | - | - | - | - | - | |
| 2A | Cerignola | Brassica oleracea var. italica | Mugnoli | 2022 | Leaves | - | - | - | - | - | |
| 2AR | Cerignola | Brassica oleracea var. italica | Mugnoli | 2022 | Leaves | PV872871 | PV952644 | PV929799 | PV934161 | - | |
| 3B | Cerignola | Brassica oleracea var. italica | Mugnoli | 2022 | Leaves | - | - | - | - | - | |
| 3BC | Cerignola | Brassica oleracea var. italica | Mugnoli | 2023 | Leaves | - | - | - | - | - | |
| 3BD | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Leaves | PV872874 | PV952647 | PV929802 | PV934165 | - | |
| 4AA | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Leaves | - | - | - | - | - | |
| 4BD | Cerignola | Brassica rapa var. cymosa (Turnip) | Centoventina | 2023 | Leaves | - | - | - | - | - | |
| 6AA | Foggia | Brassica rapa var. cymosa (Turnip) | Centoventina | 2023 | Leaves | - | - | - | - | - | |
| 6BB | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Leaves | PV872870 | PV952643 | PV929798 | PV934162 | - | |
| 10BA | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Leaves | PV872875 | PV952648 | PV929803 | PV934166 | - | |
| 3A | Cerignola | Brassica oleracea var. italica | Mugnoli | 2022 | Leaves | - | - | - | - | - | |
| 3Bs | Cerignola | Brassica rapa var. cymosa (Turnip) | Centoventina | 2022 | Leaves | PV872869 | PV952642 | PV929797 | PV934160 | - | |
| A. brassicicola | ALT5 | Lucera | Brassica oleracea var. botrytis | Akinen | 2023 | Leaves | - | - | - | - | - |
| 2AA | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Stem | - | - | - | - | - | |
| 2AB | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Stem | - | - | - | - | - | |
| 2AC | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Stem | PV920688 | PV952661 | PV934179 | PV942081 | - | |
| 4AB | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Leaves | - | - | - | - | - | |
| 7AB | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Leaves | - | - | - | - | - | |
| 7AD | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Leaves | PV920689 | PV952662 | PV934180 | PV942082 | - | |
| 10AC | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Leaves | PV920690 | PV952663 | PV934181 | PV942083 | - | |
| A. japonica | 1BB | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Stem | PV920679 | PV952652 | PV934170 | PV942072 | - |
| Alternaria sp. | 1B1 | Lucera | Brassica oleracea var. botrytis | Akinen | 2023 | Leaves | - | - | - | - | - |
| 1B2 | Lucera | Brassica oleracea var. botrytis | Akinen | 2023 | Leaves | - | - | - | - | - | |
| 2C | Cerignola | Brassica oleracea var. italica | Mugnoli | 2022 | Leaves | PV920684 | PV952657 | PV934175 | PV942077 | - | |
| 5A | Cerignola | Brassica oleracea var. italica | Mugnoli | 2022 | Leaves | PV920680 | PV952653 | PV934171 | PV942073 | - | |
| 3AB | Foggia | Brassica oleracea var. italica | Parthenon | 2023 | Leaves | - | - | - | - | - | |
| 4BB | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Stem | PV920685 | PV952658 | PV934176 | PV942078 | - | |
| 6AB | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Stem | - | - | - | - | - | |
| 7AC | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Stem | PV920686 | PV952659 | PV934177 | PV942079 | - | |
| 7AE | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Corymb | - | - | - | - | - | |
| 10AA | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Leaves | PV920683 | PV952656 | PV934174 | PV942076 | - | |
| 10AB | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Leaves | - | - | - | - | - | |
| 10BB | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Corymb | PV920682 | PV952655 | PV934173 | PV942075 | - | |
| 10BC | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Leaves | - | - | - | - | - | |
| 3BG | Cerignola | Brassica rapa var. cymosa (Turnip) | Centoventina | 2023 | Leaves | PV920681 | PV952654 | PV934172 | PV942074 | - | |
| 4B | Foggia | Brassica oleracea var. italica | Parthenon | 2023 | Leaves | - | - | - | - | - | |
| 4BA | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Leaves | PV920687 | PV952660 | PV934178 | PV942080 | - | |
| Fusarium solani species complex | 1G | Lucera | Brassica oleracea var. botrytis | Akinen | 2022 | Stem | - | - | - | - | - |
| 1F | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Stem | PV920568 | PV952664 | - | - | - | |
| 1H | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Stem | - | - | - | - | - | |
| 2G | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Stem | - | - | - | - | - | |
| 3F | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Stem | - | - | - | - | - | |
| 3GA | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Stem | - | - | - | - | - | |
| 3H | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Stem | - | - | - | - | - | |
| 5E | Cerignola | Brassica oleracea var. italica | Mugnoli | 2023 | Stem | PV920570 | PV952666 | - | - | - | |
| 6D | Cerignola | Brassica oleracea var. italica | Mugnoli | 2023 | Stem | - | - | - | - | - | |
| 6ED | Foggia | Brassica oleracea var. italica | Parthenon | 2022 | Leaves | - | - | - | - | - | |
| 7GA | Cerignola | Brassica oleracea var. italica | Mugnoli | 2023 | Stem | - | - | - | - | - | |
| 7BC | Cerignola | Brassica oleracea var. italica | Mugnoli | 2023 | Leaves | - | - | - | - | - | |
| 7D | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Leaves | - | - | - | - | - | |
| 7F | Cerignola | Brassica oleracea var. italica | Mugnoli | 2023 | Leaves | - | - | - | - | - | |
| 9D | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Leaves | - | - | - | - | - | |
| 9EF | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Leaves | - | - | - | - | - | |
| 10H | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Leaves | - | - | - | - | - | |
| 11A | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Corymb | - | - | - | - | - | |
| 11DE | Foggia | Brassica oleracea var. italica | Parthenon | 2022 | Corymb | - | - | - | - | - | |
| 19A | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Corymb | - | - | - | - | - | |
| 4FG | Cerignola | Brassica rapa var. cymosa | Centoventina | 2022 | Stem | PV920569 | PV952665 | - | - | - | |
| 10C | Cerignola | Brassica rapa var. cymosa | Centoventina | 2022 | Stem | - | - | - | - | - | |
| 18B | Cerignola | Brassica rapa var. cymosa | Centoventina | 2022 | Stem | - | - | - | - | - | |
| 19E | Cerignola | Brassica rapa var. cymosa | Centoventina | 2022 | Leaves | - | - | - | - | - | |
| 23A | Cerignola | Brassica rapa var. cymosa | Centoventina | 2022 | Leaves | - | - | - | - | - | |
| Plectosphaerella cucumerina | 4F | Lucera | Brassica oleracea var. botrytis | Akinen | 2023 | Corymb | - | - | - | - | - |
| 6F | Lucera | Brassica oleracea var. botrytis | Akinen | 2023 | Stem | - | - | - | - | - | |
| 11F | Lucera | Brassica oleracea var. botrytis | Akinen | 2023 | Leaves | - | - | - | - | - | |
| 15F | Lucera | Brassica oleracea var. botrytis | Akinen | 2023 | Leaves | - | - | - | - | - | |
| 19C | Lucera | Brassica oleracea var. botrytis | Akinen | 2023 | Leaves | - | - | - | - | - | |
| 2G | Foggia | Brassica oleracea var. italica | Parthenon | 2022 | Leaves | - | - | - | - | - | |
| 4G | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Leaves | - | - | - | - | - | |
| 7BC | Cerignola | Brassica oleracea var. italica | Mugnoli | 2023 | Leaves | - | - | - | - | - | |
| 11G | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Leaves | - | - | - | - | - | |
| 12B | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Leaves | - | - | - | - | - | |
| 13B | Cerignola | Brassica oleracea var. italica | Mugnoli | 2023 | Leaves | PV920675 | PV952639 | - | PV929767 | - | |
| 14B | Cerignola | Brassica oleracea var. italica | Mugnoli | 2023 | Stem | - | - | - | - | - | |
| 15AB | Cerignola | Brassica oleracea var. italica | Mugnoli | 2023 | Stem | - | - | - | - | - | |
| 1E | Cerignola | Brassica rapa var. cymosa | Centoventina | 2023 | Stem | - | - | - | - | - | |
| 2EF | Cerignola | Brassica rapa var. cymosa | Centoventina | 2023 | Stem | PV920674 | PV952638 | - | PV929766 | - | |
| 7E | Cerignola | Brassica rapa var. cymosa | Centoventina | 2023 | Stem | - | - | - | - | - | |
| 10E | Cerignola | Brassica rapa var. cymosa | Centoventina | 2023 | Leaves | - | - | - | - | - | |
| 10EF | Cerignola | Brassica rapa var. cymosa | Centoventina | 2023 | Leaves | - | - | - | - | - | |
| 12E | Cerignola | Brassica rapa var. cymosa | Centoventina | 2023 | Leaves | - | - | - | - | - | |
| 13D | Foggia | Brassica rapa var. cymosa | Centoventina | 2023 | Corymb | - | - | - | - | - | |
| P. pauciseptata | 6BF | Lucera | Brassica oleracea var. botrytis | Aprilia | 2022 | Stem | - | - | - | - | - |
| 9BC | Lucera | Brassica oleracea var. botrytis | Akinen | 2022 | Leaves | - | - | - | - | - | |
| C1AB | Lucera | Brassica oleracea var. botrytis | Akinen | 2022 | Leaves | - | - | - | - | - | |
| 2AD | Cerignola | Brassica oleracea var. italica | Mugnoli | 2023 | Leaves | - | - | - | - | - | |
| 2AE | Cerignola | Brassica oleracea var. italica | Mugnoli | 2023 | Leaves | - | - | - | - | ||
| 3BE | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Stem | - | - | - | - | - | |
| 3F | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Stem | - | - | - | - | - | |
| 4BE | Cerignola | Brassica oleracea var. italica | Mugnoli | 2023 | Stem | - | - | - | - | - | |
| 5AF | Cerignola | Brassica oleracea var. italica | Mugnoli | 2023 | Leaves | PV920673 | PV952637 | - | PV929765 | - | |
| 5BC | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Leaves | - | - | - | - | - | |
| 6BC | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Leaves | - | - | - | - | - | |
| 7BF | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Leaves | - | - | - | - | - | |
| 10AE | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Leaves | - | - | - | - | - | |
| C1A | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Leaves | - | - | - | - | - | |
| C3A | Cerignola | Brassica oleracea var. italica | Parthenon | 2023 | Leaves | - | - | - | - | - | |
| 1C | Cerignola | Brassica rapa var. cymosa | Centoventina | 2022 | Stem | PV920672 | PV952636 | - | PV929764 | - | |
| 3CA | Cerignola | Brassica rapa var. cymosa | Centoventina | 2022 | Leaves | - | - | - | - | - | |
| 9C | Foggia | Brassica rapa var. cymosa | Centoventina | 2022 | Leaves | - | - | - | - | - | |
| 16A | Cerignola | Brassica oleracea var. botrytis | Akinen | 2022 | Stem | PV920676 | PV952640 | PV929768 | |||
| 18A | Lucera | Brassica oleracea var. botrytis | Akinen | 2022 | Leaves | - | - | - | - | - | |
| 21A | Lucera | Brassica oleracea var. botrytis | Akinen | 2022 | Leaves | - | - | - | - | - | |
| P. plurivora | 14AC | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Stem | - | - | - | - | - |
| 14C | Cerignola | Brassica oleracea var. italica | Mugnoli | 2023 | Leaves | - | - | - | - | - | |
| 17C | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Leaves | - | - | - | - | - | |
| 20C | Cerignola | Brassica oleracea var. italica | Mugnoli | 2023 | Leaves | - | - | - | - | - | |
| 18D | Cerignola | Brassica rapa var. cymosa | Centoventina | 2022 | Stem | PV920677 | PV952641 | - | PV929769 | ||
| 22A | Cerignola | Brassica rapa var. cymosa | Centoventina | 2022 | Leaves | - | - | - | - | - | |
| Sclerotinia sclerotiorum | 1C | Cerignola | Brassica oleracea var. botrytis | Akinen | 2022 | Corymb | - | - | - | - | - |
| 3E | Lucera | Brassica oleracea var. botrytis | Akinen | 2022 | Corymb | - | - | - | - | - | |
| 6C | Lucera | Brassica oleracea var. botrytis | Akinen | 2022 | Corymb | - | - | - | - | - | |
| 12AB | Lucera | Brassica oleracea var. botrytis | Akinen | 2022 | Corymb | - | - | - | - | - | |
| 14D | Lucera | Brassica oleracea var. botrytis | Akinen | 2022 | Corymb | - | - | - | - | - | |
| 15B | Lucera | Brassica oleracea var. botrytis | Akinen | 2022 | Corymb | - | - | - | - | - | |
| 17AC | Lucera | Brassica oleracea var. botrytis | Akinen | 2022 | Corymb | - | - | - | - | - | |
| 18E | Lucera | Brassica oleracea var. botrytis | Akinen | 2022 | Corymb | - | - | - | - | - | |
| 2F | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Corymb | - | - | - | - | - | |
| 3D | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Corymb | - | - | - | - | - | |
| 4C | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Corymb | - | - | - | - | - | |
| 11C | Foggia | Brassica oleracea var. italica | Parthenon | 2022 | Corymb | - | - | - | - | - | |
| 13F | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Corymb | PV920572 | - | - | - | - | |
| 16D | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Corymb | - | - | - | - | - | |
| 4D | Cerignola | Brassica rapa var. cymosa | Centoventina | 2023 | Corymb | - | - | - | - | - | |
| 5CD | Cerignola | Brassica rapa var. cymosa | Centoventina | 2023 | Corymb | PV920571 | - | - | - | - | |
| 7B | Cerignola | Brassica rapa var. cymosa | Centoventina | 2023 | Corymb | - | - | - | - | - | |
| Stemphylium vesicarium | 1BA | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Leaves | - | - | - | - | - |
| 2BA | Cerignola | Brassica oleracea var. italica | Mugnoli | 2023 | Leaves | - | - | - | - | - | |
| 3C | Lucera | Brassica oleracea var. botrytis | Akinen | 2022 | Stem | - | - | - | - | - | |
| 3BA | Foggia | Brassica oleracea var. italica | Parthenon | 2022 | Leaves | - | - | - | - | - | |
| 6B | Cerignola | Brassica oleracea var. italica | Mugnoli | 2023 | Leaves | PV920566 | - | - | - | PV942084 | |
| 7C | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Stem | PV920567 | - | - | - | PV942085 | |
| 7DB | Cerignola | Brassica oleracea var. italica | Parthenon | 2022 | Stem | - | - | - | - | - | |
| No. of Isolates (IF, %) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fungal Species Isolated | Brassica oleracea var. botrytis “Cauliflower” (Sample Number = 4) | Brassica oleracea var. italica “Broccoli” (n = 8) | Brassica oleracea var. italica “Mugnoli” (n = 5) | Brassica rapa var. cymosa “Turnip” (n = 5) | Total (n = 22) | ||||||||||||
| Stem | Leaf | Corymb | Subtotal | Stem | Leaf | Corymb | Subtotal | Stem | Leaf | Corymb | Subtotal | Stem | Leaf | Corymb | Subtotal | ||
| Alternaria alternata | 3 (7.5) | 3 (7.5) | 0 (0.0) | 6 (15.0) | 1 (0.7) | 5 (3.3) | 0 (0.0) | 6 (4.0) | 0 (0.0) | 6 (9.0) | 0 (0.0) | 6 (9.0) | 0 (0.0) | 3 (4.2) | 0 (0.0) | 3 (4.2) | 21 (6.4) |
| A. brassicicola | 0 (0.0) | 1 (2.5) | 0 (0.0) | 1 (2.5) | 3 (2.0) | 4 (2.6) | 0 (0.0) | 7 (4.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 8 (2.4) |
| A. japonica | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.7) | 0 (0.0) | 0 (0.0) | 1 (0.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.3) |
| Alternaria spp. | 0 (0.0) | 2 (5.0) | 0 (0.0) | 2 (5.0) | 3 (2.0) | 6 (4.0) | 2 (1.3) | 11 (7.3) | 0 (0.0) | 2 (3.0) | 0 (0.0) | 2 (3.0) | 0 (0.0) | 1 (1.4) | 0 (0.0) | 1 (1.4) | 16 (4.8) |
| Stemphylium vesicarium | 1 (2.5) | 0 (0.0) | 0 (0.0) | 1 (2.5) | 2 (1.3) | 2 (1.3) | 0 (0.0) | 4 (2.6) | 0 (0.0) | 2 (3.0) | 0 (0.0) | 2 (3.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 7 (2.1) |
| Subtotal Alternaria/Stemphylium ssp. | 4 (10.0) | 6 (15.0) | 0 (0.0) | 10 (25.0) | 10 (6.7) | 17 (11.2) | 2 (1.3) | 29 (19.2) | 0 (0.0) | 10 (15.0) | 0 (0.0) | 10 (15.0) | 0 (0.0) | 4 (5.6) | 0 (0.0) | 4 (5.6) | 53 (16.0) |
| Plectosphaerella cucumerina | 1 (2.5) | 3 (7.5) | 1 (2.5) | 5 (12.5) | 0 (0.0) | 4 (2.6) | 0 (0.0) | 4 (2.6) | 2 (3.0) | 2 (3.0) | 0 (0.0) | 4 (6.0) | 3 (4.2) | 3 (4.2) | 1 (1.4) | 7 (9.7) | 20 (6.1) |
| P. pauciseptata | 1 (2.5) | 2 (5.0) | 0 (0.0) | 3 (7.5) | 2 (1.3) | 6 (4.0) | 0 (0.0) | 8 (5.3) | 1 (1.5) | 3 (4.5) | 0 (0.0) | 4 (6.0) | 1 (1.4) | 2 (2.8) | 0 (0.0) | 3 (4.2) | 18 (5.5) |
| P. plurivora | 1 (2.5) | 2 (5.0) | 0 (0.0) | 3 (7.5) | 1 (0.7) | 1 (0.7) | 0 (0.0) | 2 (1.3) | 1 (1.5) | 1 (1.5) | 0 (0.0) | 2 (3.0) | 1 (1.4) | 1 (1.4) | 0 (0.0) | 2 (2.8) | 9 (2.7) |
| Subtotal Plectosphaerella spp. | 3 (7.5) | 7 (17.5) | 1 (2.5) | 11 (27.5) | 3 (2.0) | 11 (7.3) | 0 (0.0) | 14 (9.2) | 3 (4.5) | 6 (9.0) | 0 (0.0) | 9 (13.5) | 5 (6.6) | 6 (7.0) | 1 (1.4) | 12 (16.7) | 47 (14.2) |
| Fusarium solani species complex | 1 (2.5) | 0 (0.0) | 0 (0.0) | 1 (2.5) | 6 (4.0) | 5 (3.3) | 3 (2.0) | 14 (9.3) | 3 (4.5) | 2 (3.0) | 0 (0.0) | 5 (7.5) | 3 (4.2) | 2 (2.8) | 0 (0.0) | 5 (6.9) | 25 (7.6) |
| Sclerotinia sclerotiorum | 0 (0.0) | 0 (0.0) | 8 (20.0) | 8 (20.0) | 0 (0.0) | 0 (0.0) | 6 (4.0) | 6 (4.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (4.2) | 3 (4.2) | 17 (5.2) |
| Aspergillus spp. | 0 (0.0) | 1 (2.5) | 1 (2.5) | 2 (5.0) | 5 (3.3) | 8 (5.3) | 9 (6.0) | 22 (14.6) | 0 (0.0) | 3 (4.5) | 3 (4.5) | 6 (9.0) | 1 (1.4) | 3 (4.2) | 2 (2.8) | 6 (8.3) | 36 (10.9) |
| Epicoccum spp. | 1 (2.5) | 1 (2.5) | 0 (0.0) | 2 (5.0) | 3 (2.0) | 7 (4.6) | 6 (4.0) | 16 (10.6) | 0 (0.0) | 4 (6.0) | 3 (4.5) | 7 (10.4) | 1 (1.4) | 5 (6.9) | 2 (2.8) | 8 (11.1) | 33 (10.0) |
| Penicillium spp. | 0 (0.0) | 1 (2.5) | 2 (5.0) | 3 (7.5) | 7 (4.6) | 8 (5.3) | 11 (7.3) | 26 (17.2) | 0 (0.0) | 4 (6.0) | 4 (6.0) | 8 (11.9) | 2 (2.8) | 3 (4.2) | 6 (8.3) | 11 (15.3) | 48 (14.5) |
| Total fungi | 9 (22.5) | 16 (40.0) | 12 (30.0) | 37 (92.5) | 34 (22.5) | 56 (37.1) | 37 (24.5) | 127 (84.1) | 6 (8.9) | 29 (43.2) | 11 (16.5) | 46 (68.6) | 12 (16.6) | 23 (32.0) | 14 (19.5) | 49 (68.1) | 259 (78.5) |
| Opportunistic bacteria | 1 (2.5) | 0 (0.0) | 1 (2.5) | 2 (5.0) | 0 (0.0) | 5 (3.3) | 2 (1.3) | 7 (4.6) | 0 (0.0) | 6 (9.0) | 0 (0.0) | 6 (9.0) | 3 (4.2) | 5 (6.9) | 0 (0.0) | 8 (11.1) | 23 (7.0) |
| No growth | 0 (0.0) | 0 (0.0) | 1 (2.5) | 1 (2.5) | 2 (1.3) | 7 (4.6) | 8 (5.3) | 17 (11.3) | 2 (3.0) | 5 (7.5) | 8 (11.9) | 15 (22.4) | 3 (4.2) | 5 (6.9) | 7 (9.7) | 15 (20.8) | 48 (14.5) |
| Total | 10 (25.0) | 16 (40.0) | 14 (35.0) | 40 (100.0) | 36 (23.8) | 68 (45.0) | 47 (31.1) | 151 (100.0) | 8 (11.9) | 40 (59.7) | 19 (28.4) | 67 (100.0) | 18 (25.0) | 33 (45.8) | 21 (29.2) | 72 (100.0) | 330 (100.0) |
| Locus | No. of Sequences | No. of Characters | Parsimony-Informative | Constant | Unique | Model | |
|---|---|---|---|---|---|---|---|
| Alternaria | ITS | 144 | 570 | 97 | 430 | 43 | GTR + I + G |
| tef-1α | 140 | 344 | 117 | 184 | 43 | SYM + G | |
| Alt a1 | 109 | 147 | 85 | 46 | 16 | HKY + I + G | |
| rpb2 | 146 | 780 | 247 | 463 | 70 | SYM + I + G | |
| Total | 1841 | 546 | 1123 | 172 | - | ||
| Stemphylium | ITS | 30 | 564 | 65 | 489 | 10 | K2P + I |
| gapdh | 30 | 582 | 117 | 458 | 7 | K2P + G4 | |
| Total | 1146 | 182 | 947 | 17 | - | ||
| Plectospaerella | ITS | 44 | 529 | 67 | 446 | 16 | TN + F + I + G4 |
| tef-1α | 44 | 749 | 177 | 526 | 46 | TIM3 + F + I + G4 | |
| rpb2 | 44 | 1284 | 221 | 946 | 117 | TN + F + I + G4 | |
| Total | 2562 | 465 | 1918 | 179 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Mourou, M.; Raimondo, M.L.; Spetik, M.; Lops, F.; Ricciardi, G.; Morea, M.G.; Eichmeier, A.; Carlucci, A. Diversity of Fungi Associated with Diseases of Cultivated Brassicaceae in Southern Italy. J. Fungi 2026, 12, 13. https://doi.org/10.3390/jof12010013
Mourou M, Raimondo ML, Spetik M, Lops F, Ricciardi G, Morea MG, Eichmeier A, Carlucci A. Diversity of Fungi Associated with Diseases of Cultivated Brassicaceae in Southern Italy. Journal of Fungi. 2026; 12(1):13. https://doi.org/10.3390/jof12010013
Chicago/Turabian StyleMourou, Marwa, Maria Luisa Raimondo, Milan Spetik, Francesco Lops, Gaetana Ricciardi, Maria Grazia Morea, Ales Eichmeier, and Antonia Carlucci. 2026. "Diversity of Fungi Associated with Diseases of Cultivated Brassicaceae in Southern Italy" Journal of Fungi 12, no. 1: 13. https://doi.org/10.3390/jof12010013
APA StyleMourou, M., Raimondo, M. L., Spetik, M., Lops, F., Ricciardi, G., Morea, M. G., Eichmeier, A., & Carlucci, A. (2026). Diversity of Fungi Associated with Diseases of Cultivated Brassicaceae in Southern Italy. Journal of Fungi, 12(1), 13. https://doi.org/10.3390/jof12010013







