AoChk1 Is Required for Sporulation, Trap Formation, and Metabolic Process in Arthrobotrys oligospora
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Phylogenetic Tree Construction and Sequence Analysis
2.3. Deletion of Aochk1
2.4. Assays of Mycelial Growth and Spore Yield
2.5. Comparison of Trap Formation and Pathogenicity
2.6. Staining and Observation of Mycelial Structures
2.7. RT-qPCR Analysis
2.8. Analysis of Metabolomics
2.9. Transcriptomics Analysis
2.10. Data Analysis
3. Results
3.1. Bioinformatics Analysis and Knockout of the Aochk1 Gene
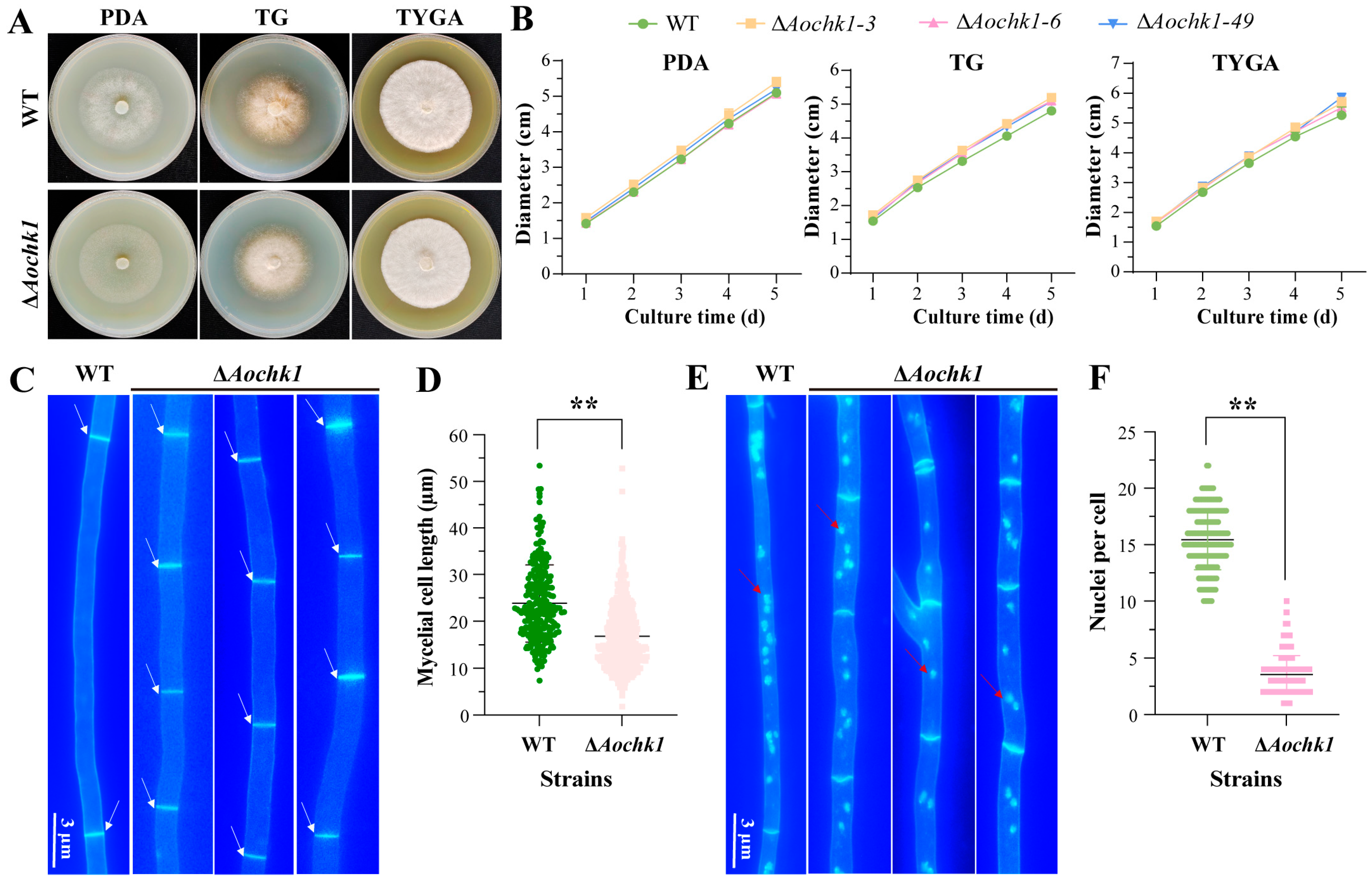
3.2. Aochk1 Gene Affects the Length of Mycelial Cells and the Number of Nuclei
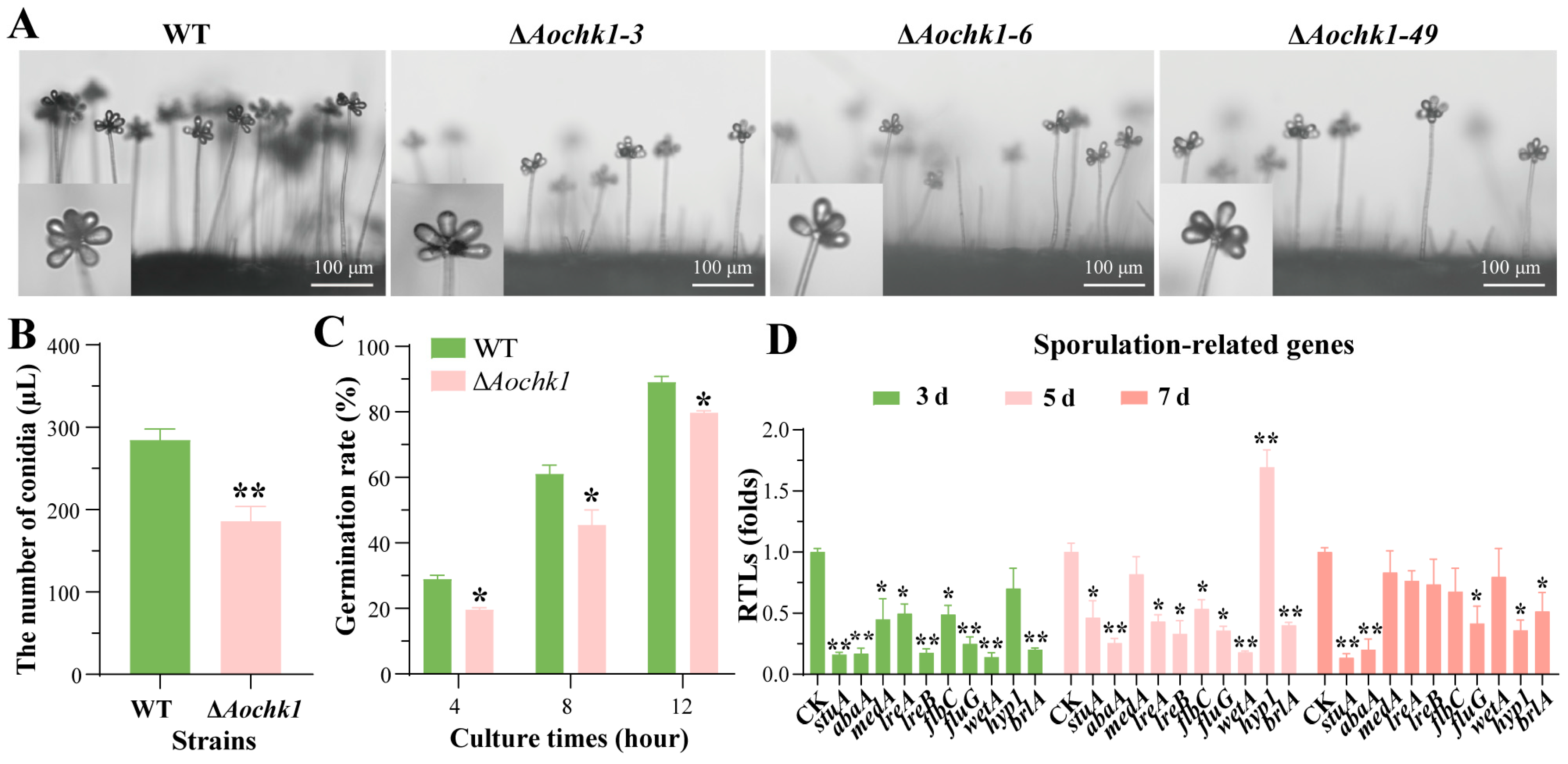
3.3. Deletion of Aochk1 Impairs Sporulation and Germination Rate
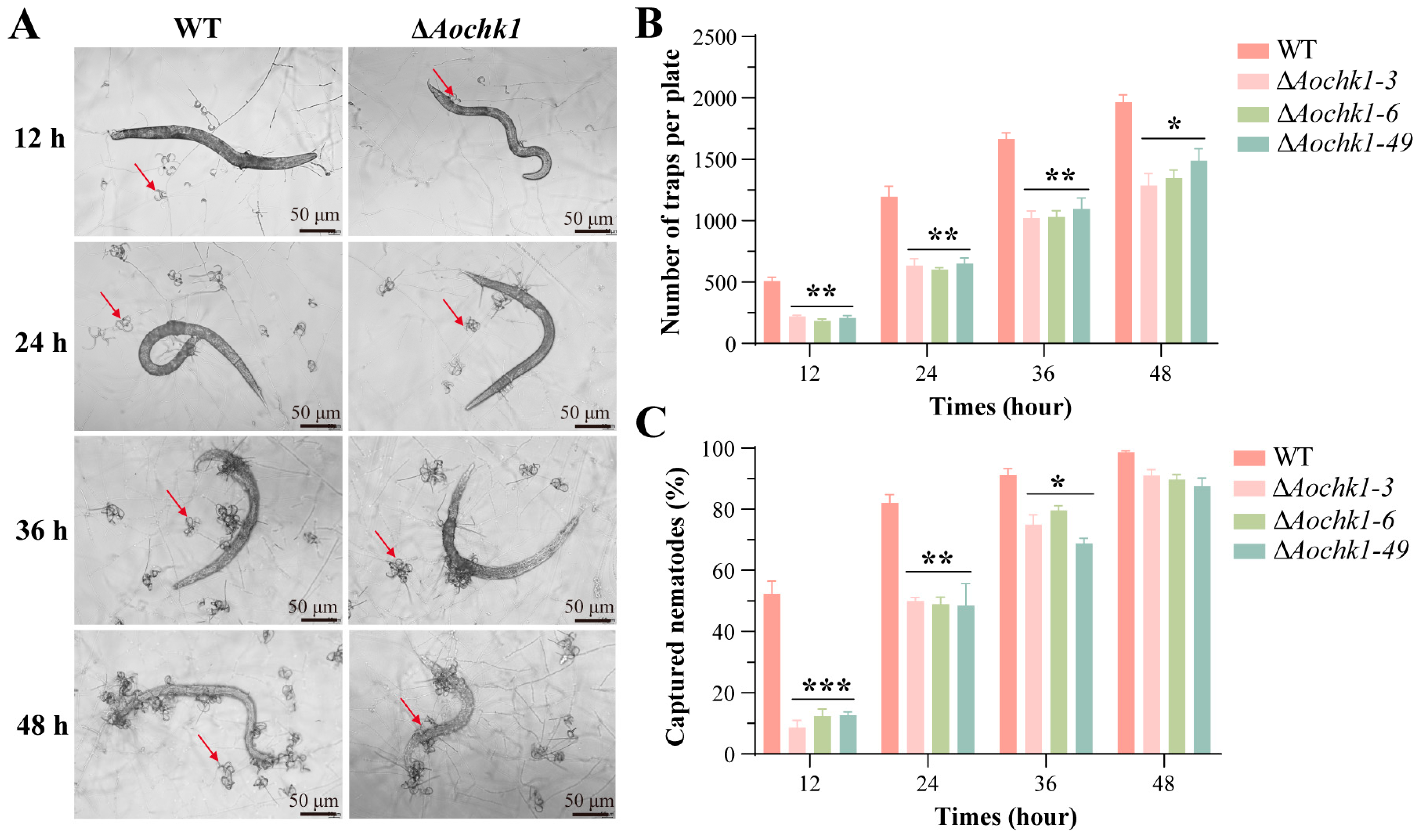
3.4. Aochk1 Is Involved in Regulating Trap Formation and Pathogenicity
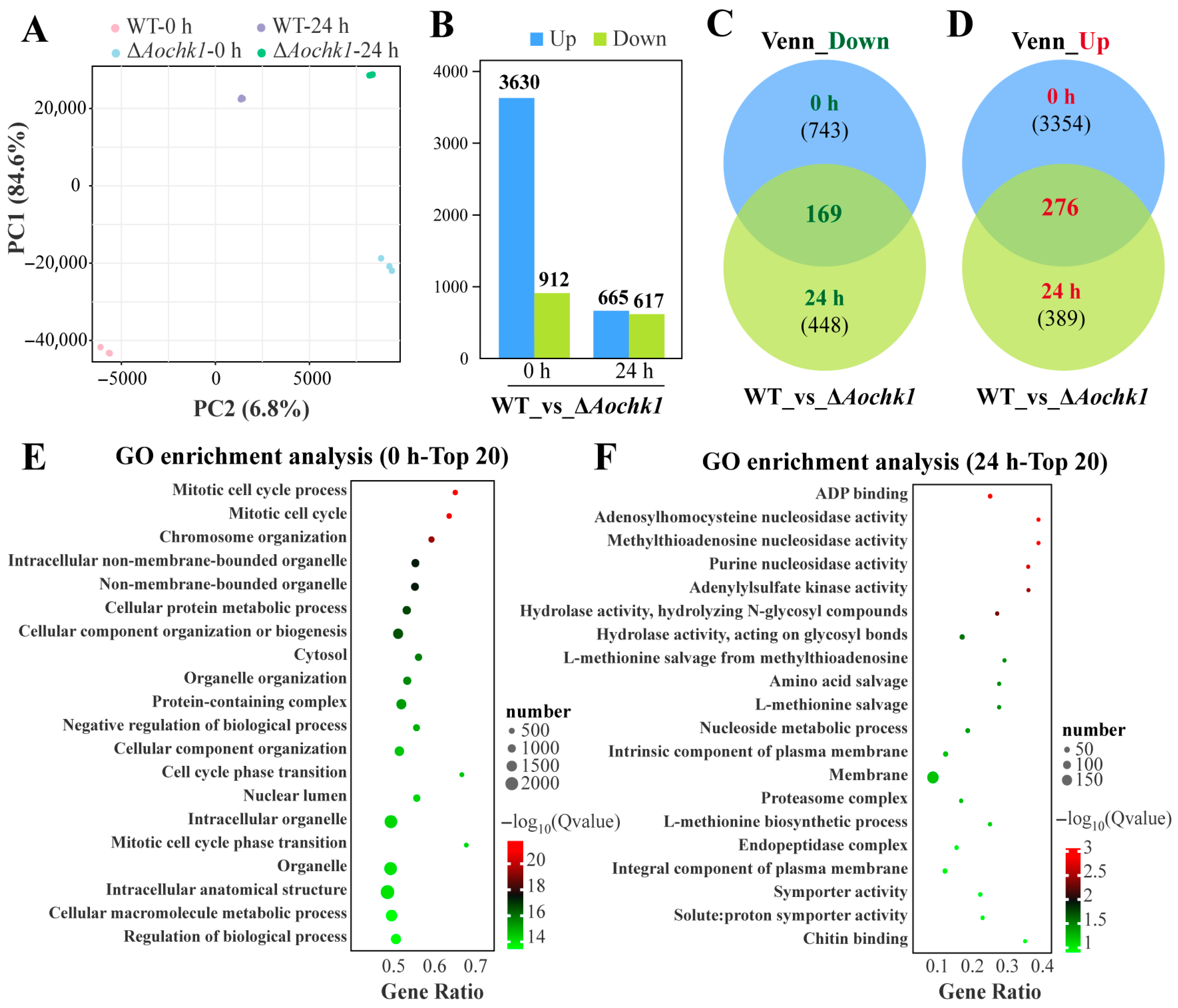
3.5. Transcriptomic Analysis of the Aochk1 Gene
3.6. Aochk1 Is Involved in Regulating Lipid Metabolism and Autophagy Levels
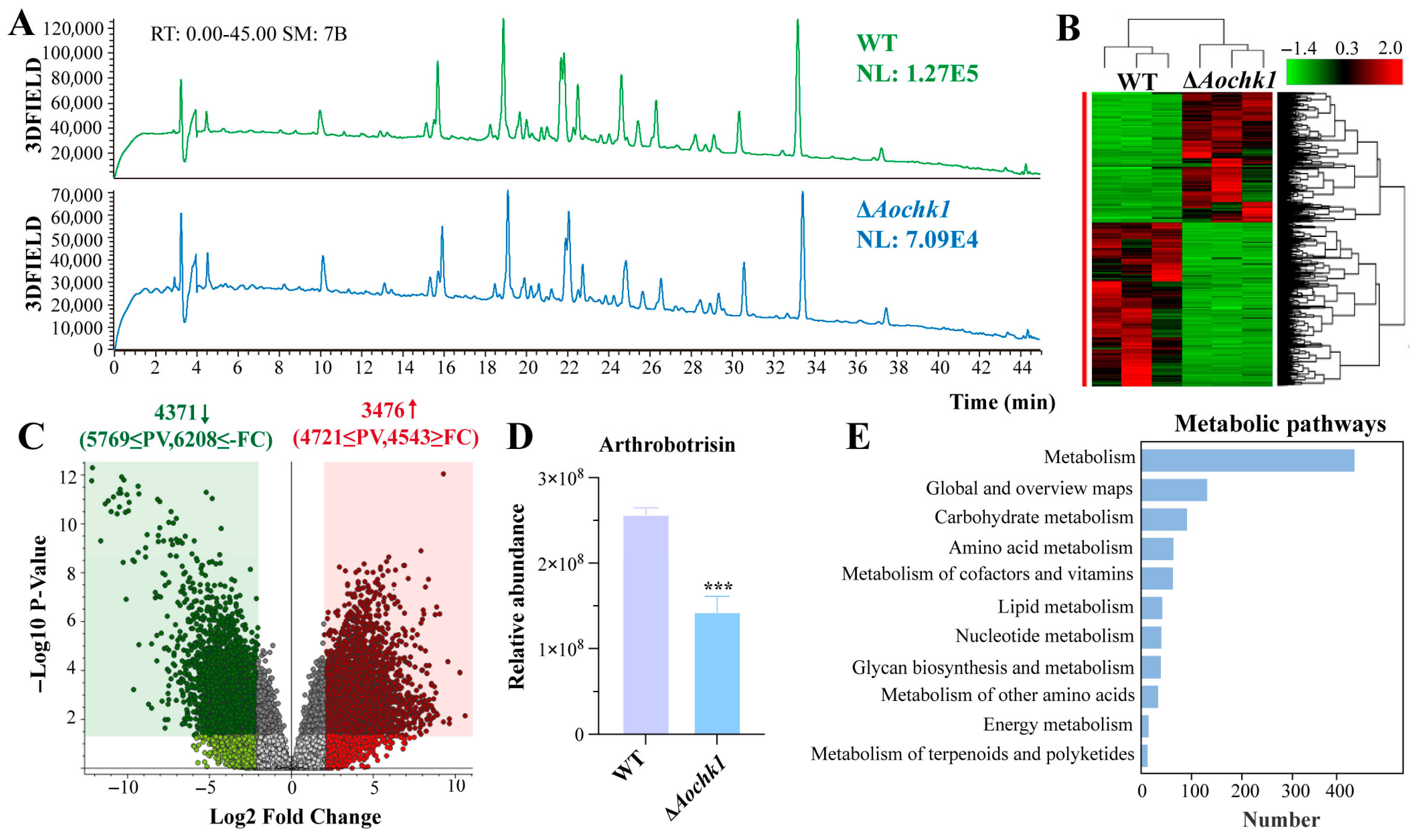
3.7. Aochk1 Is Involved in the Regulation of Secondary Metabolism
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Y.; Sanchez, Y. Chk1 in the DNA damage response: Conserved roles from yeasts to mammals. DNA Repair 2004, 3, 1025–1032. [Google Scholar] [CrossRef]
- DeStephanis, D.; McLeod, M.; Yan, S. REV1 is important for the ATR-Chk1 DNA damage response pathway in Xenopus egg extracts. Biochem. Biophys. Res. Commun. 2015, 460, 609–615. [Google Scholar] [CrossRef]
- Kizhedathu, A.; Kunnappallil, R.S.; Bagul, A.V.; Verma, P.; Guha, A. Multiple Wnts act synergistically to induce Chk1/Grapes expression and mediate G2 arrest in Drosophila tracheoblasts. eLife 2020, 9, e57056. [Google Scholar] [CrossRef]
- Collart, C.; Smith, J.C.; Zegerman, P. Chk1 inhibition of the replication factor Drf1 guarantees cell-cycle elongation at the Xenopus laevis mid-blastula transition. Dev. Cell 2017, 42, 82–96.e3. [Google Scholar] [CrossRef]
- Petrus, M.J.; Wilhelm, D.E.; Murakami, M.; Kappas, N.C.; Carter, A.D.; Wroble, B.N.; Sible, J.C. Altered expression of Chk1 disrupts cell cycle remodeling at the midblastula transition in Xenopus laevis embryos. Cell Cycle 2004, 3, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, Y.; Wong, C.; Thoma, R.S.; Richman, R.; Wu, Z.; Piwnica-Worms, H.; Elledge, S.J. Conservation of the Chk1 checkpoint pathway in mammals: Linkage of DNA damage to Cdk regulation through Cdc25. Science 1997, 277, 1497–1501. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, L.; Xu, X.; Yuan, Y.; Zhang, B.; Li, Z.; Xie, Y.; Yan, R.; Zheng, Z.; Ji, J.; et al. The intra-S phase checkpoint directly regulates replication elongation to preserve the integrity of stalled replisomes. Proc. Natl. Acad. Sci. USA 2021, 118, e2019183118. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.C.; Shieh, S.Y. Determination of CHK1 cellular localization by immunofluorescence microscopy. Methods Mol. Biol. 2021, 2267, 1–6. [Google Scholar] [CrossRef]
- Zhang, Y.; Hunter, T. Roles of Chk1 in cell biology and cancer therapy. Int. J. Cancer 2014, 134, 1013–1023. [Google Scholar] [CrossRef]
- Palermo, C.; Hope, J.C.; Freyer, G.A.; Rao, H.; Walworth, N.C. Importance of a C-terminal conserved region of Chk1 for checkpoint function. PLoS ONE 2008, 3, e1427. [Google Scholar] [CrossRef]
- Kosoy, A.; O’Connell, M.J. Regulation of Chk1 by its C-terminal domain. Mol. Biol. Cell 2008, 19, 4546–4553. [Google Scholar] [CrossRef] [PubMed]
- Leman, A.R.; Noguchi, E. The replication fork: Understanding the eukaryotic replication machinery and the challenges to genome duplication. Genes 2013, 4, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.W.; Lee, Y.; Huh, E.Y.; Lee, S.C.; Lim, S.; Bahn, Y.S. Rad53- and Chk1-dependent DNA damage response pathways cooperatively promote fungal pathogenesis and modulate antifungal drug susceptibility. mBio 2019, 10, e01726-18. [Google Scholar] [CrossRef]
- Rocha, D.; Castro, T.L.P.; Aguiar, E.; Pacheco, L.G.C. Gene expression analysis in bacteria by RT-qPCR. Methods Mol. Biol. 2020, 2065, 119–137. [Google Scholar] [CrossRef]
- Michelena, J.; Gatti, M.; Teloni, F.; Imhof, R.; Altmeyer, M. Basal CHK1 activity safeguards its stability to maintain intrinsic S-phase checkpoint functions. J. Cell Biol. 2019, 218, 2865–2875. [Google Scholar] [CrossRef]
- Smits, V.A.; Gillespie, D.A. DNA damage control: Regulation and functions of checkpoint kinase 1. FEBS J. 2015, 282, 3681–3692. [Google Scholar] [CrossRef]
- Syljuåsen, R.G.; Sørensen, C.S.; Hansen, L.T.; Fugger, K.; Lundin, C.; Johansson, F.; Helleday, T.; Sehested, M.; Lukas, J.; Bartek, J. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol. Cell. Biol. 2005, 25, 3553–3562. [Google Scholar] [CrossRef]
- Furnari, B.; Rhind, N.; Russell, P. Cdc25 mitotic inducer targeted by chk1 DNA damage checkpoint kinase. Science 1997, 277, 1495–1497. [Google Scholar] [CrossRef]
- Myers, K.; Gagou, M.E.; Zuazua-Villar, P.; Rodriguez, R.; Meuth, M. ATR and Chk1 suppress a caspase-3-dependent apoptotic response following DNA replication stress. PLoS Genet. 2009, 5, e1000324. [Google Scholar] [CrossRef]
- Stolz, A.; Ertych, N.; Bastians, H. Tumor suppressor CHK2: Regulator of DNA damage response and mediator of chromosomal stability. Clin. Cancer Res. 2011, 17, 401–405. [Google Scholar] [CrossRef] [PubMed]
- Shimada, M.; Niida, H.; Zineldeen, D.H.; Tagami, H.; Tanaka, M.; Saito, H.; Nakanishi, M. Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell 2008, 132, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Goto, H.; Natsume, T.; Kanemaki, M.T.; Kaito, A.; Wang, S.; Gabazza, E.C.; Inagaki, M.; Mizoguchi, A. Chk1-mediated Cdc25A degradation as a critical mechanism for normal cell cycle progression. J. Cell Sci. 2019, 132, jcs223123. [Google Scholar] [CrossRef]
- Qu, M.; Yang, B.; Tao, L.; Yates, J.R., 3rd; Russell, P.; Dong, M.Q.; Du, L.L. Phosphorylation-dependent interactions between Crb2 and Chk1 are essential for DNA damage checkpoint. PLoS Genet. 2012, 8, e1002817. [Google Scholar] [CrossRef]
- Cheetham, J.; Smith, D.A.; da Silva Dantas, A.; Doris, K.S.; Patterson, M.J.; Bruce, C.R.; Quinn, J. A single MAPKKK regulates the Hog1 MAPK pathway in the pathogenic fungus Candida albicans. Mol. Biol. Cell 2007, 18, 4603–4614. [Google Scholar] [CrossRef]
- Li, D.; Gurkovska, V.; Sheridan, M.; Calderone, R.; Chauhan, N. Studies on the regulation of the two-component histidine kinase gene CHK1 in Candida albicans using the heterologous lacZ reporter gene. Microbiology 2004, 150, 3305–3313. [Google Scholar] [CrossRef]
- Basso, V.; Znaidi, S.; Lagage, V.; Cabral, V.; Schoenherr, F.; LeibundGut-Landmann, S.; d’Enfert, C.; Bachellier-Bassi, S. The two-component response regulator Skn7 belongs to a network of transcription factors regulating morphogenesis in Candida albicans and independently limits morphogenesis-induced ROS accumulation. Mol. Microbiol. 2017, 106, 157–182. [Google Scholar] [CrossRef]
- Kruppa, M.; Goins, T.; Cutler, J.E.; Lowman, D.; Williams, D.; Chauhan, N.; Menon, V.; Singh, P.; Li, D.; Calderone, R. The role of the Candida albicans histidine kinase [CHK1) gene in the regulation of cell wall mannan and glucan biosynthesis. FEMS Yeast Res. 2003, 3, 289–299. [Google Scholar] [CrossRef][Green Version]
- Ahamad, N.; Verma, S.K.; Ahmed, S. Activation of checkpoint kinase Chk1 by reactive oxygen species resulting from disruption of wat1/pop3 in Schizosaccharomyces pombe. Genetics 2016, 204, 1397–1406. [Google Scholar] [CrossRef]
- Lev, S.; Sharon, A.; Hadar, R.; Ma, H.; Horwitz, B.A. A mitogen-activated protein kinase of the corn leaf pathogen Cochliobolus heterostrophus is involved in conidiation, appressorium formation, and pathogenicity: Diverse roles for mitogen-activated protein kinase homologs in foliar pathogens. Proc. Natl. Acad. Sci. USA 1999, 96, 13542–13547. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Duan, Z.; Liu, X.L.; Li, Z.; Shen, Z.; Gong, S.; Lu, Q.; Hu, Y.; Song, L.; Wang, Z.; et al. Checkpoint kinases regulate the circadian clock after DNA damage by influencing chromatin dynamics. Nucleic Acids Res. 2025, 53, gkaf162. [Google Scholar] [CrossRef] [PubMed]
- Tarsha, W. Chk1 prevents abnormal mitosis of S-phase HeLa cells containing DNA damage. Chin. Sci. Bull. 2009, 54, 4205–4213. [Google Scholar]
- Jung, K.W.; Kwon, S.; Jung, J.H.; Bahn, Y.S. Essential roles of ribonucleotide reductases under DNA damage and replication stresses in cryptococcus neoformans. Microbiol. Spectr. 2022, 10, e0104422. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Xiang, M.; Liu, X. Nematode-Trapping Fungi. Microbiol. Spectr. 2017, 5, 10. [Google Scholar] [CrossRef]
- Li, L.; Ma, M.; Liu, Y.; Zhou, J.; Qu, Q.; Lu, K.; Fu, D.; Zhang, K. Induction of trap formation in nematode-trapping fungi by a bacterium. FEMS Microbiol. Lett. 2011, 322, 157–165. [Google Scholar] [CrossRef]
- Su, H.; Zhao, Y.; Zhou, J.; Feng, H.; Jiang, D.; Zhang, K.Q.; Yang, J. Trapping devices of nematode-trapping fungi: Formation, evolution, and genomic perspectives. Biol. Rev. Camb. Philos. Soc. 2017, 92, 357–368. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, E.; An, Z.; Liu, X. Evolution of nematode-trapping cells of predatory fungi of the Orbiliaceae based on evidence from rRNA-encoding DNA and multiprotein sequences. Proc. Natl. Acad. Sci. USA 2007, 104, 8379–8384. [Google Scholar] [CrossRef] [PubMed]
- Ahren, D.; Tunlid, A. Evolution of parasitism in nematode-trapping fungi. J. Nematol. 2003, 35, 194–197. [Google Scholar]
- Dijksterhuis, J.; Veenhuis, M.; Harder, W.; Nordbring-Hertz, B. Nematophagous fungi: Physiological aspects and structure-function relationships. Adv. Microb. Physiol. 1994, 36, 111–143. [Google Scholar]
- Yang, J.; Wang, L.; Ji, X.; Feng, Y.; Li, X.; Zou, C.; Xu, J.; Ren, Y.; Mi, Q.; Wu, J.; et al. Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation. PLoS Pathog. 2011, 7, e1002179. [Google Scholar] [CrossRef]
- Zhu, M.C.; Zhao, N.; Liu, Y.K.; Li, X.M.; Zhen, Z.Y.; Zheng, Y.Q.; Zhang, K.Q.; Yang, J.K. The cAMP-PKA signalling pathway regulates hyphal growth, conidiation, trap morphogenesis, stress tolerance, and autophagy in Arthrobotrys oligospora. Environ. Microbiol. 2022, 24, 6524–6538. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.A.; Lin, H.C.; Hsueh, Y.P. The cAMP-PKA pathway regulates prey sensing and trap morphogenesis in the nematode-trapping fungus Arthrobotrys oligospora. G3 2022, 12, jkac217. [Google Scholar] [CrossRef]
- Kuo, C.Y.; Chen, S.A.; Hsueh, Y.P. The high osmolarity glycerol (HOG) pathway functions in osmosensing, trap morphogenesis and conidiation of the Nematode-trapping fungus Arthrobotrys oligospora. J. Fungi 2020, 6, 191. [Google Scholar] [CrossRef]
- Xie, M.; Ma, N.; Bai, N.; Yang, L.; Yang, X.; Zhang, K.Q.; Yang, J. PKC-SWI6 signaling regulates asexual development, cell wall integrity, stress response, and lifestyle transition in the nematode-trapping fungus Arthrobotrys oligospora. Sci. China Life Sci. 2022, 65, 2455–2471. [Google Scholar] [CrossRef]
- Xie, M.; Bai, N.; Yang, X.; Liu, Y.; Zhang, K.Q.; Yang, J. Fus3 regulates asexual development and trap morphogenesis in the nematode-trapping fungus Arthrobotrys oligospora. iScience 2023, 26, 107404. [Google Scholar] [CrossRef]
- Kuo, C.Y.; Tay, R.J.; Lin, H.C.; Juan, S.C.; Vidal-Diez de Ulzurrun, G.; Chang, Y.C.; Hoki, J.; Schroeder, F.C.; Hsueh, Y.P. The nematode-trapping fungus Arthrobotrys oligospora detects prey pheromones via G protein-coupled receptors. Nat. Microbiol. 2024, 9, 1738–1751. [Google Scholar] [CrossRef]
- Xie, M.; Ma, N.; Bai, N.; Zhu, M.; Zhang, K.Q.; Yang, J. Phospholipase C (AoPLC2) regulates mycelial development, trap morphogenesis, and pathogenicity of the nematode-trapping fungus Arthrobotrys oligospora. J. Appl. Microbiol. 2022, 132, 2144–2156. [Google Scholar] [CrossRef]
- Park, G.; Colot, H.V.; Collopy, P.D.; Krystofova, S.; Crew, C.; Ringelberg, C.; Litvinkova, L.; Altamirano, L.; Li, L.; Curilla, S.; et al. High-throughput production of gene replacement mutants in Neurospora crassa. Methods Mol. Biol. 2011, 722, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Wang, Y.; Tang, L.; Yang, L.; Zhou, D.; Li, Q.; Niu, X.; Zhang, K.Q.; Yang, J. AoStuA, an APSES transcription factor, regulates the conidiation, trap formation, stress resistance and pathogenicity of the nematode-trapping fungus Arthrobotrys oligospora. Environ. Microbiol. 2019, 21, 4648–4661. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.A.; Lin, H.C.; Schroeder, F.C.; Hsueh, Y.P. Prey sensing and response in a nematode-trapping fungus is governed by the MAPK pheromone response pathway. Genetics 2021, 217, iyaa008. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Colot, H.V.; Park, G.; Turner, G.E.; Ringelberg, C.; Crew, C.M.; Litvinkova, L.; Weiss, R.L.; Borkovich, K.A.; Dunlap, J.C. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 2006, 103, 10352–10357. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Hoffmann, D.S.; Wang, M.; Schuhmacher, L.; Stroe, M.C.; Schreckenberger, B.; Elstner, M.; Fischer, R. GprC of the nematode-trapping fungus Arthrobotrys flagrans activates mitochondria and reprograms fungal cells for nematode hunting. Nat. Microbiol. 2024, 9, 1752–1763. [Google Scholar] [CrossRef]
- Bong, D.; Sohn, J.; Lee, S.V. Brief guide to RT-qPCR. Mol. Cells 2024, 47, 100141. [Google Scholar] [CrossRef]
- Bustin, S.A.; Shipley, G.L.; Kirvell, S.; Mueller, R.; Nolan, T. RT-qPCR detection of SARS-CoV-2: No need for a dedicated reverse transcription step. Int. J. Mol. Sci. 2022, 23, 1303. [Google Scholar] [CrossRef]
- Zhao, N.; Zhu, M.; Liu, Q.; Shen, Y.; Duan, S.; Zhu, L.; Yang, J. AoPrdx2 Regulates Oxidative Stress, Reactive Oxygen Species, Trap Formation, and Secondary Metabolism in Arthrobotrys oligospora. J. Fungi 2024, 10, 110. [Google Scholar] [CrossRef]
- Zhu, L.; Zhu, M.; Li, X.; Shen, Y.; Duan, S.; Yang, J. Functional Characterization of Ao4g24: An Uncharacterized Gene Involved in Conidiation, Trap Formation, Stress Response, and Secondary Metabolism in Arthrobotrys oligospora. Microorganisms 2024, 12, 1532. [Google Scholar] [CrossRef]
- Youssar, L.; Wernet, V.; Hensel, N.; Yu, X.; Hildebrand, H.G.; Schreckenberger, B.; Kriegler, M.; Hetzer, B.; Frankino, P.; Dillin, A.; et al. Intercellular communication is required for trap formation in the nematode-trapping fungus Duddingtonia flagrans. PLoS Genet. 2019, 15, e1008029. [Google Scholar] [CrossRef]
- Lin, H.C.; de Ulzurrun, G.V.; Chen, S.A.; Yang, C.T.; Tay, R.J.; Iizuka, T.; Huang, T.Y.; Kuo, C.Y.; Gonçalves, A.P.; Lin, S.Y.; et al. Key processes required for the different stages of fungal carnivory by a nematode-trapping fungus. PLoS Biol. 2023, 21, e3002400. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.T.; Vidal-Diez de Ulzurrun, G.; Gonçalves, A.P.; Lin, H.C.; Chang, C.W.; Huang, T.Y.; Chen, S.A.; Lai, C.K.; Tsai, I.J.; Schroeder, F.C.; et al. Natural diversity in the predatory behavior facilitates the establishment of a robust model strain for nematode-trapping fungi. Proc. Natl. Acad. Sci. USA 2020, 117, 6762–6770. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Melesse, M.; Zhang, S.; Hao, C.; Wang, C.; Zhang, H.; Hall, M.C.; Xu, J.R. FgCDC14 regulates cytokinesis, morphogenesis, and pathogenesis in Fusarium graminearum. Mol. Microbiol. 2015, 98, 770–786. [Google Scholar] [CrossRef] [PubMed]
- Emser, J.; Wernet, N.; Hetzer, B.; Wohlmann, E.; Fischer, R. The cysteine-rich virulence factor NipA of Arthrobotrys flagrans interferes with cuticle integrity of Caenorhabditis elegans. Nat. Commun. 2024, 15, 5795. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, X.; Ma, Y.; Zhu, M.; Zhang, K.Q.; Yang, J. The Arf-GAPs, AoAge1 and AoAge2, regulate diverse cellular processes, conidiation, trap formation, and secondary metabolism in Arthrobotrys oligospora. Microbiol. Res. 2024, 285, 127779. [Google Scholar] [CrossRef]
- He, Z.Q.; Tan, J.L.; Li, N.; Zhang, H.X.; Chen, Y.H.; Wang, L.J.; Zhang, K.Q.; Niu, X.M. Sesquiterpenyl epoxy-cyclohexenoids and their signaling functions in nematode-trapping fungus Arthrobotrys oligospora. J. Agric. Food Chem. 2019, 67, 13061–13072. [Google Scholar] [CrossRef]
- Zhao, N.; Liu, Q.; Zhu, M.; Zhu, L.; Yang, J. The Hog1-Nmd5 signaling pathway regulates asexual development, lipid metabolism, stress response, trap morphogenesis, and secondary metabolism of Arthrobotrys oligospora. Virulence 2025, 16, 2468294. [Google Scholar] [CrossRef]
- Wang, R.; Li, B.; Lam, S.M.; Shui, G. Integration of lipidomics and metabolomics for in-depth understanding of cellular mechanism and disease progression. J. Genet. Genom. 2020, 47, 69–83. [Google Scholar] [CrossRef]
- Bauermeister, A.; Mannochio-Russo, H.; Costa-Lotufo, L.V.; Jarmusch, A.K.; Dorrestein, P.C. Mass spectrometry-based metabolomics in microbiome investigations. Nat. Rev. Microbiol. 2022, 20, 143–160. [Google Scholar] [CrossRef] [PubMed]
- Westphal, C.H. Cell-cycle signaling: Atm displays its many talents. Curr. Biol. 1997, 7, R789–R792. [Google Scholar] [CrossRef]
- Zhao, H.; Piwnica-Worms, H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell. Biol. 2001, 21, 4129–4139. [Google Scholar] [CrossRef]
- Suzuki, M.; Yamamori, T.; Bo, T.; Sakai, Y.; Inanami, O. MK-8776, a novel Chk1 inhibitor, exhibits an improved radiosensitizing effect compared to UCN-01 by exacerbating radiation-induced aberrant mitosis. Transl. Oncol. 2017, 10, 491–500. [Google Scholar] [CrossRef]
- de Sena-Tomás, C.; Navarro-González, M.; Kües, U.; Pérez-Martín, J. A DNA damage checkpoint pathway coordinates the division of dikaryotic cells in the ink cap mushroom Coprinopsis cinerea. Genetics 2013, 195, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Maksimov, V.; Wäneskog, M.; Rodriguez, A.; Bjerling, P. Stress sensitivity of a fission yeast strain lacking histidine kinases is rescued by the ectopic expression of Chk1 from Candida albicans. Curr. Genet. 2017, 63, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Cheng, L.; Liao, B.; Shi, Y.; Niu, Y.; Zhu, C.; Ye, X.; Zhou, X.; Ren, B. Candida albicans CHK1 gene from two-component system is essential for its pathogenicity in oral candidiasis. Appl. Microbiol. Biotechnol. 2021, 105, 2485–2496. [Google Scholar] [CrossRef]
- Klippel, N.; Cui, S.; Groebe, L.; Bilitewski, U. Deletion of the Candida albicans histidine kinase gene CHK1 improves recognition by phagocytes through an increased exposure of cell wall beta-1,3-glucans. Microbiology 2010, 156, 3432–3444. [Google Scholar] [CrossRef] [PubMed]
- Mielnichuk, N.; Sgarlata, C.; Pérez-Martín, J. A role for the DNA-damage checkpoint kinase Chk1 in the virulence program of the fungus Ustilago maydis. J. Cell Sci. 2009, 122, 4130–4140. [Google Scholar] [CrossRef]
- Huang, X.; Zhao, N.; Zhang, K. Extracellular enzymes serving as virulence factors in nematophagous fungi involved in infection of the host. Res. Microbiol. 2004, 155, 811–816. [Google Scholar] [CrossRef]
- Yang, J.; Liang, L.; Li, J.; Zhang, K.Q. Nematicidal enzymes from microorganisms and their applications. Appl. Microbiol. Biotechnol. 2013, 97, 7081–7095. [Google Scholar] [CrossRef]
- Ovejero, S.; Soulet, C.; Kumanski, S.; Moriel-Carretero, M. Coordination between phospholipid pools and DNA damage sensing. Biol. Cell 2022, 114, 211–219. [Google Scholar] [CrossRef]
- Liou, J.S.; Wu, Y.C.; Yen, W.Y.; Tang, Y.S.; Kakadiya, R.B.; Su, T.L.; Yih, L.H. Inhibition of autophagy enhances DNA damage-induced apoptosis by disrupting CHK1-dependent S phase arrest. Toxicol. Appl. Pharmacol. 2014, 278, 249–258. [Google Scholar] [CrossRef]
- Bu, W.; Hao, X.; Yang, T.; Wang, J.; Liu, Q.; Zhang, X.; Li, X.; Gong, Y.; Shao, C. Autophagy Contributes to the Maintenance of Genomic Integrity by Reducing Oxidative Stress. Oxidative Med. Cell. Longev. 2020, 2020, 2015920. [Google Scholar] [CrossRef]
- Chen, S.; Wang, C.; Sun, L.; Wang, D.L.; Chen, L.; Huang, Z.; Yang, Q.; Gao, J.; Yang, X.B.; Chang, J.F.; et al. RAD6 promotes homologous recombination repair by activating the autophagy-mediated degradation of heterochromatin protein HP1. Mol. Cell. Biol. 2015, 35, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, D.A.; Ryan, K.M. Autophagy is critically required for DNA repair by homologous recombination. Mol. Cell. Oncol. 2016, 3, e1030538. [Google Scholar] [CrossRef]
- Nam, H.J.; Han, J.H.; Yu, J.; Cho, C.S.; Kim, D.; Kim, Y.E.; Kim, M.J.; Kim, J.H.; Jo, D.H.; Bae, S. Autophagy induction enhances homologous recombination-associated CRISPR-Cas9 gene editing. Nucleic Acids Res. 2025, 53, gkaf258. [Google Scholar] [CrossRef]
- Chen, P.; De Winne, N.; De Jaeger, G.; Ito, M.; Heese, M.; Schnittger, A. KNO1-mediated autophagic degradation of the Bloom syndrome complex component RMI1 promotes homologous recombination. EMBO J. 2023, 42, e111980. [Google Scholar] [CrossRef]
- Zhang, D.; Tang, B.; Xie, X.; Xiao, Y.F.; Yang, S.M.; Zhang, J.W. The interplay between DNA repair and autophagy in cancer therapy. Cancer Biol. Ther. 2015, 16, 1005–1013. [Google Scholar] [CrossRef]
- Gomes, L.R.; Menck, C.F.M.; Leandro, G.S. Autophagy roles in the modulation of DNA repair pathways. Int. J. Mol. Sci. 2017, 18, 2351. [Google Scholar] [CrossRef]
- Liu, E.Y.; Xu, N.; O’Prey, J.; Lao, L.Y.; Joshi, S.; Long, J.S.; O’Prey, M.; Croft, D.R.; Beaumatin, F.; Baudot, A.D.; et al. Loss of autophagy causes a synthetic lethal deficiency in DNA repair. Proc. Natl. Acad. Sci. USA 2015, 112, 773–778. [Google Scholar] [CrossRef]
- Yamamoto, H.; Hirasawa, A. Homologous recombination deficiencies and hereditary tumors. Int. J. Mol. Sci. 2021, 23, 348. [Google Scholar] [CrossRef]
- Do, K.T.; Kochupurakkal, B.; Kelland, S.; de Jonge, A.; Hedglin, J.; Powers, A.; Quinn, N.; Gannon, C.; Vuong, L.; Parmar, K.; et al. Phase 1 combination study of the CHK1 inhibitor prexasertib and the PARP inhibitor olaparib in high-grade serous ovarian cancer and other solid tumors. Clin. Cancer Res. 2021, 27, 4710–4716. [Google Scholar] [CrossRef] [PubMed]
- Francesconi, S.; Grenon, M.; Bouvier, D.; Baldacci, G. p56(chk1) protein kinase is required for the DNA replication checkpoint at 37 degrees C in fission yeast. EMBO J. 1997, 16, 1332–1341. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, H.; Liu, Q.; Chen, S.; Li, X.; Chen, H.; Xia, Y.; Yang, J. AoChk1 Is Required for Sporulation, Trap Formation, and Metabolic Process in Arthrobotrys oligospora. J. Fungi 2025, 11, 602. https://doi.org/10.3390/jof11080602
Luo H, Liu Q, Chen S, Li X, Chen H, Xia Y, Yang J. AoChk1 Is Required for Sporulation, Trap Formation, and Metabolic Process in Arthrobotrys oligospora. Journal of Fungi. 2025; 11(8):602. https://doi.org/10.3390/jof11080602
Chicago/Turabian StyleLuo, Huan, Qianqian Liu, Si Chen, Xiaoli Li, Haitao Chen, Yuanyuan Xia, and Jinkui Yang. 2025. "AoChk1 Is Required for Sporulation, Trap Formation, and Metabolic Process in Arthrobotrys oligospora" Journal of Fungi 11, no. 8: 602. https://doi.org/10.3390/jof11080602
APA StyleLuo, H., Liu, Q., Chen, S., Li, X., Chen, H., Xia, Y., & Yang, J. (2025). AoChk1 Is Required for Sporulation, Trap Formation, and Metabolic Process in Arthrobotrys oligospora. Journal of Fungi, 11(8), 602. https://doi.org/10.3390/jof11080602






