Is the Co-Occurrence of Neophysopella meliosmae-myrianthae and N. montana (Pucciniales) Common on Grapevines in Japan?
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Okane, I.; Ono, Y. Phylogenetic study of indigenous grapevine leaf rust fungi in North America and biological identity of an invasive grapevine leaf rust fungus in Brazil. Mycoscience 2018, 59, 99–104. [Google Scholar] [CrossRef]
- Ono, Y.; Okane, I.; Chatasiri, S.; Pota, S.; Unartngam, J.; Ayawong, C.; Nguyen, H.D.; Le, T.M.C. Taxonomy of Southeast Asian-Australasian grapevine leaf rust fungus and its close relatives. Mycol. Prog. 2020, 19, 905–919. [Google Scholar] [CrossRef]
- Pota, S.; Chatasiri, S.; Unartngam, J.; Yamaoka, Y.; Hosaka, K.; Ono, Y. Taxonomic identity of a Phakopsora fungus causing the grapevine leaf rust disease in Southeast Asia and Australasia. Mycoscience 2014, 56, 198–204. [Google Scholar] [CrossRef]
- Ono, Y.; Chatasiri, S.; Pota, S.; Yamaoka, Y. Phakopsora montana, another grapevine leaf rust pathogen in Japan. J. Gen. Plant Pathol. 2012, 78, 338–347. [Google Scholar] [CrossRef]
- Santos, R.F.; Primiano, I.V.; Amorim, L. Identification and pathogenicity of Neophysopella species associated with Asian grapevine leaf rust in Brazil. Plant Pathol. 2021, 70, 74–86. [Google Scholar] [CrossRef]
- Ono, Y. Taxonomy of the Phakopsora ampelopsidis species complex on vitaceous hosts in Asia including a new species P. euvitis. Mycologia 2000, 92, 154–173. [Google Scholar] [CrossRef]
- Ono, Y. Mixed infections of grapevine leaf rusts Phakopsora meliosmae-myrianthae and P. montana in Japan. J. Gen. Plant Pathol. 2016, 82, 149–153. [Google Scholar] [CrossRef]
- Production-Table Grapes. Available online: https://fas.usda.gov/data/production/commodity/0575100 (accessed on 25 November 2024).
- Nakagawa, S. Origin, history, and the history of viticulture and cultivars. In Vitiology of Japan; Horiuchi, S., Matsui, H., Eds.; Yokendo Ltd.: Tokyo, Japan, 1996; pp. 1–57. [Google Scholar]
- Suyama, Y.; Kawamuro, K.; Kinoshita, I.; Yoshimura, K.; Tsumura, Y.; Takahara, H. DNA sequence from a fossil pollen of Abies spp. from pleistocene peat. Genes Genet. Syst. 1996, 71, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Virtudazo, E.V.; Nakamura, H.; Kakishima, M. Phylogenetic analysis of sugarcane rusts based on sequences of ITS, 5.8 S rDNA and D1/D2 regions of LSU rDNA. J. Gen. Plant Pathol. 2001, 67, 28–36. [Google Scholar] [CrossRef]
- O’Donnell, K. Fusarium and its near relatives. In The fungal holomorph: Mitotic, Meiotic and Pleomorphic Speciation in Fungal Systematics; Reynolds, D.R., Taylor, J.W., Eds.; CABI: Wallingford, UK, 1993; pp. 225–233. [Google Scholar]
- Daly, A.; Hennessy, D. Grapevine Leaf Rust Project Final Report-Assessment of Cultivars for Resistance or Immunity and Fungicides Useful for Control (Project1B) and Molecular Characterisation, Host Range and Biological Studies (Project 1C); Grape and Wine Research & Development Corporation: Canberra, Australia, 2007; p. 36. [Google Scholar]
- Ozoe, S.; Kadowaki, Y. Ecology and control of grape rust disease. Plant Prot. 1971, 25, 401–440. (In Japanese) [Google Scholar]
- Ono, Y.; Weng, Y.T.; Xu, Y. Species of Phragmidiaceae (Pucciniales) collected in the Liupanshan, Ningxia Hui Autonomous Region, China. Bull. Coll. Educ. Ibaraki Univ. 2018, 67, 27–36. [Google Scholar]
- Lamichhane, J.R.; Venturi, V. Synergisms between microbial pathogens in plant disease complexes: A growing trend. Front. Plant Sci. 2015, 6, 385. [Google Scholar] [CrossRef] [PubMed]
- Saharan, G.S.; Mehta, N.K.; Meena, P.D. Association or mixed infection of downy mildew and white rust disease complex. In Downy Mildew Disease of Crucifers: Biology, Ecology and Disease Management; Saharan, G.S., Mehta, N.K., Meena, P.D., Eds.; Springer: Singapore, 2017; pp. 199–213. [Google Scholar] [CrossRef]
- Horikawa, Y. Atlas of the Japanese Flora: An Introduction to Plant Sociology of East Asia; Gakken: Tokyo, Japan, 1972. [Google Scholar]

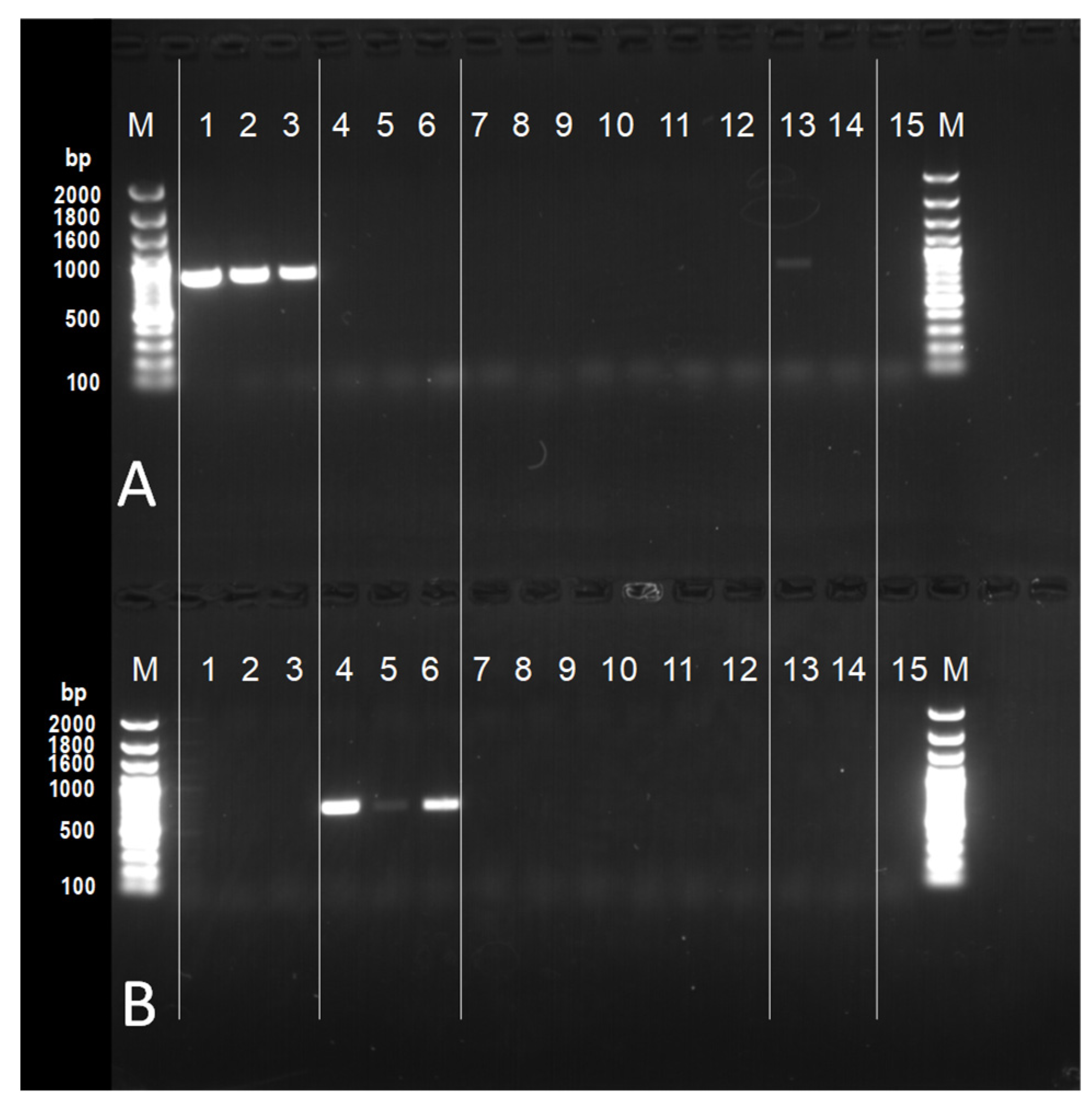
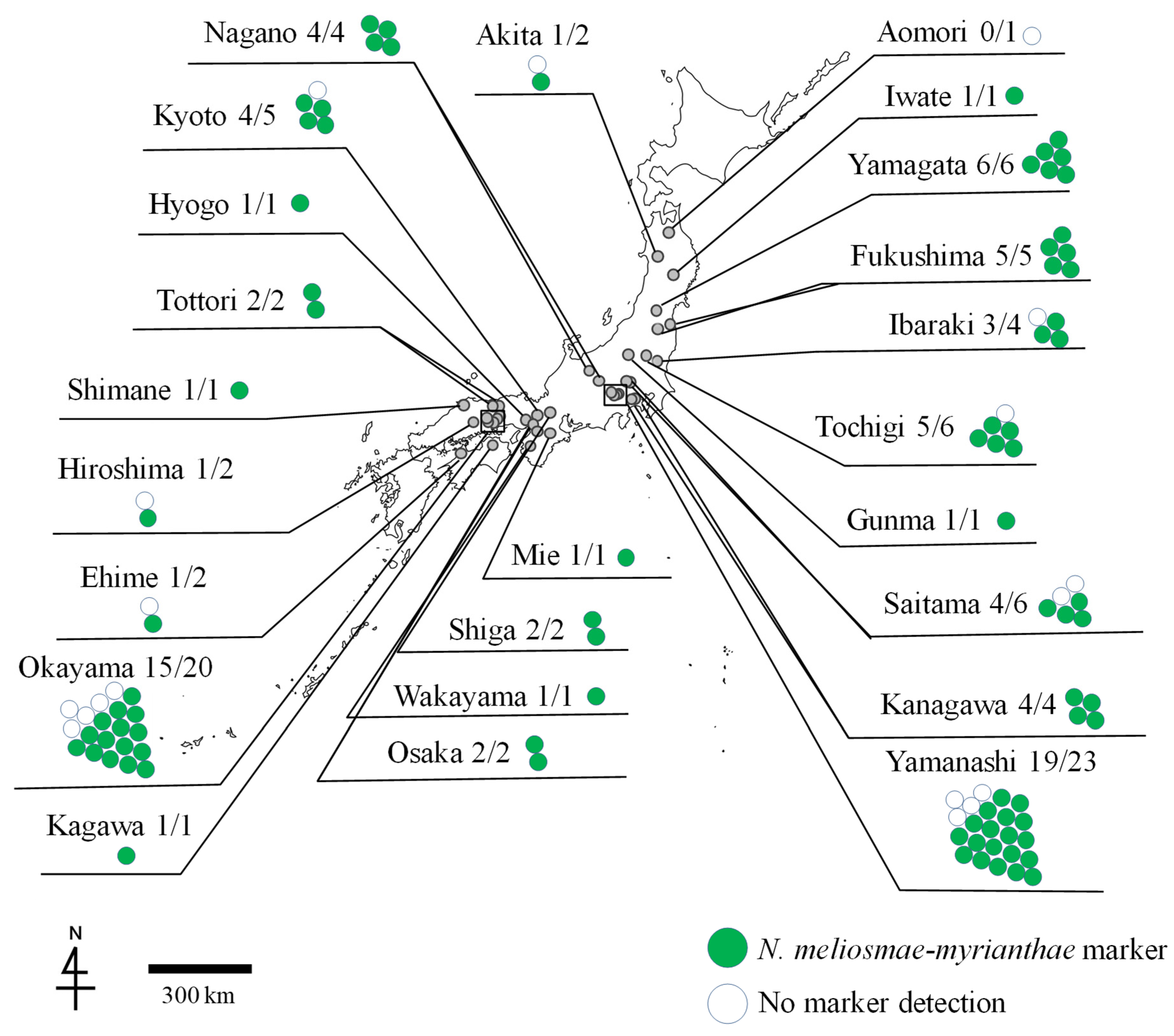

| Specimen Number | Location | Grapevine Cultivar (V. ×: Hybridization with V. vinifera) | Detection of N. meliosmae-myrianthae (No. of Target DNA Samples Detected/Total No. of DNA Samples) | Detection of N. montana (No. of Target DNA Samples Detected/Total No. of DNA Samples) |
|---|---|---|---|---|
| TSH-R58051 (=IBAR 10118) | Arakawa, Chichibu City, Saitama Pref. | Unknown | Negative (0/6) | Negative (0/6) |
| TSH-R58052 (=IBAR 10119) | Arakawa, Chichibu City, Saitama Pref. | Unknown | Negative (0/4) | Negative (0/4) |
| TSH-R58171 (=IBAR 10241) | Hiroshima Agricultural Technology Center, Higashihiroshima City, Hiroshima Pref. | V. vinifera × V. labrascana ‘Shine Muscat’ | Positive (2/3) | Negative (0/3) |
| TSH-R58172 (=IBAR 10242) | Hiroshima Agricultural Technology Center, Higashihiroshima City, Hiroshima Pref. | V. × labruscana ‘Muscat Bailey A’ | Negative (0/3) | Negative (0/3) |
| TSH-R58387 (=IBAR 10466) | Hojo, Hokuei Town, Tohaku, Tottori Pref. | V. × labruscana ‘Kyoho’ | Positive (6/6) | Negative (0/6) |
| TSH-R58388 (=IBAR 10467) | Kose, Mimasaka City, Okayama Pref. | Unknown | Negative (0/6) | Negative (0/6) |
| TSH-R58389 (=IBAR 10468) | Kose, Mimasaka City, Okayama Pref. | Unknown | Negative (0/5) | Negative (0/5) |
| TSH-R58390 (=IBAR 10469) | Kose, Mimasaka City, Okayama Pref. | Unknown | Positive (6/6) | Negative (0/6) |
| TSH-R58391 (=IBAR 10470) | Kose, Mimasaka City, Okayama Pref. | Unknown | Positive (4/6) | Negative (0/6) |
| TSH-R58392 (=IBAR 10471) | Saeki Town, Wake, Okayama Pref. | Unknown | Negative (0/5) | Negative (0/5) |
| TSH-R58393 (=IBAR 10472) | Saeki Town, Wake, Okayama Pref. | Unknown | Positive (4/5) | Negative (0/5) |
| TSH-R58394 (=IBAR 10473) | Saeki Town, Wake, Okayama Pref. | Unknown | Positive (6/6) | Negative (0/6) |
| TSH-R58395 (=IBAR 10474) | Saeki Town, Wake, Okayama Pref. | Unknown | Positive (6/6) | Negative (0/6) |
| TSH-R58397 (=IBAR 10476) | National Route 429, Kita City, Okayama Pref. | Unknown | Negative (0/6) | Negative (0/6) |
| TSH-R58398 (=IBAR 10477) | Prefectural Route 66, Katta, Maniwa City, Okayama Pref. | Unknown | Positive (4/5) | Negative (0/5) |
| TSH-R58399 (=IBAR 10478) | Yoshikawa, Kibichuo Town, Kaga, Okayama Pref. | Unknown | Positive (3/3) | Negative (0/3) |
| TSH-R58400 (=IBAR 10479) | Yoshikawa, Kibichuo Town, Kaga, Okayama Pref. | Unknown | Positive (6/6) | Negative (0/6) |
| TSH-R58401 (=IBAR 10480) | Yoshikawa, Kibichuo Town, Kaga, Okayama Pref. | Unknown | Positive (6/6) | Negative (0/6) |
| TSH-R58402 (=IBAR 10481) | Shirochi, Ochiai Town, Takahashi City, Okayama Pref. | Unknown | Positive (6/6) | Negative (0/6) |
| TSH-R58403 (=IBAR 10482) | Higashikarube, Akaiwa City, Okayama Pref. | Unknown | Negative (0/6) | Negative (0/6) |
| TSH-R58404 (=IBAR 10483) | Higashikarube, Akaiwa City, Okayama Pref. | Unknown | Positive (2/2) | Negative (0/2) |
| TSH-R58405 (=IBAR 10484) | Ashigakubo, Yokoze Town, Chichibu, Saitama Pref. | V. × labruscana ‘Kyoho’ | Positive (6/6) | Negative (0/6) |
| TSH-R58406 (=IBAR 10485) | Ashigakubo, Yokoze Town, Chichibu, Saitama Pref. | V. × labruscana ‘Kyoho’ | Positive (6/6) | Negative (0/6) |
| TSH-R58407 (=IBAR 10486) | Ashigakubo, Yokoze Town, Chichibu, Saitama Pref. | V. × labruscana ‘Kyoho’ | Positive (6/6) | Negative (0/6) |
| TSH-R58408 (=IBAR 10487) | Yokoze, Yokoze Town, Chichibu, Saitama Pref. | V. × labruscana ‘Kyoho’ | Positive (6/6) | Negative (0/6) |
| TSH-R58409 (=IBAR 10488) | Makiokacho Kurashina, Yamanashi City, Yamanashi Pref. | V. × labruscana ‘Kyoho’ | Positive (6/6) | Negative (0/6) |
| TSH-R58410 (=IBAR 10489) | Higashi, Yamanashi City, Yamanashi Pref. | Unknown | Positive (5/6) | Negative (0/6) |
| TSH-R58411 (=IBAR 10490) | Ochiai, Yamanashi City, Yamanashi Pref. | Unknown | Positive (6/6) | Negative (0/6) |
| TSH-R58412 (=IBAR 10491) | Kasugai, Fuefuki City, Yamanashi Pref. | Unknown | Negative (0/6) | Negative (0/6) |
| TSH-R58413 (=IBAR 10492) | Sakurai, Fuefuki City, Yamanashi Pref. | Unknown | Positive (6/6) | Negative (0/6) |
| TSH-R58414 (=IBAR 10493) | Isawa Town, Fuefuki City, Yamanashi Pref. | Unknown | Negative (0/6) | Negative (0/6) |
| TSH-R58415 (=IBAR 10494) | Enzan Kaminishi, Koshu City, Yamanashi Pref. | Unknown | Positive (6/6) | Negative (0/6) |
| TSH-R58416 (=IBAR 10495) | Enzan Kamioso, Koshu City, Yamanashi Pref. | Unknown | Positive (6/6) | Negative (0/6) |
| TSH-R58417 (=IBAR 10496) | Yama, Katsunuma Town, Koshu City, Yamanashi Pref. | Unknown | Negative (0/6) | Negative (0/6) |
| TSH-R58418 (=IBAR 10497) | Osade, Katsunuma Town, Koshu City, Yamanashi Pref. | Unknown | Positive (6/6) | Negative (0/6) |
| TSH-R58419 (=IBAR 10498) | Katsunuma, Katsunuma Town, Koshu City, Yamanashi Pref. | Unknown | Positive (6/6) | Negative (0/6) |
| TSH-R58420 (=IBAR 10499) | Katsunuma, Katsunuma Town, Koshu City, Yamanashi Pref. | V. × labruscana ‘Pione’ | Positive (6/6) | Negative (0/6) |
| TSH-R58421 (=IBAR 10500) | Kamiyama Town, Nirasaki City, Yamanashi Pref. | Unknown | Positive (3/3) | Negative (0/3) |
| TSH-R58422 (=IBAR 10501) | Kamiyama Town, Nirasaki City, Yamanashi Pref. | Unknown | Positive (4/4) | Negative (0/4) |
| TSH-R58427 (=IBAR 10506) | Kamishiroi, Shibukawa City, Gunma Pref. | Unknown | Positive (1/3) | Negative (0/3) |
| TSH-R58430 (=IBAR 10509) | Shimotsubara, Iwafune Town, Shimotsuga, Tochigi Pref. | V. × labruscana ‘Kyoho’ | Positive (6/6) | Negative (0/6) |
| TSH-R58431 (=IBAR 10510) | Shimotsubara, Iwafune Town, Shimotsuga, Tochigi Pref. | V. × labruscana ‘Kyoho’ | Positive (6/6) | Negative (0/6) |
| TSH-R58432 (=IBAR 10511) | Shimotsubara, Iwafune Town, Shimotsuga, Tochigi Pref. | V. × labruscana ‘Kyoho’ | Positive (5/6) | Negative (0/6) |
| TSH-R58433 (=IBAR 10512) | Nishiyamada, Oohira Town, Tochigi City, Tochigi Pref. | V. × labruscana ‘Kyoho’ | Positive (6/6) | Negative (0/6) |
| TSH-R58434 (=IBAR 10513) | Nishiyamada, Oohira Town, Tochigi City, Tochigi Pref. | V. × labruscana ‘Kyoho’ | Negative (0/6) | Negative (0/6) |
| TSH-R58435 (=IBAR 10514) | Nishiyamada, Oohira Town, Tochigi City, Tochigi Pref. | V. × labruscana ‘Kyoho’ | Positive (6/6) | Negative (0/6) |
| TSH-R30450 | Ezohara, Yamanashi City, Yamanashi Pref. | V. × labruscana ‘Kyoho’ | Positive (6/6) | Negative (0/6) |
| TSH-R30451 | Nanokaichiba, Yamanashi City, Yamanashi Pref. | V. × labruscana ‘Kyoho’ | Positive (6/6) | Negative (0/6) |
| TSH-R30453 | Oyama, Misaka Town, Fuefuki City, Yamanashi Pref. | V. × labruscana ‘Kyoho’ | Positive (5/5) | Negative (0/5) |
| TSH-R30455 | Katsunuma Town, Koshu City, Yamanashi Pref. | V. vinifera × V. labrascana ‘Queen Nina’ | Negative (0/6) | Negative (0/6) |
| TSH-R30456 | Nanokaichiba, Yamanashi City, Yamanashi Pref. | V. vinifera × V. labrascana ‘Oriental Star’ | Positive (6/6) | Negative (0/6) |
| TSH-R30457 | Manriki, Yamanashi City, Yamanashi Pref. | V. × labruscana ‘Kyoho’ | Positive (6/6) | Negative (0/6) |
| TSH-R30461 | Shimoidai Town, Matsuyama City, Ehime Pref. | V. × labruscana ‘Fujiminori’ | Negative (0/6) | Negative (0/6) |
| TSH-R30462 | Shimoidai Town, Matsuyama City, Ehime Pref. | V. × labruscana ‘Black Beet’ | Positive (6/6) | Negative (0/6) |
| TSH-R30463 | Fruit-Tree Experiment Station, Tenno, Katagami City, Akita Pref. | V. × labruscana ‘Campbell Early’? | Negative (0/6) | Negative (0/6) |
| TSH-R30464 | Fruit-Tree Experiment Station, Tenno, Katagami City, Akita Pref. | V. × labruscana ‘Black Beet’? | Positive (6/6) | Negative (0/6) |
| TSH-R30466 | Kuroki, Soma City, Fukushima Pref. | V. vinifera × V. labrascana ‘Shine Muscat’? | Positive (6/6) | Negative (0/6) |
| TSH-R30467 | Yokone Town, Kofu City, Yamanashi Pref. | V. × labruscana ‘Delaware’ | Positive (6/6) | Negative (0/6) |
| TSH-R30468 | Yokone Town, Kofu City, Yamanashi Pref. | V. vinifera ‘Koshu’ or V. × labruscana ‘Pione’ | Positive (6/6) | Negative (0/6) |
| TSH-R30469 | Yokone Town, Kofu City, Yamanashi Pref. | V. vinifera ‘Koshu’ | Positive (6/6) | Negative (0/6) |
| TSH-R30470 | Pre. Res. Ins. for the Environ., Agricul., For. and Fisher., Habikino City, Osaka Pref. | V. × labruscana ‘Delaware’ | Positive (6/6) | Negative (0/6) |
| TSH-R30472 | Akaiwa City, Okayama Pref. | V. × labruscana ‘Pione’ | Positive (6/6) | Negative (0/6) |
| TSH-R30473 | Aono, Ibara City, Okayama Pref. | V. × labruscana ‘Pione’ | Positive (6/6) | Negative (0/6) |
| TSH-R30475 | Tsuchida, Okayama City, Okayama Pref. | V. × labruscana ‘Pione’ | Positive (6/6) | Negative (0/6) |
| TSH-R30476 | Nagasaki, Funao Town, Kurashiki City, Okayama Pref. | V. × labruscana ‘Pione’ | Positive (5/6) | Negative (0/6) |
| TSH-R30477 | Izumo City, Shimane Pref. | V. × labruscana ‘Delaware’ | Positive (2/2) | Negative (0/2) |
| HHUF 10428 | Sakaimatsu, Kuroishi City, Aomori Pref. | Unknown | Negative (0/6) | Negative (0/6) |
| HHUF 15175 | Oohasama Town, Hanamaki City, Iwate Pref. | Unknown | Positive (2/6) | Negative (0/6) |
| TSH-R30478 | Hashimoto, Yazu Town, Yazu, Tottori Pref. | Unknown | Positive (5/6) | Negative (0/6) |
| TSH-R30517 | Atsugi City, Kanagawa Pref. | Unknown | Positive (6/6) | Negative (0/6) |
| TSH-R30480 | Kita Town, Kobe City, Hyogo Pref. | V. × labruscana ‘Pione’ | Positive (1/1) | Negative (0/1) |
| TSH-R30482 | Kyoto City, Kyoto Pref. | V. vinifera × V. labrascana ‘Shine Muscat’ | Negative (0/5) | Negative (0/5) |
| TSH-R30483 | Kyoto City, Kyoto Pref. | V. × labruscana ‘Delaware’ | Positive (4/4) | Negative (0/4) |
| TSH-R30484 | Kyoto City, Kyoto Pref. | V. × labruscana ‘Pione’ | Positive (3/3) | Negative (0/3) |
| TSH-R30485 | Kyoto City, Kyoto Pref. | V. × labruscana ‘Fujiminori’ | Positive (4/5) | Negative (0/5) |
| TSH-R30487 | Ryuo Town, Shiga Pref. | V. × labruscana ‘Kyoho’ | Positive (4/4) | Negative (0/4) |
| TSH-R30488 | Ryuo Town, Shiga Pref. | V. × labruscana ‘Kyoho’ | Positive (1/2) | Negative (0/2) |
| TSH-R30489 | Nabari City, Mie Pref. | V. × labruscana ‘Kyoho’ | Positive (5/5) | Negative (0/5) |
| TSH-R30491 | Kashiwara City, Osaka Pref. | V. × labruscana ‘Delaware’ | Positive (5/5) | Negative (0/5) |
| TSH-R30493 | Aridagawa Town, Wakayama Pref. | V. × labruscana ‘Kyoho’ | Positive (4/4) | Negative (0/4) |
| TSH-R30495 | Okaya City, Nagano Pref. | V. × labruscana ‘Kyoho’ | Positive (1/1) | Negative (0/1) |
| TSH-R30496 | Okaya City, Nagano Pref. | V. × labrusca ‘Niagara’ | Positive (1/2) | Negative (0/2) |
| TSH-R30497 | Ikusaka Village, Higashichikuma, Nagano Pref. | V. vinifera × V. labrascana ‘Shine Muscat’ | Positive (1/1) | Negative (0/1) |
| TSH-R30499 | Ikusaka Village, Higashichikuma, Nagano Pref. | V. × labruscana ‘Kyoho’ | Positive (1/2) | Negative (0/2) |
| TSH-R30505 | Kasumigaura City, Ibaraki Pref. | V. × labruscana ‘Kyoho’ | Negative (0/5) | Negative (0/5) |
| TSH-R30507 | Kasumigaura City, Ibaraki Pref. | V. × labruscana ‘Kyoho’ | Positive (6/6) | Negative (0/6) |
| TSH-R30515 | Kasumigaura City, Ibaraki Pref. | V. × labruscana ‘Muscat Bailey A’ | Positive (6/6) | Negative (0/6) |
| TSH-R30516 | Kasumigaura City, Ibaraki Pref. | V. × labruscana ‘Takatsuma’ | Positive (4/6) | Negative (0/6) |
| TSH-R30518 | Tokyo Univ. of Agricul. and Tech. Isehara Farm, Sannomiya, Isehara City, Kanagawa Pref. | V. × labruscana ‘Fujiminori’ | Positive (6/6) | Negative (0/6) |
| TSH-R30519 | Tokyo Univ. of Agricul. and Tech. Isehara Farm, Sannomiya, Isehara City, Kanagawa Pref. | V. vinifera × V. labrascana ‘Shine Muscat’ | Positive (2/2) | Negative (0/2) |
| TSH-R30520 | Tokyo Univ. of Agricul. and Tech. Isehara Farm, Sannomiya, Isehara City, Kanagawa Pref. | V. vinifera × V. labrascana ‘Queen Nina’ | Positive (3/6) | Negative (0/6) |
| TSH-R30521 | Kaminoyama City, Yamagata Pref. | V. × labruscana ‘Kyoho’? | Positive (3/5) | Negative (0/5) |
| TSH-R30522 | Kaminoyama City, Yamagata Pref. | V. × labruscana ‘Pione’ | Positive (5/5) | Negative (0/5) |
| TSH-R30523 | Kaminoyama City, Yamagata Pref. | V. × labruscana ‘Tenshu’ | Positive (3/4) | Negative (0/4) |
| TSH-R30526 | Kaminoyama City, Yamagata Pref. | V. × labruscana ‘Fujiminori’ | Positive (3/5) | Negative (0/5) |
| TSH-R30527 | Kaminoyama City, Yamagata Pref. | V. × labruscana ‘Takao’ | Positive (5/5) | Negative (0/5) |
| TSH-R30530 | Kaminoyama City, Yamagata Pref. | Unknown | Positive (4/6) | Negative (0/6) |
| TSH-R30531 | Nihonmatsu City, Fukushima Pref. | V. coignetiae × V. vinifera ‘Yama Sauvignon’ | Positive (2/2) | Negative (0/2) |
| TSH-R30532 | Nihonmatsu City, Fukushima Pref. | V. × labruscana ‘Steuben’ | Positive (6/6) | Negative (0/6) |
| TSH-R30533 | Nihonmatsu City, Fukushima Pref. | V. × labruscana ‘Kai Noir’ | Positive (1/6) | Negative (0/6) |
| TSH-R30534 | Nihonmatsu City, Fukushima Pref. | Unknown | Positive (6/6) | Negative (0/6) |
| TSH-R30486 | Kyoto City, Kyoto Pref. | Unknown | Positive (6/6) | Negative (0/6) |
| TSH-R30543 | Takamatsu City, Kagawa Pref. | Unknown | Positive (5/6) | Negative (0/6) |
| Specimen Number | Location | Detection of N. meliosmae-myrianthae (No. of Target DNA Samples Detected/Total No. of DNA Samples) | Detection of N. montana (No. of Target DNA Samples Detected/Total No. of DNA Samples) |
|---|---|---|---|
| TSH-R51432 (=IBAR 1817) | Mt. Owasezawasan, Yama, Fukushima Pref. | Negative (0/6) | Negative (0/6) |
| TSH-R52161 (=IBAR 2782) | Takizawa forest road, Mt. Fuji, Yamanashi Pref. | Negative (0/4) | Negative (0/4) |
| TSH-R52162 (=IBAR 2783) | Takizawa forest road, Mt. Fuji, Yamanashi Pref. | Negative (0/6) | Negative (0/6) |
| TSH-R53179 (=IBAR 3930) | Nasu Town, Nasu, Tochigi Pref. | Negative (0/6) | Negative (0/6) |
| TSH-R54801 (=IBAR 6279) | Mt. Yahikoyama, Yahiko Village, Nishikanbara, Niigata Pref. | Negative (0/6) | Negative (0/6) |
| TSH-R54804 (=IBAR 6282) | Mt. Yahikoyama, Yahiko Village, Nishikanbara, Niigata Pref. | Positive (6/6) | Negative (0/6) |
| TSH-R58370 (=IBAR 10449) | Nakamiyori, Nikko City, Tochigi Pref. | Positive (2/2) | Negative (0/2) |
| TSH-R58371 (=IBAR 10450) | Yunishigawa, Nikko City, Tochigi Pref. | Negative (0/4) | Positive (1/4) |
| TSH-R58372 (=IBAR 10451) | Yunishigawa, Nikko City, Tochigi Pref. | Positive (1/6) | Positive (2/6) |
| TSH-R58373 (=IBAR 10452) | Yunishigawa, Nikko City, Tochigi Pref. | Negative (0/4) | Positive (1/4) |
| TSH-R58374 (=IBAR 10453) | Yunishigawa, Nikko City, Tochigi Pref. | Negative (0/6) | Negative (0/6) |
| TSH-R58375 (=IBAR 10454) | Yunishigawa, Nikko City, Tochigi Pref. | Negative (0/6) | Negative (0/6) |
| TSH-R58376 (=IBAR 10455) | Yunishigawa, Nikko City, Tochigi Pref. | Negative (0/3) | Positive (2/3) |
| TSH-R58377 (=IBAR 10456) | Yasugamori Forest Road, Minamiaizu, Fukushima Pref. | Negative (0/6) | Negative (0/6) |
| TSH-R58378 (=IBAR 10457) | Yasugamori Forest Road, Minamiaizu, Fukushima Pref. | Negative (0/5) | Negative (0/5) |
| TSH-R58379 (=IBAR 10458) | Mt. Daisen, Daisen Town, Saihaku, Tottori Pref. | Negative (0/4) | Positive (2/4) |
| TSH-R58380 (=IBAR 10459) | Mt. Daisen, Daisen Town, Saihaku, Tottori Pref. | Negative (0/6) | Negative (0/6) |
| TSH-R58381 (=IBAR 10460) | Mt. Daisen, Daisen Town, Saihaku, Tottori Pref. | Negative (0/6) | Positive (6/6) |
| TSH-R58384 (=IBAR 10463) | Mt. Daisen, Daisen Town, Saihaku, Tottori Pref. | Negative (0/5) | Positive (3/5) |
| TSH-R58385 (=IBAR 10464) | Mt. Daisen, Daisen Town, Saihaku, Tottori Pref. | Positive (1/5) | Positive (2/5) |
| TSH-R58386 (=IBAR 10465) | Okudaisen, Kofu Town, Hino, Tottori Pref. | Negative (0/6) | Positive (2/6) |
| TSH-R58428 (=IBAR 10507) | Mt. Omineyama, Minakami Town, Tone, Gunma Pref. | Negative (0/6) | Positive (3/6) |
| TSH-R58429 (=IBAR 10508) | Mt. Omineyama, Minakami Town, Tone, Gunma Pref. | Positive (1/2) | Positive (1/2) |
| HHUF 3963 | Hengasa Forest Road, Hirosaki City, Aomori Pref. | Negative (0/3) | Negative (0/3) |
| HHUF 11069 | Jizotai, Gonohe Town, Sannohe, Aomori Pref. | Negative (0/6) | Positive (1/6) |
| TSH-R30536 | Mt. Daisen, Daisen Town, Saihaku, Tottori Pref. | Negative (0/1) | Negative (0/1) |
| TSH-R30537 | Mt. Daisen, Daisen Town, Saihaku, Tottori Pref. | Negative (0/1) | Positive (1/1) |
| TSH-R30538 | Mt. Daisen, Daisen Town, Saihaku, Tottori Pref. | Positive (1/3) | Positive (2/3) |
| TSH-R30539 | Mt. Daisen, Daisen Town, Saihaku, Tottori Pref. | Negative (0/5) | Positive (5/5) |
| TSH-R30540 | Mt. Daisen, Daisen Town, Saihaku, Tottori Pref. | Positive (1/1) | Positive (1/1) |
| TSH-R30541 | Mt. Daisen, Daisen Town, Saihaku, Tottori Pref. | Negative (0/2) | Positive (2/2) |
| TSH-R30542 | Mt. Daisen, Daisen Town, Saihaku, Tottori Pref. | Negative (0/1) | Positive (1/1) |
| TSH-R2204 | Nobeyama, Minamimaki Village, Minamisaku, Nagano Pref. | Negative (0/1) | Negative (0/1) |
| TSH-R2205 | Nobeyama, Minamimaki Village, Minamisaku, Nagano Pref. | Negative (0/1) | Negative (0/1) |
| TSH-R2206 | Nobeyama, Minamimaki Village, Minamisaku, Nagano Pref. | Negative (0/1) | Negative (0/1) |
| TSH-R2207 | Nobeyama, Minamimaki Village, Minamisaku, Nagano Pref. | Negative (0/1) | Negative (0/1) |
| TSH-R2803 | Nobeyama, Minamimaki Village, Minamisaku, Nagano Pref. | Negative (0/1) | Negative (0/1) |
| TSH-R2804 | Nobeyama, Minamimaki Village, Minamisaku, Nagano Pref. | Negative (0/1) | Negative (0/1) |
| TSH-R2805 | Nobeyama, Minamimaki Village, Minamisaku, Nagano Pref. | Negative (0/1) | Negative (0/1) |
| TSH-R2806 | Nobeyama, Minamimaki Village, Minamisaku, Nagano Pref. | Negative (0/1) | Negative (0/1) |
| TSH-R3452 | Nobeyama, Minamimaki Village, Minamisaku, Nagano Pref. | Negative (0/5) | Negative (0/5) |
| TSH-R13225 | Lake Yamanakako, Yamanashi Pref. | Negative (0/4) | Negative (0/4) |
| TSH-R1606 | Niigata Pref. | Negative (0/6) | Negative (0/6) |
| TSH-R30544 | Nobeyama, Minamimaki Village, Minamisaku, Nagano Pref. | Positive (4/4) | Negative (0/4) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okane, I.; Kurita, A.; Ono, Y. Is the Co-Occurrence of Neophysopella meliosmae-myrianthae and N. montana (Pucciniales) Common on Grapevines in Japan? J. Fungi 2025, 11, 193. https://doi.org/10.3390/jof11030193
Okane I, Kurita A, Ono Y. Is the Co-Occurrence of Neophysopella meliosmae-myrianthae and N. montana (Pucciniales) Common on Grapevines in Japan? Journal of Fungi. 2025; 11(3):193. https://doi.org/10.3390/jof11030193
Chicago/Turabian StyleOkane, Izumi, Akiko Kurita, and Yoshitaka Ono. 2025. "Is the Co-Occurrence of Neophysopella meliosmae-myrianthae and N. montana (Pucciniales) Common on Grapevines in Japan?" Journal of Fungi 11, no. 3: 193. https://doi.org/10.3390/jof11030193
APA StyleOkane, I., Kurita, A., & Ono, Y. (2025). Is the Co-Occurrence of Neophysopella meliosmae-myrianthae and N. montana (Pucciniales) Common on Grapevines in Japan? Journal of Fungi, 11(3), 193. https://doi.org/10.3390/jof11030193









